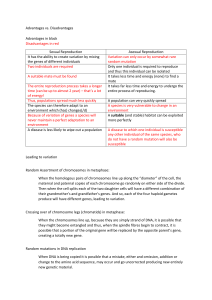
3.1 Genes Essential idea: Every living organism inherits a blueprint for life from its parents. Genes and hence genetic information is inherited from parents, but the combination of genes inherited from parents by each offspring will be different. In sexual reproduction each parent can only pass on 50% of there genes as the other 50% comes from the second parent. By Chris Paine http://www.nature.com/scitable/content/ne0000/ne0000/ne0000/ne0000/5 6538/shaw_family_FULL.jpg http://publications.nigms.nih.gov/insidethecell/images/ch4_meiosissex.jpg https://bioknowledgy.weebly.com/ Understandings, Applications and Skills Statement 3.1.U1 3.1.U2 3.1.U3 3.1.U4 3.1.U5 3.1.U6 3.1.U7 3.1.A1 3.1.A2 3.1.S1 Guidance A gene is a heritable factor that consists of a length of DNA and influences a specific characteristic. A gene occupies a specific position on a chromosome. The various specific forms of a gene are alleles. Alleles differ from each other by one or only a few bases. Deletions, insertions and frame shift mutations do New alleles are formed by mutation. not need to be included. The genome is the whole of the genetic information of an organism. The entire base sequence of human genes was sequenced in the Human Genome Project. Students should be able to recall one specific The causes of sickle cell anemia, including a base substitution mutation, a change to the base sequence base substitution that causes glutamic acid to be substituted by valine as the sixth amino acid in the of mRNA transcribed from it and a change to the hemoglobin polypeptide. sequence of a polypeptide in hemoglobin. The number of genes in a species should not be Comparison of the number of genes in humans with referred to as genome size as this term is used other species. for the total amount of DNA. At least one plant and one bacterium should be included in the comparison and at least one species with more genes and one with fewer genes than a human. Use of a database to determine differences in the base The Genbank® database can be used to search for DNA base sequences. The cytochrome C sequence of a gene in two species. gene sequence is available for many different organisms and is of particular interest because of its use in reclassifying organisms into three domains. 3.1.U1 A gene is a heritable factor that consists of a length of DNA and influences a specific characteristic. AND 3.1.U2 A gene occupies a specific position on a chromosome. AND 3.1.U3 The various specific forms of a gene are alleles. AND 3.1.U4 Alleles differ from each other by one or only a few bases. A gene is a heritable factor that controls or influences a specific characteristic, consisting of a length of DNA occupying a particular position on a chromosome (locus) http://learn.genetics.utah.edu/content/m olecules/gene/ 3.1.U1 A gene is a heritable factor that consists of a length of DNA and influences a specific characteristic. AND 3.1.U2 A gene occupies a specific position on a chromosome. AND 3.1.U3 The various specific forms of a gene are alleles. AND 3.1.U4 Alleles differ from each other by one or only a few bases. A gene is a heritable factor that controls or influences a specific characteristic, consisting of a length of DNA occupying a particular position on a chromosome (locus) http://learn.genetics.utah.edu/content/m olecules/gene/ 3.1.A2 Comparison of the number of genes in humans with other species. Humans see themselves as being more complex and evolved than other species. Therefore you might well expect to see a larger number of genes in humans than in other organisms. It is not just plants such as the grapevine that have large numbers of genes; water fleas are an animal example of an organism with more genes than humans. Q - When analysing an organisms’ complexity, what other than the count of an organisms’ genes needs to be considered? https://www.sciencenews.org/sites/default/files/storyone_backstory_2.gif http://learn.genetics.utah.edu/content/chromosomes/in tro/ DNA Supercoiling: https://youtu.be/AF2wwMReTf8 3.1.U6 The genome is the whole of the genetic information of an organism. AND 3.1.U7 The entire base sequence of human genes was sequenced in the Human Genome Project. http://web.ornl.gov/sci/techresources/Human_Genome/index.shtml The Human Genome* Project (HGP) was an international 13-year effort, 1990 to 2003. Primary goals were to discover the complete set of human genes and make them accessible for further biological study, and determine the complete sequence of DNA bases in the human genome. http://www.ncbi.nlm.nih.gov/geno me/guide/human/ https://www.dnalc.org/view/15477-The-publicHuman-Genome-Project-mapping-the-genomesequencing-and-reassembly-3D-animation-.html *The genome is the entire genetic material of an organism. It consists of DNA (or RNA in RNA viruses) and includes both the genes and the non-coding sequences. Nature of Science: Developments in scientific research follow improvements in technology - gene sequencers are used for the sequencing of genes. (1.8) http://web.ornl.gov/sci/techresources/Human_Genome/index.shtml “The first methods for sequencing DNA were developed in the mid-1970s. At that time, scientists could sequence only a few base pairs per year, not nearly enough to sequence a single gene, much less the entire human genome. By the time the HGP began in 1990, only a few laboratories had managed to sequence a mere 100,000 bases, and the cost of sequencing remained very high. Since then, technological improvements and automation have increased speed and lowered cost to the point where individual genes can be sequenced routinely, and some labs can sequence well over 100 million bases per year.” (https://www.genome.gov/10001177) Key advances in technology: • Biotechnology techniques such as PCR are used to prepare samples: the DNA needs to be copied to prepare a sufficiently large pure samples to sequence • Computers automate the sequencing process • Fluorescent labeling techniques enable all four nucleotides to be analysed together • Lasers are used to fluoresce the dye markers • Digital camera technology reads the dye markers • Computers are used to assemble the base sequence 3.1.S1 Use of a database to determine differences in the base sequence of a gene in two species. One use of aligning base sequences is to determine the differences between species: this can be used to help determine evolutionary relationships. Your task is to analyse the differences between three or more species (the skill asks for two species, but the online Clustal tool works better with a minimum of three). GenBank http://www.ncbi.nlm .nih.gov/genbank For each chosen species retrieve the base sequence: • Go to GenBank website http://www.ncbi.nlm.nih.gov/genbank • Select ‘Gene’ from the search bar • Enter the name of a gene (e.g. AMY1A for salivary amylase 1A or COX1 for cytochrome oxidase 1) AND the organism (use the binomial) and press ‘Search’ n.b. if you are comparing species the gene chosen needs to be the same for each species • Select the ‘Name/Gene ID’ to get a detailed view • Scroll down to the ‘Genomic regions, transcripts, and products’ section and click on ‘FASTA’ • Copy the entire sequence from ‘>’ onwards • Save the sequence – you will need to align with the other species next http://bitesizebio.s3.amazonaws.com/wp-content/uploads/2012/10/header-image-copy18.jpg 3.1.S1 Use of a database to determine differences in the base sequence of a gene in two species. To align the sequences: • Go to the Clustal Omega website http://www.ebi.ac.uk/Tools/msa/clustalo/ • In STEP 1 Select ‘DNA’ under ‘a set of’ • Paste the chosen sequences into the box (each sequence must start on a new line) • Press ‘Submit’ (and wait – depending on the size of the sequences you may have to wait for a couple of minutes) http://www.ebi.ac.uk/Tools/msa/clustalo/ Analysis: • ‘Alignments’ allows you to visually check the results – this is easier if the chosen gene has a short base sequence • Under ‘Results Summary’ use the ‘Percent Identity Matrix’ to quantify the overall similarity (0 = no similarity, 100 = identical) • Under ‘Phylogenic Tree’ chose the ‘Real’ option for the Phylogram to get a visual representation of how similar the species are (based on the chosen gene). http://bitesizebio.s3.amazonaws.com/wp-content/uploads/2012/10/header-image-copy18.jpg Bibliography / Acknowledgments Bob Smullen 3.2 Chromosomes Essential idea: Chromosomes carry genes in a linear sequence that is shared by members of a species. The asian rice (Oryza sativa) genome can be seen illustrated above. Rice possesses up 63,000 genes divided up between 12 chromosomes. Below is a map of part of the first chromosome showing the gene loci present on it. Although different varieties (estimated 40,000 worldwide) will possess different alleles for genes, all individuals will share the same twelve chromosomes and the alleles of each variety will occur at the same position on same chromosome, i.e. at the same gene loci. By Chris Paine http://www.cambia.org/daisy/RiceGenome/2959/version/default/part/Ima geData/data/Rice%20chromosomes.png http://rgp.dna.affrc.go.jp/E/publicdata/naturegenetics/chr01.gif https://bioknowledgy.weebly.com/ Understandings Statement 3.2.U1 Guidance Prokaryotes have one chromosome consisting of a circular DNA molecule. 3.2.U2 Some prokaryotes also have plasmids but eukaryotes do not. 3.2.U3 Eukaryote chromosomes are linear DNA molecules associated with histone proteins. 3.2.U4 In a eukaryote species there are different chromosomes that carry different genes. 3.2.U5 Homologous chromosomes carry the same sequence of genes but not necessarily the same alleles of those genes. 3.2.U6 Diploid nuclei have pairs of homologous chromosomes. The two DNA molecules formed by DNA 3.2.U7 Haploid nuclei have one chromosome of each pair. replication prior to cell division are considered to be sister chromatids until the splitting of the centromere at the start of anaphase. After this, they are individual chromosomes. 3.2.U8 The number of chromosomes is a characteristic feature of members of a species. 3.2.U9 A karyogram shows the chromosomes of an organism The terms karyotype and karyogram have in homologous pairs of decreasing length. different meanings. Karyotype is a property of a cell - the number and type of chromosomes present in the nucleus, not a photograph or diagram of them. 3.2.U10 Sex is determined by sex chromosomes and autosomes are chromosomes that do not determine sex. Applications and Skills Statement 3.2.A1 3.2.A2 3.2.A3 3.2.A4 3.2.S1 Guidance Cairns’ technique for measuring the length of DNA molecules by autoradiography. Comparison of genome size in T2 phage,Escherichia Genome size is the total length of DNA in an coli, Drosophila melanogaster, Homo sapiens and Paris organism. The examples of genome and japonica. chromosome number have been selected to allow points of interest to be raised. Comparison of diploid chromosome numbers of Homo sapiens, Pan troglodytes, Canis familiaris, Oryza sativa, Parascaris equorum. Use of karyograms to deduce sex and diagnose Down syndrome in humans. Use of databases to identify the locus of a human gene and its polypeptide product. Review: 1.2.U1 Prokaryotes have a simple cell structure without compartmentalization. Ultrastructure of E. coli as an example of a prokaryote http://www.tokresource.org/tok_classes/biobiobio/biomenu/metathink/required_drawings/index.htm • E. Coli is a model organism used in research and teaching. Some strains are toxic to humans and can cause food poisoning. • We refer to the cell parts/ultrastructure of prokaryotes rather than use the term organelle as very few structures in prokaryotes are regarded as organelles. 3.2.U1 Prokaryotes have one chromosome consisting of a circular DNA molecule. 3.2.U2 Some prokaryotes also have plasmids but eukaryotes do not. Prokaryotes have two types of DNA: • single chromosome • plasmids The single prokaryotic chromosome is coiled up and concentrated in the nucleoid region. Because there is only a single chromosome there is only one copy of each gene. A copy of the chromosome is made just before cell division (by binary fission). https://commons.wikimedia.org/wiki/File:Plasmid_%28english%29.svg https://commons.wikimedia.org/wiki/File:Average_prokaryote_cell-_en.svg 3.2.U2 Some prokaryotes also have plasmids but eukaryotes do not. Prokaryote bacteria may have plasmids, but these structures are not found in eukaryotes.* Features of Plasmids: • Naked DNA - not associated with histone proteins • Small circular rings of DNA • Not responsible for normal life processes – these are controlled by the nucleoid chromosome • Commonly contain survival characteristics, e.g. antibiotic resistance • Can be passed between prokaryotes • Can be incorporated into the nucleoid chromosome n.b. Plasmid characteristics mean that Scientists have found them useful in genetic engineering. Plasmids can be used to transfer genes into bacteria. *Scientists have found plasmids in archea and eukaryota, but very rarely. https://commons.wikimedia.org/wiki/File:Plasmid_%28english%29.svg https://en.wikipedia.org/wiki/File:PBR322_plasmid_showing_restriction_sites_and_resistance_genes.jpg Review: 3.1.U1 A gene is a heritable factor that consists of a length of DNA and influences a specific characteristic. AND 3.1.U2 A gene occupies a specific position on a chromosome. AND 3.1.U3 The various specific forms of a gene are alleles. AND 3.1.U4 Alleles differ from each other by one or only a few bases. A gene is a heritable factor that controls or influences a specific characteristic, consisting of a length of DNA occupying a particular position on a chromosome (locus) http://learn.genetics.utah.edu/content/m olecules/gene/ 3.2.A1 Cairns’ technique for measuring the length of DNA molecules by autoradiography. AND Nature of Science: Developments in research follow improvements in techniques - autoradiography was used to establish the length of DNA molecules in chromosomes. (1.8) John Cairns produced images of DNA molecules from Escherichia coli (E.coli) • E. Coli was grown with thymidine containing a radioactive isotope of hydrogen (the DNA was labelled). • The E. Coli cells were broken open by enzymes to release the cell contents • The cell contents were applied to a photographic emulsion and placed in the dark (for two months) • The radioative isotopes reacted with the emulsion (similarly to light does) • Dark areas on the photographic emulsion indicated the presence of DNA • The images showed that E. coli possesses a single circular chromosome which is 1,100 μm long (E. coli cells have a length of only 2 μm) • Cairns images also provided evidence to support the theory of semi-conservative replication n.b. The insights and improvements in theory would not have been possible without the development and use of autoradiography (exposure of photographic emulsion by radioactive isotopes). http://schaechter.asmblog.org/.a/6a00d8341c5e1453ef017c37accbac970b-300wi 3.2.U3 Eukaryote chromosomes are linear DNA molecules associated with histone proteins. Linear strands of DNA held in a helix Eukaryotic chromosomes may be up to 85mm in length. To fit such a length of DNA into a nucleus with a diameter of 10 μm it has to be coiled in a predictable fashion that still allows for processes, such as replication and protein synthesis, to occur. Nucleosomes are formed by wrapping DNA around histone proteins n.b. Prokaryotic DNA is, like eukaryotic DNA, supercoiled, but differently: Prokaryotic DNA maybe associated with proteins, but it is not organised by histones and is therefore sometimes referred as being ‘naked’. http://en.wikipedia.org/wiki/File:DNA_to_Chromatin_Formation.jpg 3.2.U4 In a eukaryote species there are different chromosomes that carry different genes. Eukaryotes possess multiple chromosomes. All individuals of a species possess the same chromosomes, with the same gene loci. For example all humans have twenty three pairs. Chromosomes can vary by: • Length – the number of base pairs in the DNA molecule • Position of the centromere • Genes occur at a specific locus (location), i.e. it is always found at the same position on the same chromosome (the locus and genes possessed vary between species) https://public.ornl.gov/site/gallery/originals/ 3.2.S1 Use of databases to identify the locus of a human gene and its polypeptide product. Use the online database (http://www.genecards.org/) to search for the genes and the loci responsible for synthesising the following polypeptides: • • • • Rhodopsin 3 different types of Collagen Insulin One other protein of your choice n.b. the list of polypeptides reflects the examples you were required to learn for 2.4.A1 3.2.U8 The number of chromosomes is a characteristic feature of members of a species. The chromosome number is an important characteristic of the species. Organisms with different numbers of chromosomes are unlikely to be able to interbreed successfully Chromosomes can fuse or spit during evolution – these are rare events and chromosome numbers tend to stay the same for millions of years. The number of chromosomes possessed by a species is known as the N number, for example humans have 23 different chromosomes. A chromosome number does reflect the complexity of an organism https://commons.wikimedia.org/wiki/File:NHGRI_human_male_karyotype.png 3.2.U6 Diploid nuclei have pairs of homologous chromosomes. AND 3.2.U7 Haploid nuclei have one chromosome of each pair. A diploid nucleus has two of each chromosome (2N). Therefore diploid nuclei have two copies of every gene, apart from the genes on the sex chromosomes. For example the Diploid nuclei in humans contain 46 chromosomes. The fertilised egg cell (Zygote) therefore is a diploid (2N) cell containing two of each chromosome. Gametes are the sex cells that fuse together during sexual reproduction. Gametes have haploid nuclei, so in humans both egg and sperm cells contain 23 chromosomes. n.b. Diploid nuclei are less susceptible to genetic diseases: have two copies of a gene means organisms are more likely to possess at least one healthy copy. A haploid nucleus has one of each chromosome (N). Haploid nuclei in humans have 23 different chromosomes. http://www.biologycorner.com/resources/diploid_life_cycle.gif 3.2.A3 Comparison of diploid chromosome numbers of Homo sapiens, Pan troglodytes, Canis familiaris, Oryza sativa, Parascaris equorum. Humans (Homo sapiens) 46 46 is the number of diploid chromosomes in each human cell. https://upload.wikimedia.org/wikipedia/commons/f/f6/Usain_Bolt_100_m_Daegu_2011.jpg 3.2.A3 Comparison of diploid chromosome numbers of Homo sapiens, Pan troglodytes, Canis familiaris, Oryza sativa, Parascaris equorum. Asian rice (Oryza sativa) Equine roundworm (Parascaris equorum http://pic20.picturetrail.com/VOL176/4853602/20795519/357799225.j How many diploid chromosomes does each https://commons.wikimedia.org/wiki/File:Hinohikari.jpg species possess? Domestic Dog (Canis familiaris) Chimpanzee (Pan troglodytes) https://commons.wikimedia.org/wiki/File:Dog_%28Canis_lupus_fa miliaris%29_%281%29.jpg https://commons.wikimedia.org/wiki/File:Pan_troglodytes_Sweetwaters_Ch panzee_Sanctuary,_Kenya.jpg 3.2.A3 Comparison of diploid chromosome numbers of Homo sapiens, Pan troglodytes, Canis familiaris, Oryza sativa, Parascaris equorum. Asian rice (Oryza sativa) 24 2 Equine roundworm (Parascaris equorum http://pic20.picturetrail.com/VOL176/4853602/20795519/357799225.j How many diploid chromosomes does each https://commons.wikimedia.org/wiki/File:Hinohikari.jpg species possess? Domestic Dog (Canis familiaris) 78 48 https://commons.wikimedia.org/wiki/File:Dog_%28Canis_lupus_fa miliaris%29_%281%29.jpg Chimpanzee (Pan troglodytes) https://commons.wikimedia.org/wiki/File:Pan_troglodytes_Sweetwaters_Ch panzee_Sanctuary,_Kenya.jpg 3.2.U5 Homologous chromosomes carry the same sequence of genes but not necessarily the same alleles of those genes. 3.2.U5 Homologous chromosomes carry the same sequence of genes but not necessarily the same alleles of those genes. 3.2.U10 Sex is determined by sex chromosomes and autosomes are chromosomes that do not determine sex. Sex Determination: It’s all about X and Y… Humans have 23 pairs of chromosomes in diploid somatic cells (n=2). 22 pairs of these are autosomes, which are homologous pairs. One pair is the sex chromosomes. XX gives the female gender, XY gives male. Karyotype of a human male, showing X and Y chromosomes: http://en.wikipedia.org/wiki/Karyotype SRY The X chromosome is much larger than the Y. X carries many genes in the non-homologous region which are not present on Y. The presence and expression of the SRY gene on Y leads to male development. Chromosome images from Wikipedia: http://en.wikipedia.org/wiki/Y_chromosome 3.2.U10 Sex is determined by sex chromosomes and autosomes are chromosomes that do not determine sex. Sex Determination: It’s all about X and Y… Chromosome pairs segregate in meiosis. Females (XX) produce only eggs containing the X chromosome. Males (XY) produce sperm which can contain either X or Y chromosomes. Segregation of the sex chromosomes in meiosis. SRY gene determines maleness. gametes X Y X XX XY X XX XY Therefore there is an even chance* of the offspring being male or female. Find out more about its role and just why do men have nipples? http://www.hhmi.org/biointeractive/gender/lectures.html Chromosome images from Wikipedia: http://en.wikipedia.org/wiki/Y_chromosome 3.2.A2 Comparison of genome size in T2 phage, Escherichia coli, Drosophila melanogaster, Homo sapiens and Paris japonica. Humans (Homo sapiens) 3.2 billion base pairs Genome size is the total number of DNA base pairs in one copy of a haploid genome. https://upload.wikimedia.org/wikipedia/commons/f/f6/Usain_Bolt_100_m_Daegu_2011.jpg 3.2.A2 Comparison of genome size in T2 phage, Escherichia coli, Drosophila melanogaster, Homo sapiens and Paris japonica. Canopy plant (Paris japonica) T2 phage Escherichia coli https://s-media-cacheak0.pinimg.com/736x/2d/0e/3e/2d0e3ea8ddf652f25a5f2c3b1050 af79.jpg n.b. T2 phage (orange) is a virus that attacks E. Coli bacterium (green and white). What is the genome size of each species? Fruit fly (Drosophila melanogaster) https://commons.wikimedia.org/wiki/File:Dr osophila_melanogaster__side_%28aka%29.jpg https://commons.wikimedia.org/wiki/File:Paris_japonica_Kinugasasou_in_Hakusan_2003_7_27.jpg 3.2.A2 Comparison of genome size in T2 phage, Escherichia coli, Drosophila melanogaster, Homo sapiens and Paris japonica. Canopy plant (Paris japonica) 150 billion base pairs T2 phage 164 thousand base pairs Escherichia coli 4.6 million base pairs https://s-media-cacheak0.pinimg.com/736x/2d/0e/3e/2d0e3ea8ddf652f25a5f2c3b1050 af79.jpg n.b. T2 phage (orange) is a virus that attacks E. Coli bacterium (green and white). What is the genome size of each species? Fruit fly (Drosophila melanogaster) 130 million base pairs https://commons.wikimedia.org/wiki/File:Dr osophila_melanogaster__side_%28aka%29.jpg https://commons.wikimedia.org/wiki/File:Paris_japonica_Kinugasasou_in_Hakusan_2003_7_27.jpg 3.2.U9 A karyogram shows the chromosomes of an organism in homologous pairs of decreasing length. Karyogram is a diagram or photograph of the chromosomes present in a nucleus (of a eukaryote cell) arranged in homologous pairs of decreasing length. The chromosomes are visible in cells that are undergoing mitosis – most clearly in metaphase. Stains used to make the chromosomes visible also give each chromosome a distinctive banding pattern. A micrograph are taken and the chromosomes are arranged according to their size, shape and banding pattern. They are arranged by size, starting with the longest pair and ending with the smallest. https://commons.wikimedia.org/wiki/File:NHGRI_human_male_karyotype.png 3.2.U9 A karyogram shows the chromosomes of an organism in homologous pairs of decreasing length. Karyogram is a diagram or photograph of the chromosomes present in a nucleus (of a eukaryote cell) arranged in homologous pairs of decreasing length. Karyotype is a property of the cell described by the number and type of chromosomes present in the nucleus (of a eukaryote cell). a Karyogram is a diagram that shows, or can be used to determine, the karyotype. http://learn.genetics.utah.edu/content/chromosomes/karyotype/ Bibliography / Acknowledgments Bob Smullen