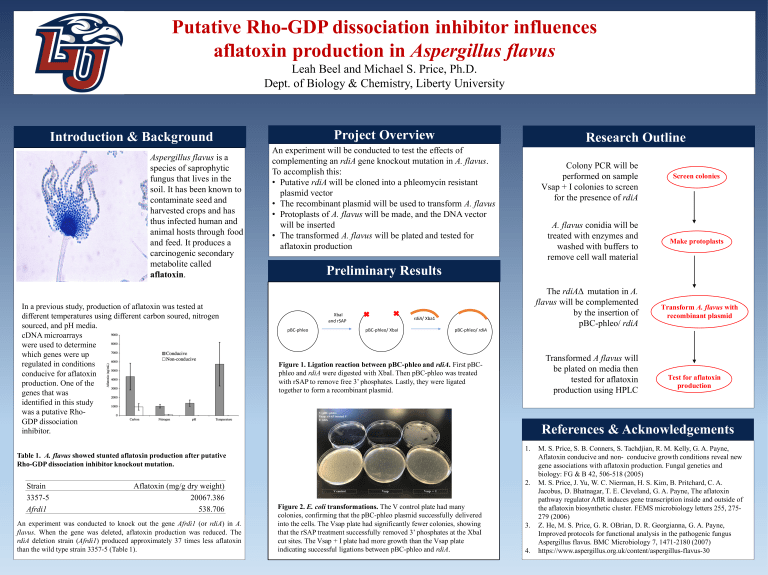
Putative Rho-GDP dissociation inhibitor influences aflatoxin production in Aspergillus flavus Leah Beel and Michael S. Price, Ph.D. Dept. of Biology & Chemistry, Liberty University Project Overview Introduction & Background Aspergillus flavus is a species of saprophytic fungus that lives in the soil. It has been known to contaminate seed and harvested crops and has thus infected human and animal hosts through food and feed. It produces a carcinogenic secondary metabolite called aflatoxin. j In a previous study, production of aflatoxin was tested at different temperatures using different carbon soured, nitrogen sourced, and pH media. cDNA microarrays were used to determine which genes were up regulated in conditions conducive for aflatoxin production. One of the genes that was identified in this study was a putative RhoGDP dissociation inhibitor. Table 1. A. flavus showed stunted aflatoxin production after putative Rho-GDP dissociation inhibitor knockout mutation. Strain 3357-5 Afrdi1 Aflatoxin (mg/g dry weight) 20067.386 538.706 An experiment was conducted to knock out the gene Afrdi1 (or rdiA) in A. flavus. When the gene was deleted, aflatoxin production was reduced. The rdiA deletion strain (Afrdi1) produced approximately 37 times less aflatoxin than the wild type strain 3357-5 (Table 1). Research Outline An experiment will be conducted to test the effects of complementing an rdiA gene knockout mutation in A. flavus. To accomplish this: • Putative rdiA will be cloned into a phleomycin resistant plasmid vector • The recombinant plasmid will be used to transform A. flavus • Protoplasts of A. flavus will be made, and the DNA vector will be inserted • The transformed A. flavus will be plated and tested for aflatoxin production Colony PCR will be performed on sample Vsap + I colonies to screen for the presence of rdiA A. flavus conidia will be treated with enzymes and washed with buffers to remove cell wall material Screen colonies Make protoplasts Preliminary Results ! ! The rdiAΔ mutation in A. flavus will be complemented by the insertion of pBC-phleo/ rdiA ! ! XbaI! and!rSAP! pBC%phleo! ! ! ! ! !! ! rdiA/!Xba1! pBC%phleo/!XbaI! ! pBC%phleo/!rdiA! ! ! Figure! 1. Ligation reaction between pBC-phleo and rdiA. First pBC! phleo and rdiA were digested with XbaI. Then pBC-phleo was treated ! with rSAP to remove free 3’ phosphates. Lastly, they were ligated ! ! to form a recombinant plasmid. together ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! Figure 2. E. coli transformations. The V control plate had many colonies, confirming that the pBC-phleo plasmid successfully delivered into the cells. The Vsap plate had significantly fewer colonies, showing that the rSAP treatment successfully removed 3’ phosphates at the XbaI cut sites. The Vsap + I plate had more growth than the Vsap plate indicating successful ligations between pBC-phleo and rdiA. Transformed A flavus will be plated on media then tested for aflatoxin production using HPLC Transform A. flavus with recombinant plasmid Test for aflatoxin production References & Acknowledgements 1. 2. 3. 4. M. S. Price, S. B. Conners, S. Tachdjian, R. M. Kelly, G. A. Payne, Aflatoxin conducive and non- conducive growth conditions reveal new gene associations with aflatoxin production. Fungal genetics and biology: FG & B 42, 506-518 (2005) M. S. Price, J. Yu, W. C. Nierman, H. S. Kim, B. Pritchard, C. A. Jacobus, D. Bhatnagar, T. E. Cleveland, G. A. Payne, The aflatoxin pathway regulator AflR induces gene transcription inside and outside of the aflatoxin biosynthetic cluster. FEMS microbiology letters 255, 275279 (2006) Z. He, M. S. Price, G. R. OBrian, D. R. Georgianna, G. A. Payne, Improved protocols for functional analysis in the pathogenic fungus Aspergillus flavus. BMC Microbiology 7, 1471-2180 (2007) https://www.aspergillus.org.uk/content/aspergillus-flavus-30