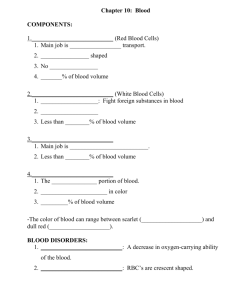
Diagram of eukaryotic cells Structure and function – cell-surface membrane - Phospholipid bilayer with embedded proteins etc. Selectively permeable – enables control of passage of substances in and out of cell Barrier between internal and external environment of cell Structure and function – nucleus - Nuclear envelope, nuclear pores, nucleolus, DNA / chromatin Controls the cells activity though transcription on mRNA Nuclear pores allow substances e.g. mRNA to move between the nucleus and cytoplasm Nucleolus makes ribosomes which are made up of proteins and ribosomal RNA Structure and function – mitochondria - Double membrane – inner membrane folded to form cristae. Matrix containing small 70S ribosomes, small circular DNA and enzymes involved in aerobic respiration (glycolysis). Site of aerobic respiration producing ATP for energy release Structure and function – Golgi apparatus - 3 or more fluid filled membrane bound sacs with vesicles at edge Receives protein from rough endoplasmic reticulum Modifies/processes protein e.g. add carbohydrates/sugars Packages into vesicles e.g. for transport to cell surface membrane for exocytosis Also makes lysosomes Structure and function – lysosomes - Type of Golgi vesicle containing lysozymes – hydrolytic enzymes Release of lysozymes to break down / hydrolyse pathogens or worn out cell components Structure and function – ribosomes - Float free in cytoplasm or bound to rER. Not membrane bound. Made from 1 large and 1 small subunit. Site of protein synthesis, specifically, translation Structure and function – rough endoplasmic reticulum - Ribosomes bound by a system of membranes Folds polypeptides to secondary / tertiary structure Packages to vesicles, transport to the Golgi apparatus etc. Structure and function – smooth endoplasmic reticulum - Similar to rER but without ribosomes – system of membranes Synthesises and processes lipids Structure and function – chloroplasts (plants and algae) - Thylakoid membranes are stacked up in some parts to form grana, which are linked by lamellae. These sit in the stroma (fluid) and are surrounded by a double membrane. Also contains starch granules and circular DNA. (Chlorophyll) absorbs light for photosynthesis to produce organic substances Structure and function – cell wall (plants, algae and fungi) - Made mainly of cellulose in plants and algae, and of chitin in fungi Rigid structure surrounding cells in plants, algae and fungi. Prevents the cell changing shape and bursting (lysis) Structure and function – cell vacuole (plants) - Contains cell sap – a weak solution of sugars and salts. Surrounding membrane is called the tonoplast. Maintains pressure in the cell (stop wilting) Stores/isolates unwanted chemicals in the cell Organisation of specialised cells in complex multicellular organisms - Specialised cell – the most basic structural/functional subunit in all living organisms; specialised for a particular function Tissue – Group of organised specialised cells; joined and working together to perform a particular function; often with the same origin Organ – Group of organised different tissues; joined and working together to perform a particular function Organ system – Group of organised organs; working together to perform a particular function You should be able to apply your knowledge to explain adaptations of eukaryotic cells with particular functions - Example: epithelial cells in the small intestine are specialised for efficient absorption. Villi and microvilli increase surface area. Lots of mitochondria to provide energy e.g. for active transport Structure of prokaryotic cell How prokaryotic cells differ from eukaryotic cells - Prokaryotic cell cytoplasm contains no membrane bound organelles e.g. mitochondria WHEREAS eukaryotic cell contains membrane bound organelles Prokaryotic cell has no nucleus / contains free floating DNA WHEREAS eukaryotic cell has a nucleus containing DNA Prokaryotic DNA is circular and isn’t associated with proteins WHEREAS eukaryotic DNA is linear and is associated with proteins Prokaryotic cell wall contains murein and peptidoglycan WHEREAS eukaryotic cell wall is made of cellulose Prokaryotic cells have smaller 70s ribosomes WHEREAS eukaryotic cells have larger ribosomes Prokaryotic cells may have…. one or more plasmid, a capsule, and/or one or more flagella Viruses - Acellular → not made of or able to be divided into cells Non-living → unable to exist/reproduce without a host cell Principles and limitations of optical microscopes, transmission electron microscopes and scanning electron microscopes Optical microscope - Use light to form a 2D image Visible light longer wavelength so lower resolution 200nm Low magnification x1500 2D image Only used on thin specimens Low resolution; can’t see internal structures of organelles or organelles smaller than 200nm e.g. ribosomes Low magnification ☺ Can see living organisms. Scanning electron microscope - Use electrons to form a 2D image - Beams of electrons scan surface, knocking off electrons from the specimen, which are gathered in a cathode ray tube to form an image - Electrons shorter wavelength (so higher resolution 0.2nm) - High magnification x1500000 Transmission electron microscope - Use electrons to form a 3D image - Electromagnets focus beam of electrons onto specimen, transmitted, more dense = more absorbed = darker appearance - Electrons shorter wavelength (so higher resolution 0.2nm) - High magnification x1500000 Vacuum; can’t see living organisms Lower resolution than TEM 2D image Only used on thin specimens Vacuum; can’t see living organisms ☺ 3D image ☺ High resolution; can see internal structures of organelles ☺ High magnification ☺ Used on thick specimens ☺ High resolution; see internal structures of organelles ☺ High magnification The difference between magnification and resolution - Magnification - how much bigger the image of a sample is compared to the real size, 𝐒𝐢𝐳𝐞 𝐨𝐟 𝐢𝐦𝐚𝐠𝐞 measured by 𝐌𝐚𝐠𝐧𝐢𝐟𝐢𝐜𝐚𝐭𝐢𝐨𝐧 = 𝐒𝐢𝐳𝐞 𝐨𝐟 𝐫𝐞𝐚𝐥 𝐨𝐛𝐣𝐞𝐜𝐭 - Resolution - how well distinguished an image is between 2 points; shows amount of detail; limited by wavelength of radiation used e.g. light Measuring the size of an object viewed with an optical microscope - - - Line up eyepiece graticule with stage micrometer Use stage micrometer to calculate the size of divisions on eyepiece graticule at a particular magnification Take the micrometer away and use the graticule to measure how many divisions make up the object Calculate the size of the object by multiplying the number of divisions by the size of division Recalibrate eyepiece graticule at different magnifications Preparing a ‘temporary mount’ of a specimen on a slide - - Use tweezers to place a thin section of specimen e.g. tissue on a water drop on a microscope slide Add a drop of a stain e.g. iodine in potassium iodide solution used to stain starch grains in plant cells Add a cover slip by carefully tilting and lowering it, trying not to get any air bubbles Principles of cell fractionation and ultracentrifugation as used to separate cell components 1. Homogenise tissue using a blender - Disrupts cell membrane / break open cell - Release contents / organelles 2. Place in a cold, isotonic, buffered solution - Cold reduces enzyme activity so organelles aren’t broken down - Isotonic so water doesn’t move in/out of organelles by osmosis so they don’t burst / shrivel - Buffered keeps pH constant so enzymes don’t denature 3. Filter homogenate - Remove large, unwanted debris e.g. whole cells, connective tissue 4. Ultracentrifugation a) Centrifuge homogenate in a tube at a low speed b) Remove pellet of heaviest organelle and spin supernatant at a higher speed c) Repeated at higher and higher speeds until organelles separated out, each time pellet is made of lighter organelles d) Separated in order of mass/density: nuclei → chloroplasts → mitochondria → lysosomes → endoplasmic reticulum → ribosomes There was a considerable period of time during which the scientific community distinguished between artefacts and cell organelles - Repeatedly prepared specimens in different ways If an object could only be seen with one preparation technique, but not another it was more likely to be an artefact than an organelle Cell cycle - In multicellular organisms, not all cells keep their ability to divide. Eukaryotic cells that do retain the ability to divide show a cell cycle. Interphase - S phase – DNA replicates semi-conservatively leading to two sister chromatids G1 and G2 – Number of organelles and volume of cytoplasm increases; protein synthesis; ATP content increased Mitosis - Parent cell divides = two genetically identical daughter cells, containing identical/exact copies of DNA of the parent cell. Stages - ‘PMAT’ Stages of mitosis (the behaviour of chromosomes and the role of spindle fibres attached to centromeres in the separation of chromatids) Prophase - Chromosomes condense, becoming shorter and thicker = appear as two sister chromatids joined by a centromere Nuclear envelope breaks down and centrioles move to opposite poles forming spindle network Metaphase - Chromosomes align along equator - Spindle fibres attach to chromosomes by centromeres Anaphase - Spindle fibres contract, pulling sister chromatids to opposite poles of the cell Centromere divides Telophase - Chromosomes uncoil, becoming longer and thinner Nuclear envelope reforms = two nuclei Spindle fibres and centrioles break down Cytokinesis - The division of the cytoplasm, usually occurs, producing two new cells The importance of mitosis in the life of an organism Parent cell divides to produce 2 genetically identical daughter cells for… - Growth of multicellular organisms by increasing cell number Repairing damaged tissues / replacing cells Asexual reproduction Recognising the stages of the cell cycle and explaining the appearance of cells in each stage of mitosis (source: kerboodle textbook online) - Interphase – C → no chromosomes visible (visible nucleus) Prophase – B → chromosomes visible but randomly arranged Metaphase – D → chromosomes lined up on the equator Anaphase – E → chromatids (in two sets) being separated to opposite poles by spindles, V shape shows sister chromatids have been pulled apart at their centromeres Telophase – A → chromosomes in two sets, one at each pole Uncontrolled cell division can lead to the formation of tumours and of cancers - Malignant tumour – cancer – spreads and affects other tissues / organs Benign tumour – non-cancerous Many cancer treatments are directed at controlling the rate of cell division Disrupt the cell cycle – cell division / mitosis slows – tumour growth slows - Prevent DNA replication → prevent / slows down mitosis - Disrupts spindle activity / formation → chromosomes can’t attach to spindle by their centromere → sister chromatids can’t be pulled to opposite poles of the cells → prevent/slow mitosis Disrupt cell cycle of normal cells too, especially rapidly dividing ones e.g. cells in hair follicles ☺ Drugs more effective against cancer cells because dividing uncontrollably / rapidly Prokaryotic cells replicate by binary fission - Circular DNA and plasmids replicate (circular DNA replicates once, plasmids can be replicated many times) Cytoplasm expands (cell gets bigger) as each DNA molecule moves to opposite poles of the cell Cytoplasm divides = 2 daughter cells, each with a single copy of DNA and a variable number of plasmids Viral replication Viruses don’t undergo cell division because they are non-living 1. Attachment protein binds to complementary receptor protein on surface of host cell 2. Inject nucleic acid (DNA/RNA) into host cell 3. Infected host cell replicates the virus particles Fluid-mosaic model of membrane structure - Molecules within membrane can move laterally (fluid) e.g. phospholipids Mixture of phospholipids, proteins, glycoproteins and glycolipids The structure of a cell membrane - - Phospholipid bilayer - Phosphate heads are hydrophilic so attracted to water – orientate to the aqueous environment either side of the membrane - Fatty acid tails are hydrophobic so repelled by water – orientate to the inside/interior of the membrane Embedded proteins (intrinsic or extrinsic) - Channel and carrier proteins (intrinsic) Glycolipids (lipids and attached polysaccharide chain) and glycoproteins (proteins with polysaccharide chain attached) Cholesterol (binds to phospholipid hydrophobic fatty acid tails) The fluid mosaic model of membrane structure can explain how molecules can enter/leave a cell Phospholipid bilayer - Allows movement of non-polar small/lipid-soluble molecules e.g. oxygen or water, down a concentration gradient (simple diffusion) Restricts the movement of larger/polar molecules Channel proteins (some are gated) and carrier proteins - Allows movement of water-soluble/polar molecules / ions, down a concentration gradient (facilitated diffusion) Carrier proteins - Allows the movement of molecules against a concentration gradient using ATP (active transport) Features of the plasma membrane adapt it for its other functions - Phospholipid bilayer - Maintains a different environment on each side of the cell or compartmentalisation of cell Phospholipid bilayer is fluid - Can bend to take up different shapes for phagocytosis / to form vesicles Surface proteins / extrinsic / glycoproteins / glycolipids - Cell recognition / act as antigens / receptors Cholesterol - Regulates fluidity / increases stability The role of cholesterol - Makes the membrane more rigid / stable / less flexible, by restricting lateral movement of molecules making up membrane e.g. phospholipids (binds to fatty acid tails causing them to pack more closely together) Note: not present in bacterial cell membranes Movement across membranes by simple diffusion and factors affecting rate - Net movement of small, non-polar molecules e.g. oxygen or carbon dioxide, across a selectively permeable membrane, down a concentration gradient Passive / no ATP / energy required Factors affecting rate – surface area, concentration gradient, thickness of surface / diffusion distance Movement across membranes by facilitated diffusion, factors affecting rate and role of carrier/channel proteins - Net movement of larger/polar molecules e.g. glucose, across a selectively permeable membrane, down a concentration gradient Through a channel/carrier protein Passive /no ATP/energy required Factors affecting rate – surface area, concentration gradients (until the number of proteins is the limiting factor as all are in use / saturated), number of channel/carrier proteins Role of carrier and channel proteins: - Carrier proteins transport large molecules, the protein changes shape when molecule attaches - Channel proteins transport charged/polar molecules through its pore (some are gated so can open/close e.g. Voltage-gated sodium ion channels) - Different carrier and channel proteins facilitate the diffusion of different specific molecules Movement across membranes by active transport and factors affecting rate - Net movement of molecules/ions against a concentration gradient Using carrier proteins Using energy from the hydrolysis of ATP to change the shape of the tertiary structure and push the substances though Factors affecting rate – pH/temp (tertiary structure of carrier protein), speed of carrier protein, number of carrier proteins, rate of respiration (ATP production) Movement across membranes by co-transport, illustrated by the absorption of sodium ions and glucose by cells lining the mammalian ileum 1. Sodium ions actively transported out of epithelial cells lining the ileum, into the blood, by the sodium-potassium pump. Creating a concentration gradient of sodium (higher conc. of sodium in lumen than epithelial cell) 2. Sodium ions and glucose move by facilitated diffusion into the epithelial cell from the lumen, via a co-transporter protein 3. Creating a concentration gradient of glucose – higher conc. of glucose in epithelial cell than blood 4. Glucose moves out of cell into blood by facilitated diffusion through a protein channel Movement across membranes by osmosis and factors affecting rate - Net movement of water molecules across a selectively permeable membrane down a water potential gradient Water potential is the likelihood (potential) of water molecules to diffuse out of or into a solution; pure water has the highest water potential and adding solutes to a solution lowers the water potential (more negative) Passive Factors affecting rate – surface area, water potential gradient, thickness of exchange surface / diffusion distance How might cells be adapted for transport across their internal or external membranes - By an increase in surface area Increase in number of protein channels / carriers Antigen definition - Molecules which, when recognised as non-self/foreign by the immune system, can stimulate an immune response and lead to the production of antibodies Often proteins on the surface of cells Note: proteins have a specific tertiary structure / shape allowing different proteins to act as specific antigens Antigens are specific so allow the immune system to identify… - Pathogens (disease causing organisms) e.g. viruses, fungi, bacteria Cells from other organisms of the same species e.g. organ transplant, blood transfusion Abnormal body cells e.g. cancerous cells / tumours Toxins released from bacteria Phagocytosis of pathogens – non-specific immune response 1. Phagocyte e.g. macrophage recognises foreign antigens on the pathogen and binds to the antigen 2. Phagocyte engulfs pathogen by surrounding it with its cell surface membrane / cytoplasm 3. Pathogen contained in vacuole/vesicle/phagosome in cytoplasm of phagocyte 4. Lysosome fuses with phagosome and releases lysozymes (hydrolytic enzymes) into the phagosome 5. These hydrolyse / digest the pathogen 6. Phagocyte becomes antigen presenting and stimulates specific immune response The cellular response (the response of T lymphocytes to a foreign antigen e.g. infected cells, cells of the same species) 1. T lymphocytes recognises antigen presenting cells after phagocytosis (foreign antigen) 2. Specific T helper cell with receptor complementary to specific antigen binds to it, becoming activated and dividing rapidly by mitosis to form clones which: a) Stimulate B cells for the humoral response b) Stimulate cytotoxic T cells to kill infected cells by producing perforin c) Stimulate phagocytes to engulf pathogens by phagocytosis The humoral response (the response of B lymphocytes to a foreign antigen e.g. in blood/tissues) 1. Clonal selection: a) Specific B cell binds to antigen presenting cell and is stimulated by helper T cells which releases cytokines b) Divides rapidly by mitosis to form clones (clonal expansion) 2. Some become B plasma cells for the primary immune response – secrete large amounts of monoclonal antibody into blood 3. Some become B memory cells for the secondary immune response Primary response – antigen enters body for the first time (role of plasma cells) - Produces antibodies slower and at a lower concentration because - Not many B cells available that can make the required antibody - T helpers need to activate B plasma cells to make the antibodies (takes time) So infected individual will express symptoms Secondary response – same antigen enters body again (role of memory cells) - Produces antibodies faster and at a higher concentration because B and T memory cells present B memory cells undergo mitosis quicker / quicker clonal selection Antibodies - Quaternary structured protein (immunoglobin) Secreted by B lymphocytes e.g. plasma cells and produced in response to a specific antigen Binds specifically to antigens (monoclonal) forming an antigen-antibody complex Describe and explain how the structure of an antibody relates to its function - Primary structure of protein = sequence of amino acids in a polypeptide chain Determines the folds in the secondary structure as R groups interact Determines the specific shape of the tertiary structure and position of hydrogen, ionic and disulfide bonds Quaternary structure is comprised of 4 polypeptide chains (tertiary structured) held together by hydrogen, ionic and disulfide bonds - Enables the specific shaped variable region (binding site) to form which is a complementary shape to a specific antigen - Enables antigen-antibody complex to form - - How do antibodies work to destroy pathogens e.g. bacterial cells? - Binds to two pathogens at a time (at variable region/binding site) forming an antigenantibody complex Enables antibodies to clump the pathogens together – agglutination Phagocytes bind to the antibodies and phagocytose many pathogens at once Note: the hinge region means an antibody can bind to antigens / pathogens different distances part What is a vaccination? - Injection of antigens From attenuated (dead or weakened) pathogens Stimulates the formation of memory cells A vaccine can lead to symptoms because some of the pathogens might be alive / active / viable; therefore, the pathogen could reproduce and release toxins, which can kill cells The use of vaccines to provide protection for individuals against disease - Normal immune response but the important part is that memory cells are produced On reinfection / secondary exposure to the same antigen, the secondary response therefore produces antibodies faster and at a higher concentration Leading to the destruction of a pathogen/antigen (e.g. agglutination and phagocytosis) before it can cause harm / symptoms = immunity The use of vaccines to provide protection for populations against disease (herd immunity) - Large proportion but not 100% of population vaccinated against a disease – herd immunity Makes it more difficult for the pathogen to spread through the population because… - More people are immune so fewer people in the population carry the pathogen / are infected - Fewer susceptible so less likely that a susceptible / non-vaccinated individual will come into contact with an infected person and pass on the disease Differences between active and passive immunity Active immunity Initial exposure to antigen e.g. vaccine or primary infection Memory cells involved Passive immunity No exposure to antigen No memory cells involved Antibody is produced and secreted by (B) plasma cells Slow; takes time to develop Long term immunity → antibody can be produced in response to a specific antigen again Antibody introduced into body from another organism e.g. breast milk / across placenta from mother Fast acting Short term immunity (antibody broken down) Ethical issues associated with the use of vaccines - Tested on animals before use on humans → animals have a central nervous system so feel pain (some animal based substances are also used to produce vaccines) Tested on humans → volunteers may put themselves at unnecessary risk of contracting the disease because they think they’re fully protected e.g. HIV vaccine so have unprotected sex → vaccine might not work Can have side effects Expensive – less money spent on research and treatments of other diseases Antigen variability is often an explanation for why… - New vaccines against a disease need to be developed more frequently e.g. influenza Vaccines against a disease may be hard to develop or can’t be developed in the first place e.g. HIV May experience a disease more than once e.g. common cold Explain the effect of antigen variability on disease - Change in antigen shape (due to a genetic mutation) Not recognised by B memory cell → no plasma cells / antibodies Not immune Must re-undergo primary immune response → slower / releases lower concentration of antibodies Disease symptoms felt Explain the effect of antigen variability on disease prevention (vaccines) - Change in antigen shape (due to a genetic mutation) Existing antibodies with a specific shape unable to bind to changed antigens / form antigen-antibody complex Immune system i.e. memory cells won’t recognise different antigens (strain) Evaluate methodology, evidence and data relating to the use of vaccinations - - A successful vaccination programme: - Produce suitable vaccine - Effective – make memory cells - No major side effects → side effects discourage individuals from being vaccinated - Low cost / economically viable - Easily produced / transported / stored / administered - Provides herd immunity Evaluating a conclusion that’s been made from a set of data / study - If there is a scatter graph, the relationship between two variables may be a positive / negative correlation, or no correlation - But correlation between two variables doesn’t always mean there’s a causal relationship – correlation could be due to change or another variable / factor - Repeatability (when an experiment is repeated using the same method and equipment and obtains the same results) - Have there been other experiments / studies showing the same? - Validity (suitability of the investigative procedure to answer the question being asked) - Does the data answer the question set out to investigate? - Example: research project on potential vaccines to protect people against HIV used monkeys and a virus called SIV (which only infects monkeys and causes a condition similar to AIDS in them) . Scientists have questioned the value of the research because there may be differences between human and money responses / immune systems, and a vaccine developed against SIV may not work against HIV / may be (significant) differences between SIV and HIV - Potential bias? The use of monoclonal antibodies - Monoclonal antibody = antibody produced from a single group of genetically identical (clones) B cells / plasma cells - Identical structure Bind to specific complimentary antigen - Have a binding site / variable region with a specific tertiary structure / shape - Only one complementary antigen will fit Why are monoclonal antibodies useful in medicine? - Only bind to specific target molecules / antigens because… Antibodies have a specific tertiary structure (binding site / variable region) that’s complementary to a specific antigen which can bind/fit to the antibody Monoclonal antibodies: targeting medication to specific cell types by attaching a therapeutic drug to an antibody Example: cancer cell 1. Monoclonal antibodies made to be complementary to antigens specific to cancer cells → cancer cells are abnormal body cells with different antigens (tumour markers) 2. Anti-cancer drug attached to antibody 3. Antibody binds / attaches to cancer cells (forming antigen-antibody complex) 4. Delivers attached anti-cancer drug directly to specific cancer cells so drug accumulates → fewer side effects e.g. fewer normal body cells killed Exam question example: some cancer cells have a receptor protein in their cell-surface membrane that binds to a hormone called growth factor. This stimulates the cancer cells to divide. Scientists have produced a monoclonal antibody that stops this stimulation. Use your knowledge of monoclonal antibodies to suggest how this antibody stops the growth of a tumour (3 marks) ✓ ✓ ✓ Antibody has specific tertiary structure / binding site / variable region Complementary (shape / fit) to receptor protein / GF / binds to receptor protein Prevents GF binding (to receptor) Monoclonal antibodies: medical diagnosis Example: pregnancy test - - Pregnant women have the hormone hCG in their urine Urine test strip has 3 parts with 3 different antibodies - Application area, position 1: antibodies complementary to hCG (bound to a blue coloured bead) - Middle, position 2: antibodies complementary to hCG-antibody complex - End, position 3: antibodies complementary to antibody without hCG attached If pregnant - hCG binds to antibodies in application area = hCG-antibody complex - Travels up test strip, binds to antibodies at position 2 = blue line If not pregnant - No hCG in urine so hCG doesn’t bind to antibodies in application area so doesn’t bind to antibodies at position 2 = no blue line - Bind to antibodies at position 3 → blue line = control Exam question example: Malaria is a disease caused by parasites belonging to the genus Plasmodium. Two species that cause malaria are Plasmodium falciparum and Plasmodium vivax. A test strip that uses monoclonal antibodies can be used to determine whether a person is infected by Plasmodium. It can also be used to find which species of Plasmodium they are infected by. - A sample of a person’s blood is mixed with a solution containing n antibody, A, that binds to a protein found in both species of Plasmodium. This antibody has a coloured dye attached. - A test strip is then put into the mixture. The mixture moves up the test strip by capillary action to an absorbent pad. - Three other antibodies, B, C and D are attached to the test strip. The position of these antibodies and what they bind to is shown in figure 1. (a) Explain why antibody A attaches only to the protein found in species of Plasmodium. (2 marks) ✓ Antibody has tertiary structure ✓ Complementary to binding site on protein (b) Antibody B is important is this test shows a person is not infected with Plasmodium. Explain why antibody B is important. (2 marks) ✓ Prevents false negative results ✓ (Since) shows antibody A has moved up strip / has not bound to any Plasmodium protein (c) One of these test strips was used to test a sample from a person thought to be infected with Plasmodium. Figure 2 shows the result. What can you conclude from this result? Explain how you reached your conclusion. (4) ✓ Person is infected with Plasmodium / has malaria ✓ Infected with (plasmodium) vivax ✓ Coloured dye where antibody C present ✓ That only binds to protein from vivax / no reaction with antibody for falciparum The use of antibodies in the ELISA (enzyme linked immunosorbent assay) test - Can determine if a patient has - - a) Antibodies to a certain antigen b) Antigen to a certain antibody - Used to diagnose diseases or allergies (e.g. HIV / Lactose intolerance) Why use controls when performing the ELISA test? - Controls enable a comparison with the test - To show that - Only the enzyme and nothing else causes colour change - Washing is effective and all unbound antibody is washed away Explain why the secondary and detection antibody must be washed away - Enzyme attached to antibody reacts with substrate turning the solution a different colour; indicates a positive result - Not washed out → enzymes will react with the substrate - Therefore give a positive result even if no antigen present (false positive) Exam question example: A test has been developed to find out whether a person is infected with G. lambia. The test is shown in the flow chart. (i) (ii) (iii) Explain why the antibodies used in this test must be monoclonal antibodies. (1 mark) ✓ All have same shape / only binds to Giardia / one type of / specific antigen Explain why the Giardia antigen binds to the antibody in step 2. (1 mark) ✓ Has complementary 9shape) / due to (specific) tertiary structure / variable region (of antibody) The plate must be washed at the start of step 4, otherwise a positive result could be obtained when the Giardia antigen is not present. Explain why a positive result could be obtained if the plate is not washed at the start of step 4. (2 marks) ✓ Enzyme / second antibody would remain / is removed by washing ✓ Enzyme can react with substrate (when no antigen is present) Exam question example: A test has been developed to find out whether a person has antibodies against the mumps virus. The test is shown in the diagram. (a) Explain why this test will detect mumps antibodies, but not other antibodies in the blood. (1 mark) ✓ Antibodies are specific to mumps antigen ✓ 2nd antibodies specific to mumps antibody (b) Explain why it is important to wash the well at the start of step 4. (2 marks) ✓ Removes unbound 2nd antibodies ✓ Otherwise enzyme may be present / get a colour change anyway / false positive (c) Explain why there be no colour change if mumps antibodies are not present in the blood. (2 marks) ✓ No antibodies to bind (to antigen) ✓ Therefore 2nd antibody (with the enzyme) won’t bind ✓ No enzyme/enzyme-carrying antibody present (after washing in step 4) Ethical issues associated with the use of monoclonal antibody - Animals are involved in the production of monoclonal antibodies i.e. by producing cancer in mice who have a CNS so feel pain, and it is unfair to give them a disease Although effective treatment for cancer and diabetes has caused deaths when used in treatment of Multiple Sclerosis - Patients need to be informed of risk and benefits before treatment so they can make informed decisions You also need to be able to evaluate methodology, evidence and data relating to the use of monoclonal antibodies The structure of HIV – human immunodeficiency virus The replication of HIV in helper T cells 1. HIV infects T helper cells (host cell) - HIV attachment protein (GP120) attaches to a receptor on the helper T-cell membrane 2. Virus lipid envelope fuses with cell surface membrane and capsid released into cell which uncoats, releasing RNA and reverse transcriptase into cytoplasm 3. Viral DNA is made from viral RNA - Reverse transcriptase produces a complementary viral DNA strand from viral RNA template - Double stranded DNA is made from this (DNA polymerase) 4. Viral DNA integrated into host cell’s DNA (by enzyme integrase) 5. This remains latent for a long time in host cell until activated 6. Host cell enzymes used to make viral proteins from viral DNA (within human DNA) → viral proteins assembled with viral RNA to make a new virus 7. New virus bud from cell (taking some of cell surface membrane as envelope) 8. Eventually kills helper T cells 9. Most host cells are infected and process repeat How HIV causes the symptoms of AIDS – acquired immune deficiency syndrome - - Infects and kills helper T cells (host cell) as it multiplies rapidly - T helper cells then can’t stimulate cytotoxic T cells, B cells and phagocytes → impaired immune response - E.g. B plasma cells can’t secrete antibodies for agglutination and destruction of pathogens by phagocytosis Immune system deteriorates - More susceptible to infections - Diseases that wouldn’t cause serious problems in healthy immune system are deadly (opportunistic infections) e.g. pneumonia Why antibiotics are ineffective against viruses - Antibiotics can’t enter human calls – but viruses exists in its host cell (they are acellular) Viruses don’t have own metabolic reactions e.g. ribosomes (use of the host cell’s) which antibiotics target If we did use them… act as a selection pressure + gene mutation = resistant strain of bacteria via natural selection → reducing effectiveness of antibiotics and waste money