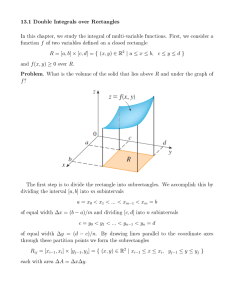
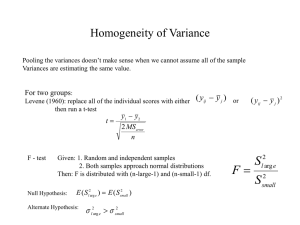
Lecture 10 Normal Theory Hypothesis tests for ANOVA Goal: Sampling distribution of F Goal: Sampling distribution of F Model Assumptions: yij = µi + ij ; i = 1, . . . , a; j = 1, . . . , n. ij ∼iid N (0, σ 2 ) (Normal; constant variance) • H0 : µi = µj for all i, j; • H1 : µi 6= µj for some i, j. As we saw last lecture, under both the null and alternative: SSE ∼ χ2a(n−1) σ2 while under the null hypothesis: SStrt /σ 2 ∼ χ2a−1 Consequently, under the null hypothesis we have F = M Strt = M SE SStrt 1 a−1 σ 2 SSE 1 a(n−1) σ 2 ∼ Fa−1,a(n−1) under H0 . Decision Procedure Reject H0 if F (y) : M Strt /M SE > c. Under H0 : P (F (Y) > c) = P (Fa−1,a(n−1) > c) we choose c = Fa−1,a(n−1),1−α , i.e. the 1 − α-quantile of Fa−1,a(n−1) distribution c can be obtained in using the R command: qf( 1 − α, a − 1, a(n − 1)). Under the normality assumptions, if the null hypothesis is true we obtain: P (F (Y) > Fa−1,a(n−1),1−α | H0 ) = α Thus theType I error-rate is α. Computation of a p-value: A p-value may be computed by finding: P r(Fa−1,a(n−1) > F (y)) where F (y) is the observed value of the F-statistic, and Fa−1,a(n−1) is a random variable with the the F-distribution on (a − 1, a(n − 1)) degrees of freedom. This may be computed via 1 - pf(F (y),a − 1,a(n − 1)). 1 Checking Model Adequacy Sensitivity of F -tests and validity of normal-theory p-values rely on several model assumptions. Model: yij = µ + τi + ij Assumptions: (i) ij are normally distributed with mean zero; (ii) V (ij ) = σ 2 for all i, j; (iii) ij are independent. Violations of assumptions can be checked via residual analysis. Model estimate: yij = ȳ·· + (ȳi· − ȳ·· ) + (yij − ȳi· ) = µ̂ + τ̂i + ˆij Our estimate of any value in group i is ŷij = µ̂ + τ̂i = ŷi· Our estimate of the error is ˆij = yij − ȳi· . ˆij is called the residual for observation i, j. Assumptions about ij can be checked by examining the values of ˆij ’s: Normality Assumptions: • Histogram: Make a histogram of ˆij ’s. This should look approximately bell-shaped; if there are enough observations compare to N (0, s2 ) distribution, graphically. (In small samples, the histograms need not look particularly bell-shaped: see examples.) • Normal probability, or qq-plot: If ij ∼ N (0, σ 2 ) then the ordered residuals (ˆ (1) , . . . , ˆ(an) ) should correspond linearly with quantiles of a standard normal distribution. Example: Hermit Crab Data yij = population total in transect j of site i. Question: Do sites vary in their population levels? Model: Yij = µ + τi + ij Note that the data are counts so they cannot be exactly normally distributed (why not?) Residuals: ˆij = yij − µ̂i = yij − ȳi· . (See histogram, qq-plots) Conclusion: Data are clearly not normally distributed. 2 Homogeneity of Variances: The ANOVA model supposes that V (ij ) = σ 2 for all i, j. Trt 1 (a) tabulate residual variance in each treatment: .. . Sample var. of (ˆ i1 , . . . , ˆin ) s21 .. . a s2a Rule of Thumb: – If s2smallest /s2largest > 1/3: don’t worry; – If s2smallest /s2largest < 1/7: worry: need to account for the non-constant variance, especially if sample sizes are different. (b) Residual vs. Fitted Value plots For many types of data there is a mean-variance relationship: typically, groups with large means tend to have larger variances. To check this: plot ˆij (residual) vs. ŷij = ȳi· (fitted value). Example: Crab data Site 4 5 6 1 3 2 Mean 9.24 10.00 12.64 33.80 50.64 68.72 SD 17.39 19.84 23.01 50.39 107.44 125.35 Here we have s2smallest /s2largest ≈ 1/50 it (See plots of fitted vs. residual.) (c) Statistical tests of equality of variance: Levene’s Test: Let dij = |yij − ỹi |, where ỹi is the sample variance from group i. These differences will be ‘large’ in group i, if group i has a ‘large’ variance; and ‘small’ in group i, if group i has a ‘small’ variance. We compute: Pa ¯ 2 /(a − 1) (di· − d) Pni=1 ¯ 2 i=1 j=1 (dij − di· ) /(a(n − 1)) F0 = Pa n which is the ratio of the between group variability of the dij to the within group variability of the dij . 3 Reject H0 : V (ij = σ 2 for all i, j if F0 > Fa−1,a(n−1),1−α Crab data: F0 = 14, 229 = 2.93 > F5,144,0.95 = 2.28 4, 860 hence we reject the null hypothesis of equal variances at the 0.05 level. See also – Bartlett’s test (p.81); – the equality test for two variances (section 2.6; F-test). So the ANOVA assumptions are violated! Now what? What should we do if the data aren’t normal, or the variances aren’t equal? Power transformations: Recall that one justification of the normal distribution model: Yij = µi + ij was that if the noise = Xij1 + Xij2 + · · · was the result of the addition of unobserved additive, independent effects then by the central limit theorem ij will be approximately normal. However, if effects are multiplicative so that in fact: Yij = µi × ij = µi × (Xij1 × Xij2 × · · · ) Then the Yij will not be normal, and the variances will not be constant: V ar(Yij ) = µ2i V ar(Xij1 × Xij2 × · · · ). Log transform: log Yij = log µi + (log Xij1 + log Xij2 + · · · ) V (log Yij ) = V (log µi + log Xij1 + log Xij2 + · · · ) = V (log Xij1 + log Xij2 + · · · ) 2 = σlog y which does not depend on the mean µi . Also note that by the central limit theorem the errors should be approximately normally distributed. Other transformations: Often we observe: σyij ∝ µα i i.e. the standard deviation of a group depends on the group mean. ∗ We would like to achieve: σyij ∝ (µ∗i )0 = 1 so that the standard deviation doesn’t depend on the mena. ∗ λ Suppose that we let yij = yij , then we obtain: α+λ−1 ∗ ∝ µ σyij i 4 hence if we take λ = 1 − α then we will succeed in removing the dependence of the group s.d. on the group mean. Of course, we typically don’t know α, but we could try to estimate it from data. Thus to summarize, our strategy is as follows: (1) Roughly estimate α from data 1−α (2) Analyse yij instead of yij . Estimation of α: σyi ∝ µα i ⇔ σ yi log σyi so log si ≈ (log θ) + α log ȳi· = θµα i = log θ + α × log µi Thus we may use the following procedure: (1) Plot log si vs. log ȳi· (2) Fit a least squares line: lm( log si ∼ log ȳi· ) (3) The slope α̂ of the line is an estimate of α. 1−α̂ ∗ (4) Analyze yij = yij . 5