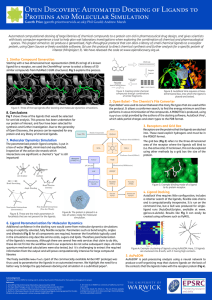
PDF - shoichetlab - University of California, San Francisco
advertisement

J. Mol. Biol. (2008) 377, 914–934 doi:10.1016/j.jmb.2008.01.049 Available online at www.sciencedirect.com Rescoring Docking Hit Lists for Model Cavity Sites: Predictions and Experimental Testing Alan P. Graves 1,2 †, Devleena M. Shivakumar 3 †, Sarah E. Boyce 1,4 , Matthew P. Jacobson 1 ⁎, David A. Case 3 ⁎ and Brian K. Shoichet 1 ⁎ 1 Department of Pharmaceutical Chemistry, University of California, San Francisco, 1700 4th Street, San Francisco, CA 94158-2330, USA 2 Graduate Group in Biophysics, University of California, San Francisco, 1700 4th Street, San Francisco, CA 94158-2330, USA 3 Department of Molecular Biology, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037, USA 4 Graduate Group in Chemistry and Chemical Biology, University of California, San Francisco, 1700 4th Street, San Francisco, CA 94158-2330, USA Received 21 September 2007; received in revised form 12 January 2008; accepted 17 January 2008 Available online 30 January 2008 Molecular docking computationally screens thousands to millions of organic molecules against protein structures, looking for those with complementary fits. Many approximations are made, often resulting in low “hit rates.” A strategy to overcome these approximations is to rescore top-ranked docked molecules using a better but slower method. One such is afforded by molecular mechanics–generalized Born surface area (MM– GBSA) techniques. These more physically realistic methods have improved models for solvation and electrostatic interactions and conformational change compared to most docking programs. To investigate MM–GBSA rescoring, we re-ranked docking hit lists in three small buried sites: a hydrophobic cavity that binds apolar ligands, a slightly polar cavity that binds aryl and hydrogen-bonding ligands, and an anionic cavity that binds cationic ligands. These sites are simple; consequently, incorrect predictions can be attributed to particular errors in the method, and many likely ligands may actually be tested. In retrospective calculations, MM–GBSA techniques with binding-site minimization better distinguished the known ligands for each cavity from the known decoys compared to the docking calculation alone. This encouraged us to test rescoring prospectively on molecules that ranked poorly by docking but that ranked well when rescored by MM– GBSA. A total of 33 molecules highly ranked by MM–GBSA for the three cavities were tested experimentally. Of these, 23 were observed to bind— these are docking false negatives rescued by rescoring. The 10 remaining molecules are true negatives by docking and false positives by MM–GBSA. X-ray crystal structures were determined for 21 of these 23 molecules. In many cases, the geometry prediction by MM–GBSA improved the initial docking pose and more closely resembled the crystallographic result; yet in several cases, the rescored geometry failed to capture large conformational changes in the protein. Intriguingly, rescoring not only rescued docking false positives, but also introduced several new false positives into the topranking molecules. We consider the origins of the successes and failures in MM–GBSA rescoring in these model cavity sites and the prospects for rescoring in biologically relevant targets. © 2008 Published by Elsevier Ltd. Edited by B. Honig Keywords: decoys; molecular docking; virtual screening; MM–GBSA; cavity *Corresponding authors. E-mail addresses: matt.jacobson@ucsf.edu; case@scripps.edu; shoichet@cgl.ucsf.edu. † A.P.G. and D.M.S. contributed equally to this work. Present address: D. M. Shivakumar, Institute for Molecular Pediatric Sciences, University of Chicago, 929 E. 57th Street, Chicago, IL 60637, USA. Abbreviations used: L99A, Leu99 → Ala mutant of T4 lysozyme; L99A/M102Q, Leu99 → Ala and Met102 → Gln double mutant of T4 lysozyme; CCP, Trp191 → Gly mutant of cytochrome c peroxidase; MM–GBSA, molecular mechanics with generalized Born surface area approximation; PLOP, Protein Local Optimization Program; ACD, Available Chemicals Directory; PDB, Protein Data Bank; SGB, surface generalized Born. 0022-2836/$ - see front matter © 2008 Published by Elsevier Ltd. Rescoring Docking Hit Lists Introduction Molecular docking computationally screens large databases of small molecules against a macromolecular binding site of defined structure. The technique is often used to find novel ligands for drug discovery. Notwithstanding important successes,1–8 docking continues to struggle with many methodological deficits. Many approximations are made to screen many molecules in a timely fashion. These include using only one conformation of the protein, neglecting the internal energies of the docking molecules, using simplified models of ligand solvation energies, typically ignoring protein desolvation, and ignoring most entropic terms entirely. These and other shortcuts lead to the high false-positive and false-negative rates for which docking screens are notorious. Docking methods are unreliable for affinity prediction and, except in domains of highly related compounds, even for rank ordering the likely hits that emerge from the virtual screens. To overcome these deficits, several groups have combined disparate scoring functions in a consensus fashion to capitalize on the strengths and overcome the deficiencies of individual methods.9–12 This “consensus scoring” approach is attractive when it has worked, but its theoretical underpinnings are slim.13 An alternative approach involves using a higher level of theory to rescore the docking hit lists after the docking calculation has completed. The goal is to reevaluate the top docking hits for energetic complementarity to the target after including more terms and degrees of freedom than modeled by the docking program. Because more terms are considered, rescoring is typically much slower than docking, so much so that only the top-scoring docking pose of the best scoring docked molecules are often considered. This approach has been adopted by versions of the program GLIDE.14 Here ligands are first docked using simplified and relaxed criteria and are then refined by more sophisticated and stringent evaluation of the energies of binding. Similarly, Wang et al. used a hierarchical technique that begins with initial database screening and progresses to molecular mechanics–Poisson–Boltzmann surface area (MM–PBSA) rescoring to find HIV-1 reverse transcriptase inhibitors. 15 The combination of an initial docking screen with subsequent rescoring by a molecular mechanics–generalized Born surface area (MM–GBSA) method has been used to improve enrichment of known ligands for several enzymes in retrospective studies and even to identify substrates.16–20 Such MM–PBSA and MM–GBSA methods involve minimization and often dynamic sampling of the protein–ligand complexes, and include ligand and receptor conformational energies and strain. They evaluate the electrostatics and solvation components of the binding energy by PB or GB methods, including both ligand and receptor desolvation. The MM–GBSA binding energy is determined by Ecomplex − Ereceptor − Eligand where E is an MM–GBSA estimate and solute configurational entropy effects 915 are ignored. In this article, we focus on relative binding energies of different ligands to the same receptor, so the free receptor energy (Ereceptor) does not affect the results. Because the MM–GBSA function includes both internal energies and solvation free energies, and because we explicitly subtract complex (Ecomplex) and ligand (Eligand) contributions, desolvation effects upon complex formation for both the ligand and the receptor are included, at least in principle. There are three main limitations: (1) the force fields and solvation energies are not uniformly accurate; (2) for reasons of computational efficiency, only a small part of configuration space near the DOCK starting pose is really explored; and (3) configurational entropy effects are ignored. Notwithstanding these limitations, the MM–GBSA methods represent a substantially higher level of theory than that encoded by most docking programs and are attractive alternatives to a more complete treatment of the energies of interaction by free-energy perturbation and thermodynamic integration,21 which remain the gold standard but are very slow. In this study, we set out to test MM–GBSA rescoring of docking hit lists in simple model cavity sites. These sites have been engineered into the buried cores of proteins and bind multiple small organic molecules. In contrast to most drug targets, these cavities are small (150–180 Å3), buried from bulk solvent, and are dominated by a single interaction term. The L99A (Leu99 → Ala) cavity in T4 lysozyme22 is almost entirely apolar, the L99A/M102Q (Leu99 → Ala/Met102 → Gln)23 cavity in the same protein has a single hydrogen-bond acceptor (the introduced Gln102), whereas the W191G (Trp191 → Gly) cavity in cytochrome c peroxidase (CCP)24,25 has a single anionic residue, Asp235 (Fig. 1). The ligands recognized by these sites correspond to these features: the hydrophobic L99A binds small, typically aromatic nonpolar molecules; the slightly polar L99A/M102Q binds not only both apolar molecules but also those bearing one or two hydrogen-bond donors, whereas the anionic W191G cavity almost exclusively binds small monocations. The simplicity of these sites is conducive to disentangling the energetic terms of ligand binding, which are so often convoluted in drug targets with their larger, more complex binding sites. It should be noted that previous work with solvent-exposed sites has suggested that a major advantage of MM–GBSA scoring functions is calculating partial receptor desolvation upon ligand binding.17 This benefit with complex solventexposed binding sites may be less relevant in the buried cavity sites, especially the hydrophobic L99A and polar L99A/M102Q sites, which are mostly desolvated. (It is our experience that the cavity sites, in fact, impose a greater strain on the GBSA solvent models to fully desolvate the pockets.) In the cavity sites, as in other simplified sites,27 an incorrect prediction is often informative, identifying a single problematic term in a scoring function; we have used these cavities as model binding sites to identify problems in molecular docking23,28–30 and, more recently, thermodynamic integration.21 Rescoring Docking Hit Lists 916 Fig. 1. The model cavity sites. (a) Cavity binding site in T4 lysozyme L99A with benzene bound. (b) Cavity binding site in T4 lysozyme L99A/M102Q with phenol bound; the hydrogen bond with the Oε2 oxygen of Gln102 is represented by a dashed line. (c) Cavity binding site of cytochrome c peroxidase W191G with aniline bound; the hydrogen bond with Asp235 is represented by a dashed line. The heme and an ordered water molecule are also depicted. In (a), (b), and (c), the cavities are represented by a tan molecular surface and the protein ribbons are green. Rendered with the program PyMOL.26 Others have found them attractive test systems for method development studies.31–34 An important advantage of these cavity sites is that they are experimentally tractable for detailed, prospective testing of ligand predictions. Because the ligands they bind are small—in the 70- to 150-amu range—many possible ligands are readily available commercially, which is rarely true of drug targets.35 The binding of these predicted ligands may be tested by direct binding assays, and the structures of the ligand– protein complexes may be routinely determined by X-ray crystallography to resolutions better than 2 Å. Extensive study in the Matthews, Goodin, and our own laboratories has resulted in many tens of diverse ligands for each cavity, as well as tens of “decoys,” which are molecules that were predicted to bind to the sites but for which no binding was observed at concentrations as high as 10 mM on experimental testing.21,23,28–30 We thus used these three simple model cavity sites, L99A, L99A/M102Q, and W191G, as templates to measure the strengths and weaknesses of MM– GBSA rescoring of docking hit lists. We used two rescoring programs: Protein Local Optimization Program (PLOP),36,37 with binding-site side-chain rotamer search and minimization, and AMBERDOCK, using short molecular dynamics (MD) steps and minimization of binding-site residues (Materials and Methods). Molecular docking was used to screen compound libraries that contained between 5000 and 60,231 fragment-like molecules from the Available Chemicals Directory (ACD); the library size was chosen to partly mitigate issues of size and charge bias from the library alone and to be consistent with earlier studies in these sites (Results).28,29 The single best pose for each compound that ranked among the top 5000 or 10,000 compounds by docking was then rescored by both MM– GBSA programs. Multiple known ligands and decoys were among the molecules rescored for all three sites' rescored sets. In retrospective calculations, MM–GBSA rescoring improved the separation of ligands from decoys in each of the cavities. We then tested 33 new ligands that were predicted to bind by the MM–GBSA methods that docking alone ranked poorly, generally much worse than the top 500. To investigate the detailed basis of the MM–GBSA predictions, we determined crystal structures for 21 of these new ligands and compared them to the geometries predicted by theory. These studies suggest areas where MM–GBSA methods can contribute to the success of virtual screening and areas where this method faces important challenges. Results Retrospective docking and rescoring in the hydrophobic cavity Approximately 60,000 small molecules were docked into the hydrophobic cavity L99A using DOCK3.5.5423,38 (Fig. 1a). The compounds in this set were selected from a much larger library so as not to exceed 25 non-hydrogen atoms, as previously described.29 This reduced the enrichment-factor bias that would have otherwise occurred by the trivial ability of the docking program to remove com- Rescoring Docking Hit Lists pounds that were simply too large to fit in the cavities. We note that reducing the number of molecules to 60,000 from the several million that are in the ACD or ZINC39 databases has the effect of reducing our enrichment factors. Among the top-scoring 10,000 molecules were 39 known ligands and 40 experimentally tested decoys. DOCK found 44% (17 molecules) of these ligands and 43% (17 molecules) of these decoys among the top 500 molecules (Fig. 2a). Ligands such as toluene (DOCK rank 32), benzene (DOCK rank 151), and ethylbenzene (DOCK rank 301) are small, aromatic, and hydrophobic compared to known decoys such as nitrosobenzene (DOCK rank 125), phenol (DOCK rank 234), and 3-methylpyrrole (DOCK rank 435). Like the ligands, these decoys are also small and aromatic, but are presumably too polar for the hydrophobic cavity to overcome their desolvation penalty (Fig. 1a). The top-ranking 10,000 docking hits for the hydrophobic cavity were re-ranked by PLOP and the top-ranking 5000 docking hits were re-ranked by AMBERDOCK; fewer molecules were treated by AMBERDOCK simply because it was much more computationally intensive than PLOP. For both methods, the enrichment of the ligands actually decreased slightly relative to that achieved by docking alone; that is to say, fewer ligands were found among the very best scoring molecules (Fig. 2a). Rescored by PLOP, 41% (16 molecules) of the known ligands were found among the top 500 molecules, whereas 28% (11 molecules) were found by AMBERDOCK. Both enrichment factors were lower than those found by docking alone. On the other hand, the enrichment of the known decoys was lower still (Fig. 2a). Only 5% of the decoys (2 molecules) were ranked among the top 500 molecules by PLOP and only 13% (5 molecules) were so ranked by AMBERDOCK. This represents a substantial improvement on docking alone, one that reflects a significant change in the relative energies of the ligands and decoys. For instance, in the L99A cavity the average differential energy between the first 10 ligands and the first 10 decoys was only 0.7 kcal/mol by docking. Meanwhile, the average total energy for the top 10 docked ligands was −15.8 kcal/mol and the difference between the 1st and the 10th ranked ligand is 2.9 kcal/ mol; the ligands and decoys were essentially indistinguishable by docking energy. For the PLOP rescored molecules, conversely, the average difference in energies for the top 10 ligands and decoys was 4.0 kcal/mol. Meanwhile, average energy for the top 10 ligands was −21.7 kcal/mol and the difference between top ranked ligand and the 10th was 5.5 kcal/mol; the best ligands and decoys are separated significantly by rescored energy. We should note that both the ligand enrichment and the decoy enrichment are strongly biased for docking—many of the ligands and almost all of the decoys were originally tested based on docking predictions23,29,30—so it is reasonable to expect that the enrichment of ligands will be higher by docking, as will the decoys. Perhaps more informative then is the separation of the ligands from the decoys, as measured by the ratios of their 917 Fig. 2. Retrospective enrichment of ligands and decoys for (a) the hydrophobic L99A cavity, (b) the polar L99A/ M102Q cavity, and (c) the anionic W191G cavity. The plots depict the percentage of known ligands (continuous lines) or decoys (dashed lines) found (y-axis) at each percentage level of the ranked database using the top 10,000 best scoring docking hits (x-axis) for L99A (a) and L99A/ M102Q (b) and the 5400 best scoring docking hits (x-axis) for CCP (c). Docking enrichment of known ligands (continuous lines) and decoys (dashed lines) are represented by the dark blue curves. PLOP enrichment of known ligands (continuous lines) and decoys (dashed lines) are represented by the pink curves. AMBERDOCK enrichment of known ligands (continuous lines) and decoys (dashed lines) are represented by green curves. enrichment factors. These were improved eightfold by PLOP and twofold for AMBERDOCK, relative to that of DOCK in this hydrophobic cavity. 918 Retrospective docking and rescoring in the polar cavity The same 60,000 molecules were docked into the polar cavity L99A/M102Q (Fig. 1b). Among the topscoring 10,000 molecules were 58 ligands and 17 experimentally tested decoys. DOCK found 45% (26 molecules) of these ligands and 35% (6 molecules) of these decoys among the top 500 molecules (Fig. 2b). The increased polarity from Oε of the Gln102 side chain in the cavity accommodates the binding of phenol (DOCK rank 354) and 3-methylpyrrole (DOCK rank 307), which are decoys for the L99A cavity, as well as hydrophobic ligands such as toluene (DOCK rank 16) and benzene (DOCK rank 78). The increased polarity of the site only goes so far, however, and it cannot accommodate decoys such as 1-vinylimidazole (DOCK rank 136) or 2-aminophenol (DOCK rank 208), whose polarity is presumably still too great for the single carbonyl oxygen of the site to overcome the attendant desolvation terms. The top 10,000 docking hits for the polar cavity were re-ranked by PLOP and the top 5000 re-ranked by AMBERDOCK. For both methods, the enrichment of the ligands again decreased slightly relative to the docking enrichment factor (Fig. 2b). Rescored by PLOP, 22% (13 molecules) of the known ligands were found among the top 500 molecules, whereas 34% (20 molecules) were found by AMBERDOCK. However, the enrichment of the known decoys was lower still. None of the decoys were ranked among the top 500 molecules by PLOP or AMBERDOCK, in contrast to DOCK where 35% (6 molecules) of the known decoys were scored among the top 500 molecules. As in the hydrophobic site, despite the decrease in overall ligand enrichment, the separation of the ligands from the decoys was improved substantially for the polar cavity: by 20-fold for PLOP and 35-fold for AMBERDOCK. Retrospective docking and rescoring in the anionic cavity Approximately 5400 molecules were docked in the charged cavity of CCP (Fig. 1c). This library was also selected from a much larger set to reduce enrichmentfactor bias from trivial physical noncomplementarity between library molecules and the CCP cavity.28 Thus, any molecules from the larger ACD that had unfavorable van der Waals scores (i.e., simply did not fit), or that bore an anionic charge, were removed from the larger library. As with the lysozyme cavities, this smaller library of more physically plausible ligands reduces the enrichment factors we would otherwise achieve with docking. Within this database were 40 known ligands and 20 experimentally tested decoys. DOCK found 78% (31 molecules) of these ligands and 20% (4 molecules) of these decoys among the top 500 molecules (Fig. 2b). The anionic cavity typically binds cationic ligands such as 2-aminopyridine (DOCK rank 6) and imidazole (DOCK rank 227). Most neutral polar compounds, such as 3,5difluoroaniline (DOCK rank 148), and apolar com- Rescoring Docking Hit Lists pounds, such as toluene (DOCK rank 411), are decoys for this cavity, as are anionic compounds or those bearing a formal charge greater than +1. All of the 5400 docking hits for the anionic cavity were re-ranked by PLOP and AMBERDOCK. Rescored by PLOP, 83% (33 molecules) of the known ligands were found among the top 500 molecules, and 80% (32 molecules) were found by AMBERDOCK (Fig. 2c). Both enrichment factors are comparable to those found by docking alone, which found 83% (33 molecules) of the known ligands among the top 500 molecules. On the other hand, fewer of the known decoys were enriched by the MM–GBSA methods. None of the known decoys were ranked among the top 500 molecules by PLOP or AMBERDOCK, and the best scoring decoy ranked 655 for PLOP and 785 for AMBERDOCK compared to 145 for docking. Thus, whereas the overall enrichment of the ligands relative to the rest of the database molecules remained unchanged, the separation of the ligands from the decoys was improved by fourfold for PLOP and AMBERDOCK. Prediction and experimental testing of new ligands A more robust test, one less biased by previous knowledge, involves prospective prediction of new ligands. For each of the three cavities, we looked for molecules that had been poorly ranked by docking but that ranked well by either PLOP or AMBERDOCK or both. We note that our use of “well” and “poorly” ranked is inexact because there is no fully reliable way to separate molecules based on docking energies alone. We therefore looked for molecules where the rankings changed substantially—typically rising from ranks lower than 1500 to ranks in the top 200. Of the 33 molecules selected, 24 were ranked worse than 1500th by docking, 7 were ranked between 500 and 1500, and 2 were ranked between 300 and 500. The rankings of all 33 rose to be among the top 200 on rescoring. Our choice of 200 was purely pragmatic, as it is a reasonable number of top-ranking hits to visualize and consider for testing, which is often done when picking docking hits; another reasonable cutoff would have been top 500. Nine compounds were picked and tested for the hydrophobic L99A cavity, 10 were tested for the polar L99A/M102Q cavity, and 14 were tested for the anionic W191G cavity. Structures for 21 of these 33 molecules in complex with the cavities were determined by protein crystallography, allowing us to compare the predicted and experimental geometries in detail. In the following discussion, we report whether binding was detected at a single concentration tested. The actual affinities were not measured but will often be substantially better than the concentration reported. New L99A ligands predicted by rescoring All of the nine ligands predicted by PLOP and AMBERDOCK were relatively large compounds Score and ranka Structure b c d ΔΔH (kcal/mol) ΔTm (°C) Binding detected Structure determined 10 3.0 31.0 6.5 Yes Yes − 16.22 (1243) b10 3.0 6.2 1.3c Yes Yes − 7.69 (5290) 10 6.8 1.6 − 0.9c No No b10 3.0 10.0 1.6c Yes Yes 5 3.0 12.0 1.2 Yes Yes − 25.19 (27) 10 3.0 8.1 1.5 Yes Yes NRd −25.17 (28) 10 3.0 5.1 1.5 Yes No NRd −24.92 (30) 10 3.0 14.9 2.8 Yes No −19.64 (55) −23.13 (68) 10 3.0 2.7 0.0 No No DOCK AMBER β-Chlorophenetole (1) − 4.89 (3786) −22.38 (5) − 26.31 (15) 4-(Methylthio)nitrobenzene (2) − 5.69 (3358) −22.36 (6) 1-Phenyl-semicarbazide (3) − 4.49 (3965) −22.03 (8) 2,6-Difluorobenzyl bromide (4) − 10.59 (1046) −22.01 (9) − 21.10 (186) 2-Ethoxyphenol (5) − 6.74 (2806) −21.54 (12) − 15.19 (1642) 3-Methyl benzylazide (6) − 10.54 (1061) −19.58 (57) cis-3-Hexenyl formate (7) 3.61 (7746) 6-Methyl-1,5-heptadiene (8) 1.78 (7035) 2-Phenoxyethanol (9) a pH Compound (ID) −5.76 (3323) PLOP |C|b (mM) Rescoring Docking Hit Lists Table 1. Compounds predicted by AMBERDOCK and PLOP to bind to T4 lysozyme L99A Compound scores and ranks (in parenthesis) for DOCK, AMBERDOCK, and PLOP. Scores and ranks in bold font indicate ligands that rank in the top 200 for the respective scoring function. Concentration at which ligand was tested. ΔTm monitored using fluorescence, exciting at λ = 283 nm and measuring the integrated emission above 300 nm. NR is not ranked. 919 920 that do not easily fit into the unminimized cavity into which they were docked, explaining their poor docking ranks, but they fit well upon receptor relaxation by MM–GBSA. Binding was detected at millimolar concentrations by temperature of melting (Tm) upshift experiments for seven of these nine compounds; however, for two no binding was detected (Table 1). AMBERDOCK correctly predicted binding for five ligands and incorrectly predicted binding for 1-phenylsemicarbazide (3) and 2-phenoxyethanol (9) (two of the prospectively tested molecules were not rescored by AMBERDOCK because docking ranked them worse than 5000). PLOP correctly predicted binding for five ligands, while Rescoring Docking Hit Lists incorrectly predicting binding for 2-phenoxyethanol (9). PLOP agreed with docking on the remaining three molecules that had been prioritized by AMBERDOCK, ranking them worse than 1000. Two of these, 4-(methylthio)nitrobenzene (2) and 2-ethoxyphenol (5), were true ligands and so are false negatives for PLOP. Five high-resolution (better than 2 Å) protein– ligand crystal structures were obtained for these new L99A ligands to compare experimental to predicted poses (Fig. 3). In each case, electron density for the ligands was unambiguous, allowing us to model their positions in the site. Docking and MM–GBSA methods predicted the binding Fig. 3. Predicted and experimental ligand orientations for the hydrophobic L99A cavity. The carbon atoms of the crystallographic pose, the DOCK predicted pose, the AMBERDOCK predicted pose, and the PLOP predicted pose are colored gray, yellow, cyan, and magenta, respectively. The Fo − Fc omit electron density maps (green mesh) are contoured at 2.5–3.0σ (a) β-chlorophenetole (1), (b) 4-(methylthio)nitrobenzene (2), (c) 2,6-difluorobenzylbromide (4), (d) 2ethoxyphenol (5), and (e) 3-methylbenzylazide (6) bound to L99A. Rendered with the program PyMOL.26 Rescoring Docking Hit Lists 1.93/1.84a NA 1.80/1.70a 1.16 0.93 1.02 1.44 0.87 1.61 0.97 NA 0.63 2.04/1.12a 2.00/0.81a 2.00/0.65a All crystals belong to space group P3221. Values in parentheses are for the highest resolution shell. NA, not available. a Two conformations of the crystallographic ligand were modeled. 1.29 0.91 1.10 1.42 0.83 1.08 0.62 0.28 0.37 1.44 1.46 1.49 0.82 0.64 0.54 0.60 0.52 0.58 1.56 (1.60) 28,172 (1930) 10.6 (36.1) 99.3 (93.6) 14.6 (5.0) 18.3 (19.6) 20.5 (25.6) 0.01 1.13 2RBS 1.43 (1.47) 35,537 (2423) 7.6 (37.6) 99.4 (93.1) 17.3 (4.2) 18.1 (23.4) 20.1 (27.1) 0.01 1.13 2RBR 1.63 (1.68) 24,034 (1662) 6.4 (45.1) 97.0 (93.1) 14.5 (2.9) 20.8 (41.4) 24.0 (55.0) 0.02 1.58 2RBQ 1.47 (1.51) 33,813 (2446) 7.1 (46.6) 99.8 (99.6) 24.3 (4.5) 18.1 (24.6) 20.8 (28.6) 0.01 1.10 2RBP 1.29 (1.32) 48,915 (3563) 6.7 (62.2) 99.9 (100.0) 26.9 (3.3) 17.2 (23.5) 19.1 (28.6) 0.01 1.22 2RBO 1.29 (1.32) 48,474 (3576) 8.0 (56.9) 98.8 (100.0) 21.0 (3.6) 17.8 (21.4) 19.1 (21.6) 0.01 1.07 2RBN 1.46 (1.50) 33,797 (2337) 9.3 (34.8) 99.5 (94.9) 13.8 (3.6) 18.0 (25.6) 21.3 (32.3) 0.01 1.08 2RB2 1.70 (1.74) 21,923 (1615) 7.4 (63.2) 99.2 (99.9) 15.2 (2.4) 19.1 (32.4) 23.0 (38.9) 0.01 1.23 2RB1 1.84 (1.89) 13,875 (772) 7.7 (38.9) 80.0 (60.6) 11.8 (2.6) 19.6 (32.3) 23.3 (34.3) 0.01 0.99 2RB0 1.64 (1.68) 24,246 (1780) 6.1 (45.4) 99.8 (99.9) 22.8 (3.3) 19.1 (34.9) 22.1 (44.3) 0.01 1.15 2RAZ 1.80 (1.84) 18,414 (1314) 7.0 (50.0) 99.7 (98.9) 23.2 (3.4) 18.7 (27.8) 21.2 (30.2) 0.01 1.25 2RAY Resolution (Å) Reflections Rmerge (%) Completeness (%) ⟨I⟩/⟨σ(I)⟩ R-factor (%) Rfree (%) Δbond lengths (Å) Δbond angles (°) PDB code RMSD (Å) DOCK AMBER PLOP 3-Chloro-1phenyl1-propanol (13) 2Phenoxy ethanol (9) 3-Methyl benzyl azide (6) 2-(n-Propyl thio) ethanol (12) n-Phenyl glycinonitrile (10) 2-Nitrothiophene (11) L99A/M102Q ligands (ID) 3-Methyl benzyl azide (6) 2-Ethoxyphenol (5) 2,6-Difluoro benzyl bromide (4) 4-(Methylthio) nitrobenzene (2) β-Chloro phenetole (1) Ten representative compounds that scored well by the MM–GBSA methods were experimentally tested for binding to the polar cavity (Table 3). These compounds were ranked poorly by docking, again typically because they were too large for the conformation of the cavity targeted by docking. Binding was detected at millimolar concentrations by Tm upshift for 6 of these 10 compounds; for the remaining 4, binding was not observed (Table 3). We note, however, that for 1 of these 4, 2-(n-propylthio) ethanol (12), we were able to determine a crystal structure in complex with the ligand by soaking a crystal of L99A/M102Q with 100 mM of compound, suggesting that it is a weak ligand for this cavity. AMBERDOCK correctly predicted binding for 4 of the 6 ligands that it suggested should bind, while incorrectly predicted binding for o-benzylhydroxylamine (14) and 1-phenylsemicarbazide (3). Of the remaining two hits tested, prioritized by a high PLOP ranking, AMBERDOCK missed 1 real ligand but correctly distinguished 1 real decoy, ranking both compounds worse than 500. Two of the prospectively tested molecules were not rescored by AMBERDOCK because docking ranked them worse L99A ligands (ID) New L99A/M102Q ligands predicted by rescoring Table 2. Crystallographic measurement and the RMSD values for predicted and crystallographic ligand geometries in the L99A and L99A/M102Q sites geometry for three of the five ligands to within 0.3 to 0.8 Å RMSD (Table 2). Conversely, the docked pose of 3-methylbenzylazide (6) was 1.4 Å RMSD from the crystallographic pose. The PLOP minimized prediction had a slightly improved RMSD of 1.1 Å, but the refined ligand also had a nonlinear azide group, highlighting a failure in ligand parameterization. In addition, docking and MM– GBSA methods predicted poses that were approximately 1.5 Å RMSD from the crystallographic pose of 4-(methylthio)nitrobenzene (2). The crystallographic poses of these two ligands would have been within 2 Å of the Val111 side chain in the conformation of the cavity used for the docking calculation, a steric conflict that is relieved by conformational expansion of the cavity in the experimental structures. Indeed, for all complexes, with the exception of β-chlorophenetole (1), the F-helix of lysozyme (residues 108–113) that forms one wall of the cavity reorients by about 2 Å and swings Val111 further out of the cavity to accommodate the ligands.40 The protein conformations seen in these structures more closely resemble the larger isobutylbenzene-bound cavity site [Protein Data Bank (PDB) ID 184L] than the smaller benzenebound cavity site (PDB ID 181L) used for docking and rescoring. Whereas the MM–GBSA methods do not capture this helix motion, receptor and ligand minimization reduces the steric clash sufficiently to improve the ranks of what were docking false negatives. Higher level calculations using free-energy methods and MD have captured the F-helix motion and explained discrepancies in free energies upon ligand binding due to its displacement.21,31 921 922 Table 3. Compounds predicted by AMBERDOCK and PLOP to bind to T4 lysozyme L99A/M102Q Score and ranka Structure Compound (ID) DOCK O-Benzylhydroxylamine (14) − 11.35 (647) − 28.05 (1) −14.14 (2271) 10 6.8 10 6.8 Binding detected Structure determined No No 0.0c No No ΔTm (°C) − 0.6 −16.42 (1354) N-Phenylglycinonitrile (10) − 8.60 (1556) − 25.47 (11) −40.17 (11)* b10 3.0 5.1 Yes Yes − 24.52 (13) −16.94 (1165) b10 3.0 4.4 Yes Yes − 24.21 (14) −15.18 (1824) 10 3.0, 6.8 Weak No NRd −27.20 (20) 10 3.0 0.1 No Yes − 12.82 (318) − 7.14 (2215) 6.02 (6847) 1.3, − 0.8 − 1.58 (4291) − 10.25 (2260) −27.19 (21) 10 3.0 0.0 No No 3-Methyl benzyl azide (6) − 5.35 (2740) − 20.51 (116) −25.87 (35) 10 3.0 1.9 Yes Yes 2-Phenoxyethanol (9) − 4.08 (3270) − 16.53 (551) −25.82 (36) 10 3.0 1.2 Yes Yes NRd −25.65 (37) 10 3.0 7.8 Yes Yes 3.6 (6074) Compound scores and ranks (in parenthesis) for DOCK, AMBERDOCK, and PLOP. Scores and ranks in bold font indicate ligands that rank in the top 200 for the respective scoring function. Concentration at which ligand was tested. ΔTm monitored using fluorescence at λ = 291.5 nm and measuring the integrated emission above 300 nm. NR, not ranked. Rescoring Docking Hit Lists cis-2-Hexen-1-ol (16) (R) (+)-3-Chloro-1-phenyl-1-propanol (13) d pH − 26.79 (4) 2-(n-Propyl-thio)ethanol (12) c (mM) − 3.76 (3783) 2-Ethoxy-3,4-dihydro-2H-pyran (15) b PLOP AMBER 1-Phenylsemi-carbazide (3) 2-Nitrothiophene (11) a |C|b Rescoring Docking Hit Lists than 5000. PLOP correctly predicted binding for 5 of the 6 ligands that it suggested should bind but incorrectly predicted binding for cis-2-hexenol (16). Of the remaining hits tested, prioritized for testing by AMBERDOCK, PLOP missed 2 true ligands but correctly distinguished 2 decoys by ranking them worse than 1000. Crystal structures of six L99A/M102Q ligand complexes were determined to compare predicted and experimental poses of these new ligands (Fig. 4). Electron density for each ligand was unambiguous and was detailed enough to suggest two binding modes for 2-nitrothiophene (11) and 3-chloro-1- 923 phenyl-1-propanol (13). Docking predicted the pose of one ligand, 2-(n-propylthio)ethanol (12), to within 1 Å RMSD, while AMBERDOCK further minimized five of its six ligands and PLOP minimized three of its six ligands to within 1 Å RMSD (Table 2). Although the MM–GBSA methods collectively improved the binding mode predictions of all but one ligand, the key hydrogen bond interaction was missed in three of these structures (Fig. 4a, e, and f). In addition, the azide group of 3-methylbenzylazide (6) was incorrectly parameterized by both AMBERDOCK and PLOP, as was also observed in the L99A cavity. Neither DOCK nor the MM–GBSA rescoring Fig. 4. Predicted and experimental ligand orientations for the polar L99A/M102Q cavity site. The carbon atoms of the crystallographic, DOCK, AMBERDOCK, and PLOP predicted poses are colored gray, yellow, cyan, and magenta, respectively. Hydrogen bonds are depicted with dashed lines. The Fo − Fc electron density omit maps (green mesh) are contoured at 2.5–3.0σ. (a) n-Phenylglycinonitrile (10), (b) 2-nitrothiophene (11), (c) 2-(n-propylthio)ethanol (12), (d) 3-methylbenzylazide (6), (e) 2-phenoxyethanol (9), and (f) (R)-(+)-3-chloro-1-phenyl-1-propanol (13) bound to L99A/ M102Q. Rendered with the program PyMOL.26 924 Table 4. Compounds predicted to bind by AMBERDOCK and PLOP to CCP W191G Score and ranka Structure |C|b (mM) Structure determined 10.0 No No − 16.21 (952) 1.0 Yes Yes 347.86 (39) − 44.39 (389) 1.0 Yes Yes − 14.74 (1830) 348.17 (49) − 31.88 (796) 0.05 Yes Yes 5-Nitro-6-aminouracil (21) − 12.14 (2435) 348.49 (62) − 31.47 (827) 1.0 No No 1,2-Dimethyl-1H-pyridin-5-amine (22) − 22.95 (362) 349.34 (87) − 54.67 (59) 0.5 Yes Yes 2-Aminobenzylamine (23) − 12.62 (2316) 349.34 (96) − 34.19 (671) 10.0 No No − 36.54 (7) 344.29 (12) − 59.87 (53) 1.0 Yes Yes DOCK N-Methyl-1,2-phenylenediamine (17) − 20.6 (618) 347.03 (30) − 38.08 (530) N-Methylbenzylamine (18) − 18.59 (942) 347.85 (38) Cyclopentane carboximidamide (19) − 13.38 (2134) (1-Methyl-1H-pyrrol-2-yl)-methylamine (20) Pyrimidine-2,4,6-triamine (24) AMBER PLOP Rescoring Docking Hit Lists Binding detected Compound (ID) a b − 8.52 (3093) 363.47 (1901) −56.65 (32) 10.0 No No 1-Methyl-5-imidazolecarboxaldehyde (26) − 21.14 (551) 358.53 (746) −57.12 (28) 10.0 Yes Yes 3-Methoxypyridine (27) − 23.05 (2665) 355.17 (393) −55.31 (44) 10.0 Yes Yes 2-Imino-4-methylpiperidine (28) − 17.30 (1695) 349.17 (82) −52.43 (119) 10.0 Yes Yes 2,4,5-Trimethyl-3-oxazoline (29) − 13.96 (1962) 355.98 (455) −52.32 (124) 0.25 Yes Yes 1-Methyl-2-vinylpyridinium (30) − 15.17 (1716) 363.60 (1938) −52.32 (125) 0.5 Yes Yes Rescoring Docking Hit Lists 1,3-Dimethyl-2-oxo-2,3-dihydro-pyrimidin-1-ium (25) Compound scores and ranks (in parenthesis) for DOCK, AMBERDOCK, and PLOP. Scores and ranks in bold font indicate ligands that rank in the top 200 for the respective scoring function. Concentration at which ligand was tested. 925 926 correctly predicted the binding mode for 3-chloro-1phenyl-1-propanol (13), with RMSD values of 1.9 and 1.7 Å, respectively. In three structures—2-nitrothiophene (11), 3-methylbenzylazide (6), and 3-chloro-1phenyl-1-propanol (13)—the F-helix of the cavity moves to accommodate the ligands while keeping the cavity still buried from solvent. In the complexes with 2-(n-propylthio)ethanol (12) and 2-phenoxyethanol (9), there is evidence of a second conformation of residue Phe114 within the cavity that rotates and opens a water channel to the surface of the protein. Neither the helix movement nor the Phe114 rotation was sampled by the MM–GBSA methods. New W191G ligands predicted by rescoring Fourteen representative compounds reprioritized to score well by the MM–GBSA methods but scored poorly by docking were experimentally tested for binding by measuring perturbation of the heme Soret band in CCP (Table 4).24 Binding was detected for 10 of these compounds at concentrations ranging from 50 μM to 10 mM. Of the 11 compounds that AMBERDOCK predicted to bind with ranks better than 500, binding was detected for 8. Of the remaining prospective hits tested, AMBERDOCK correctly distinguished 1 compound as a decoy but missed 2 ligands by ranking them worse than 500. Of the 9 compounds that PLOP predicted to bind with ranks better than 500, binding was detected for 8. Of the remaining prospective hits tested, PLOP missed 2 ligands but correctly distinguished 3 decoys, ranking them worse than 500. Crystal structures of CCP in complex with the 10 new ligands were obtained (Fig. 5). The electron density for the ligands was unambiguous. Docking predicted three structures to within 1 Å of the crystallographic result, whereas the MM–GBSA methods did so for 7 structures, typically with improved hydrogen-bonding interactions (Table 5). For 3 ligands, the docking poses were over 1.9 Å away from the crystallographic results, and MM–GBSA refinement did little to improve these structures. In 4 of the complex structures—cyclopentane carboximidamide (19), 1,2-dimethyl-1H-pyridine-5-amine (22), pyrimidine-2,4,6-triamine (24), and 1-methyl-2-vinylpyridinium (30)—the loop composed of residues 190–195 flips out by nearly 12 Å, opening the cavity to bulk solvent. This large loop motion was not sampled by MM–GBSA. Overall performance in predicting top 100 hits The simplicity of these model cavity sites, the number of known ligands and decoys, and our Rescoring Docking Hit Lists experience with their ligands21,23,28–30 often allow us to predict what turn out to be true ligands and true decoys from among top-scoring molecules, based on their physical properties. We examined the top 100 hits predicted to bind by docking and MM–GBSA, compared property distributions, and made educated guesses as to whether or not they will bind. The 100 top-ranking MM–GBSA rescored compounds for the L99A and L99A/M102Q cavities were larger, more flexible, and more polar, with more hydrogenbond acceptors and lower ClogP values per heavy atom compared to the top 100 hits from docking. For the anionic W191G cavity there was a similar trend towards larger molecules and also a drift away from the singly charged cations favored by DOCK, with more dications and neutral molecules prioritized among the top-ranking 100 molecules by the MM– GBSA methods. The increased size and greater differences in polarity of the molecules in the MM– GBSA hit lists resulted in lower mean pairwise similarities among the molecules and, consequently, an increase in the diversity of the rescored hit lists relative to the docking hit lists. Thus, using ECFP_4 fingerprints (SciTegic, Inc.), we found the average pairwise Tanimoto coefficient among the 100 topdocking molecules for the L99A cavity with DOCK, AMBERDOCK, and PLOP to be 0.17, 0.12, and 0.10, respectively (full distributions of pairwise similarities are given in Supplementary Fig. S1). Similar trends were observed in the other two cavities. The same tendencies that led to greater diversity in ligands and their properties, however, reduced the raw hit rates we anticipate from among the top 100 ranking MM–GBSA ligands compared to those predicted by docking (Table 6). For example, among the top 100 docking hits for the CCP cavity there were 29 true ligands and no experimentally determined decoys. Of the remaining molecules—all untested— were what we predict to be 79 likely ligands and 7 likely decoys, based on their similarity to known ligands and decoys and their physical properties such as size and charge complementarity. Conversely, among the top 100 PLOP hits for the anionic cavity were only 15 experimentally tested ligands and 1 experimental decoy. Among the untested molecules were what we suspect are 53 further ligands and 22 further decoys. Among the top AMBERDOCK hits for this cavity were 19 true ligands and 3 experimental decoys. Among the untested molecules prioritized by this program, we suspect that there are 67 further ligands and 14 more decoys. Similar trends were observed in the other two cavities (Table 6). Admittedly, these numbers reflect guesses only, but we suspect that the overall trends would be born out by experiment (the interested reader may draw their own conclusions from the full lists in Supple- Fig. 5. Predicted and experimental ligand orientations for the anionic CCP cavity. The carbon atoms of the crystallographic, DOCK, AMBERDOCK, and PLOP predicted poses are colored gray, green, cyan, and orange, respectively. (a) n-Methylbenzylamine (18), (b) cyclopentane carboximidamide (19), (c) (1-methyl-1H-pyrrol-2-yl)-methylamine (20), (d) 1,2-dimethyl-1H-pyridin-5-amine (22), (e) pyrimidine-2,4,6-triamine (24), (f) 1-methyl-5-imidazolecarboxaldehyde (26), (g) 3-methoxypyridine (27), (h) 2-imino-4-methylpiperdine (28), (i) 2,4,5-trimethyl-3-oxazoline (29), and (j) 1-methyl2-vinylpyridinium (30). Rendered with the program PyMOL.26 Rescoring Docking Hit Lists 927 Fig. 5 (legend on previous page) Rescoring Docking Hit Lists 2.58 2.55 2.59 0.55 0.50 0.33 1.32 0.83 0.81 L99A cavity DOCK PLOP AMBERDOCK All crystals belong to space group P212121. Values in parentheses are for the highest resolution shell. a Two conformations of the crystallographic ligand were modeled. 1.07 1.06 0.94 1.91 1.70 1.91 2.97/3.16a 2.84/3.01a 2.98/3.15a 0.58 0.34 0.30 1.34 0.31 0.99 1.05 0.37 0.45 Likely Likely True True ligands decoys ligands decoys in top in top in top in top 100 100 hits hitsb Ambiguousc hits hitsa 7 6 8 3 1 2 63 35 54 23 22 25 14 43 21 L99A/M102Q cavity DOCK 13 PLOP 5 AMBERDOCK 7 2 1 2 73 31 43 12 8 22 15 61 35 W191G cavity DOCK PLOP AMBERDOCK 0 1 3 79 53 67 7 22 14 14 25 19 29 15 19 a Molecules that, based on their physical properties and similarity to known ligands, are likely to be cavity ligands (a full list is given in Supplementary Tables S1–S9). b Molecules that, based on their physical properties and similarity to known decoys, are likely not to bind. c Molecules that are sufficiently different from known ligands and decoys and whose physical properties are not sufficiently distinctive, such that no prediction was made (for L99A and L99A/ M102Q molecules). For W191G, molecules that were misprotonated during database preparation relative to the expected protonation at pH 4.5 are not counted to measure the performance of the scoring function. 0.89 0.81 0.77 1.50 (1.54) 63,108 (3364) 4.2 (19.6) 99.3 (95.4) 39.2 (5.9) 14.6 (16.3) 16.7 (21.7) 0.01 1.14 2RC2 2.49 (2.56) 11042 (680) 2.7 (5.9) 82.3 (69.7) 26.1 (12.9) 17.7 (21.9) 22.9 (29.4) 0.02 1.95 2RC1 1.50 (1.54) 63,783 (4467) 5.0 (33.4) 99.7 (96.5) 31.0 (3.5) 14.3 (17.9) 16.9 (23.9) 0.01 1.22 2RC0 1.50 (1.54) 63,445 (4489) 3.8 (22.1) 99.7 (96.3) 37.8 (5.2) 14.5 (16.9) 16.4 (21.0) 0.01 1.24 2RBY 1.50 (1.54) 63,009 (4512) 5.2 (38.5) 99.8 (97.9) 30.6 (2.8) 15.2 (21.0) 17.3 (25.7) 0.01 1.14 2RBX 1.80 (1.85) 36,504 (2670) 5.3 (10.9) 99.4 (99.9) 51.7 (30.1) 15.9 (19.3) 19.5 (24.0) 0.02 1.39 2RBU 1.39 (1.43) 78,561 (5188) 4.5 (20.2) 99.0 (90.5) 43.2 (5.6) 13.8 (20.7) 15.4 (24.1) 0.01 1.19 2RBV 1.50 (1.54) 57,782 (4371) 5.1 (22.4) 92.0 (95.9) 41.7 (6.0) 14.4 (17.8) 16.7 (25.5) 0.01 1.22 2RBW 1.80 (1.85) 36,874 (2587) 6.4 (23.3) 99.5 (96.2) 34.8 (9.2) 15.0 (18.4) 19.1 (24.0) 0.02 1.50 2RBZ Method 1.24 (1.27) 104,081 (4744) 6.1 (40.7) 93.1 (58.0) 29.9 (1.8) 12.2 (24.8) 14.6 (24.4) 0.01 1.59 2RBT n-Methylbenzylamine (18) Table 6. Likely ligands and decoys among the top 100 ranked ligands by docking and MM–GBSA Resolution (Å) Reflections Rmerge (%) Completeness (%) ⟨I⟩/⟨σ(I)⟩ R-factor (%) Rfree (%) Δbond lengths (Å) Δbond angles (°) PDB code RMSD (Å) DOCK AMBER PLOP 1-Methyl-2vinylpyridinium (30) 1-Methyl-52,4,5imidazole 2-Imino-4carboxaldehyde 3-Methoxypyridine methylpiperidine Trimethyl-3oxazoline (29) (26) (27) (28) 1,2-Dimethyl- Pyrimidine1H-pyridin-52,4,6amine (22) triamine (24) (1-Methyl-1Hpyrrol-2-yl)methylamine (20) Cyclopentane carbox imidamide (19) Table 5. Crystallographic measurement and the RMSD values for predicted and crystallographic ligand geometries in the CCP site (compound IDs in parentheses) 928 mentary Tables 1–9) Thus, whereas the MM–GBSA methods rescued many docking false negatives and sampled a more diverse chemical space among the top hits, they also suggested more false positives among the very top scoring molecules and, we suspect, have a lower overall hit rate in this segment of the molecules prioritized for testing. Origins of false-positive hits suggested by MM–GBSA rescoring In these simple cavities, false-positive hits often identify specific pathologies in a scoring function. For example, the MM–GBSA methods seemed distracted by compounds bearing what is almost certainly the wrong net charge for the W191G cavity, which extensive testing has shown preferentially binds monocations over neutral molecules (few of which have been observed to bind, and then only weakly) and dications (none of which have been observed to bind). For instance, among the top 100 ranking molecules predicted by PLOP, there were 13 dications. Whereas AMBERDOCK predicted only one dication, it prioritized five neutral molecules among the top 100 hits. The dications will pay too high a desolvation penalty to be compensated for by the interaction with the single anion in the site (Asp235), and the neutral compounds desolvate the same aspartate without recouping enough in interaction energy. Balancing polar and ionic interactions with concomitant solvation penalties is a challenge for the field, one clearly faced by these methods as Rescoring Docking Hit Lists well. On the other hand, many of the top-ranked PLOP ligands for L99A (47 out of the top 50) and L99A/M102Q contained one or more nitriles. While some of these compounds may well be ligands, as in the case of n-phenylglycinonitrile (10) for L99A/ M102Q (Table 3), we suspect that this represents a ligand parameterization problem as opposed to a genuinely meaningful enrichment. Indeed, the PLOP solvation energies for the top 47 nitriles actually rewarded desolvation, rather than penalizing it as is almost always the case otherwise, suggesting that there is an issue with determining the correct selfenergy for this functional group. Not wishing this term to dominate our analysis, we excluded these compounds from the PLOP rescored hit lists for L99A and L99A/M102Q; they do not contribute to the accounting described in this work. This highlights the importance of good ligand parameterization for database screening—which is a considerable challenge for hundreds of thousands of molecules typically screened by docking—the lack of which can undermine any improvement in theory. Discussion In principle, the most important improvements of MM–GBSA over docking, certainly over the program used in this study, DOCK3.5.54, are the better representation of electrostatic interactions, ligand and protein desolvation energies, and relaxation of the ligand–protein complex. The simplicity of the model cavity sites allows us to explore how these terms influence docking results in detail and to make prospective predictions for ligands that we can, in fact, acquire and test. Many investigators will be unsurprised to see that the MM–GBSA methods can rescue molecules that rank poorly in the docking calculation owing to the rigid-receptor approximation used in docking. Ligands that were too big to be accommodated well in the original docking are well fit by a binding site that has been allowed to relax by energy minimization and, in the case of AMBERDOCK, short MD simulations. This was true both in retrospective calculations as well as in prospective predictions. The ability to relax the site also resulted in rescored hit lists that were more diverse with a wider range of likely ligands. Perhaps less anticipated was the cost of allowing such conformational change—some of the rescued, high-scoring molecules by MM–GBSA do not, in fact, bind to the cavity sites. These molecules are new false positives introduced by the higher level of theory. Indeed, the overall hit rates at the very top of the ranked lists are arguably better by simple docking than by MM– GBSA rescoring, at least when evaluated simplistically by the raw number of hits and likely hits (this is arguably offset by the greater diversity of the MM– GBSA hit lists). Partly this reflects problems in ligand parameterization, and partly difficulties in the treatment of the electrostatics in the binding sites. The most important challenge for MM–GBSA and for flexible receptor models in general is 929 balancing the opportunities to find new ligands as receptor geometries are relaxed with the introduction of new false positives as the need to consider large receptor internal energies is introduced. Specific examples of these opportunities and problems are apparent in the three cavity sites studied here. The principal improvement conferred by MM– GBSA rescoring in the model cavity sites over docking was the inclusion of receptor binding site relaxation, which improved the ranks of larger ligands that rigid receptor docking missed. AMBERDOCK, for example, correctly predicted 2-ethoxyphenol (5) to bind to L99A (Table 1, Fig. 3d). This compound is too large for the unrelaxed conformation of this cavity targeted by docking, but minimization and MD simulations allow the ligand to be well accommodated by effectively expanding the site. Often, this relaxation led not only to improved rankings but also improved geometries. For many ligands, RMSD values between the MM–GBSA predictions and the crystallographic results declined relative to those of the docking predictions and, especially in the W191G anionic cavity, many ligands refined by MM–GBSA had improved hydrogen bonding to the site. Examples of this include the new W191G cavity ligands n-methylbenzylamine (18) and cyclopentane carboximidamide (19) (Table 4, Fig. 5a and b, respectively). The structural relaxation with MM–GBSA performed well when the initial docking geometry resembled the crystallographic pose, but did little when large protein conformational changes were provoked by ligand binding. For instance, F-helix unwinding and rotamer change by Val111 in L99A and L99A/M102Q were never captured by the method, nor was the extensive loop flipping observed in several of the W191G–ligand complexes. When such movements occurred, MM–GBSA rescoring could not rescue substantially incorrect docking poses, such as that adopted by 3-chloro-1phenyl-propanol (13) for L99A/M102Q (Table 3, Fig. 4f) and pyrimidine-2,4,6-triamine (24) predicted for CCP (Table 4, Fig. 5e), notwithstanding the large improvement in their rankings conferred by the rescoring. These large movements are outside the radius of convergence of the local relaxation undertaken by the MM–GBSA methods. Indeed, even more time-consuming thermodynamic integration methods are hard put to sample such changes without explicit “confine-and-release” strategies, which depend on a foreknowledge that such movements are likely.41 And whereas loop sampling methods have had encouraging successes in predicting such large movements,42 this remains a frontier challenge for ligand and structure prediction methods. Pragmatically, the inability to predict the structural accommodations provoked by some large ligands is offset by the correct reprioritization of what were docking false negatives as ligands. The same comfort is not afforded by the 10 false negatives introduced by the MM–GBSA methods, nor by the lower overall hit rates compared to docking among 930 the very top scoring ligands (Table 6). By allowing the receptor to respond to ligand binding, one allows for new and potentially unfavorable receptor conformations. These must be distinguished by the MM–GBSA energy functions from the true lowenergy conformations that may be sampled in solution. This is challenging, as the receptor conformational energies are large, and the errors in these calculations are typically on the same order of the net interaction energy of the protein–ligand complex. Although some of the errors are cancelled by subtraction of the internal energies before and after ligand binding, one is still subtracting two large numbers with relatively large errors to find a small one, the net binding free energy. Consistent with this view, ligands achieved their maximal advantage over decoys on rescoring when we allowed only a 5 Å region around the binding site to relax. Allowing the full protein to relax, or even an 8 Å region around the binding site, diminished the discrimination of known ligands from decoys. Of course, relaxing the entire system is the more physically correct way to calculate these energies. Falling back on limited relaxation speaks of a larger methodological issue. The three cavity sites targeted here are contrivances of human design, and ligands discovered for them have no intrinsic value other than for testing methods. Indeed, in these simple model systems the failures are often more interesting than the successes, as they can illuminate a specific methodological problem.21,23,28–30 Examples are the 10 false positives predicted for the cavity sites by MM–GBSA rescoring. Some of these reflect ligand parameterization problems. For instance, we suspect that the many nitrile-containing decoys predicted by PLOP for L99A and L99A/M102Q reflect failures in ligand parameterization. Such mechanical failures may be addressed by close attention to particular ligand groups and improved partial atomic charge models; admittedly, this can be a daunting task for screening databases containing hundreds of thousands of disparate molecules. More interesting are the 8 false positives that are true energy function decoys. Several of these highlight difficulties in the treatment of electrostatics and solvation in the binding sites. 2Phenyoxyethanol (9), for example, was predicted to bind by both PLOP and AMBERDOCK to L99A (Table 1). This decoy has a similar topology to 2phenylpropanol, a known ligand22 (Fig. 6a); however, the ether of 2-phenoxyethanol (9) increases its polarity and presumably its solvation energy, which is not fully captured by the MM–GBSA implicit solvent model (another possibility would be that the 2-phenoxyethanol is docked in a high-energy conformation, one that is not recognized by the rescoring methods, but this turns out not to be the case, with both the decoy and the ligand 2-phenylpropanol adopting similar and low-energy conformations). Similarly, o-benzylhydroxylamine (14) was the top-ranking AMBERDOCK hit for L99A/ M102Q, but is a decoy (Table 3). The terminal -ONH2 of this compound is too polar for the site, stranding one unpaired polar hydrogen from the Rescoring Docking Hit Lists Fig. 6. The topologically similar ligands and decoys of (a) 2-phenylpropanol and 2-phenoxyethanol (9) for L99A and (b) n-phenylhydroxylamine and o-benzylhydroxylamine (14) to L99A/M102Q. NH2 group in this largely hydrophobic site. Interestingly, the polar cavity does bind n-phenylhydroxylamine (unpublished data), which has the same hydrogen bond accounting as o-benzylhydroxylamine (14) and topologically resembles it closely (Fig. 6b). The difference between these nearly identical molecules is that in the former the two hydrogenbond donors from the ligand can both be accommodated by the carbonyl of the receptor glutamine, whereas in the decoy both hydrogen-bond donors originate from the same atom—the nitrogen of the obenzylhydroxylamine (14)—and only one can be accommodated by the carbonyl oxygen. The challenges of balancing ligand electrostatic interaction energies and desolvation penalties were also apparent in the anionic W191G cavity. Most obvious were those molecules that did not bear the correct monocationic charge state. The 13 molecules that were doubly charged among the top-scoring PLOP hits are almost certainly decoys, and this is also the case for the AMBERDOCK false-positive 5nitro-6-aminouracil (21), which is neutral and cannot make the ion–pair interaction with Asp235 (Table 4). More subtly, whereas 1,3-dimethyl-2-oxo2,3-dihydropyrimidin-1-ium (25) is charged, this charge is shared between the two cyclic nitrogen atoms and results in a compound with reduced electrophilicity compared to a compound with a localized charge. The AMBERDOCK false positives n-methyl-1,2-phenylene-diamine (17) and 2-aminobenzylamine (23) (Table 4) most likely do not bind because of steric clashes that inhibit optimal positioning of the charge–charge interaction. These failures point to specific directions for improved treatment of the balance between electrostatic interaction and desolvation energies in the MM– GBSA methods. Overall, the results of MM–GBSA rescoring of docking hit lists on the model binding sites seem conflicted. On the one hand, rescoring rescued many docking false negatives, improved the geometric fidelity of most of the predicted structures, and increased the diversity of the hit lists. On the other hand, rescoring introduced more false positives, especially among the very top ranking ligands, Rescoring Docking Hit Lists compared to the simpler docking protocol. These observations may be reconciled by recognizing that what is probably the greatest advantage of the MM–GBSA methods over docking for the model sites, the relaxation of the protein–ligand complex, also presents the greatest challenge to discrimination. To allow a flexible receptor, one must consider the relative energies of the different protein conformations explored. This implicates the pairwise interactions of thousands of protein atoms, as opposed to the tens of atoms involved in the immediate protein–ligand complex. To properly rank the energies of the complexes, one must also properly account for the larger uncertainties that accompany the much higher magnitude energies of the overall system. Whereas this is the thermodynamically correct approach, it introduces many interactions that have little bearing on the intimacies of the protein–ligand complex itself. Rigid receptor docking, for all the calumny poured upon it, can ignore these large-magnitude yet low-relevance interactions. Of course, this leads to many false negatives, but it avoids many of the false positives to which the MM–GBSA methods are prone. Pragmatically, this suggests that hits derived from docking to a rigid experimental receptor conformation—and ideally more than one30,43—and hits prioritized by rescoring after MM–GBSA refinement with binding site minimization will provide good candidates for experimental testing. Despite its greater sophistication, MM–GBSA rescoring has a harder task, and its predictions will not, by every criterion, be better than those of a modern docking program; rather, our results suggest they will complement and add to them. Still, MM–GBSA is a higher level of theory, and because it is grounded in physics, they can be built upon and improved in a regular way. They are thus on a path to fundamental improvement in molecular docking and structure-based screening, which is so actively sought.44 931 Rescoring with PLOP The rescoring procedure with PLOP36,37 was essentially as described.17 Ligand parameters were calculated with IMPACT.47 The partial atomic charges of the ligands were replaced by the AM1-CM2 charges calculated by AMSOL (v6.5.3) as these were the same charges used during the initial docking.23 The same protein structure file used in docking was used for rescoring. Protein parameters were defined by IMPACT with the exception of the partial charges for the heme cofactor in CCP W191G, which were the same as used in the docking method.28 All energy minimizations were performed using PLOP with the allatom OPLS force field (OPLS-AA)48 and the surface generalized Born (SGB) implicit solvent model.49 PLOP implements a multiscale truncated-Newton minimization algorithm as described.50 For receptor minimization and calculation of Ecomplex and Ereceptor, residues in a prespecified list within 5 Å of the binding site were minimized after an initial side-chain rotamer search. (Residues 78, 84, 85, 87, 88, 91, 98–100, 102, 103, 106, 111, 118, 121, 133, and 153 for L99A and L99A/M102Q and residues 174–180, 189–192, 202, 230–232, 235, and water 308 for CCP). The rotamer search algorithm is as described in the Supplementary Material. Preliminary PLOP calculations of the hydrophobic and polar cavities were performed with a rigid receptor and resulted in very little separation of ligands and known decoys. On the other hand, PLOP calculations in which a larger set of residues (those within 8 Å of the binding site) were minimized and resulted in worse overall enrichments of known ligands and a decreased separation of known ligands and decoys relative to minimizing a smaller 5 Å pocket. To approximate a fully desolvated ligand and cavity for the hydrophobic L99A and polar L99A/M102Q sites, only the SBG solvation term of the free ligand was included in the calculation of the total PLOP binding energies. Initial PLOP calculations including the SGB solvation terms for the calculation of the complex and free protein energies resulted in poor enrichments of known ligands, decreased separation of ligands and known decoys, as well as an enrichment of hits with increased polarity and electrostatic interactions. For the more solvated CCP cavity, the SGB terms were included in the calculation of the complex, free protein, and free ligand energies for the total binding energy. Materials and Methods Rescoring with AMBERDOCK Docking against cavity sites DOCK3.5.5423,38 was used to dock a multiconformer database of small molecules into the model cavity sites. The receptors, grids, spheres, and ligand databases were prepared as described for the T4 Lysozyme23 and CCP28 cavities, respectively. Briefly, to sample ligand orientations, ligand, receptor, and overlap bins were set to 0.2 Å; the distance tolerance for matching ligand atoms to receptor was set to 0.75 Å. Each docking pose was evaluated for steric fit. Compounds passing this filter were scored for electrostatic and van der Waals complementarity and assigned the full penalty for transfer from a dielectric of 80 to a dielectric of 2, as calculated by AMSOL.45,46 Sampling and scoring required less than a second per ligand on a single 3.2-GHz Xeon processor. The best scoring conformation of each of the 10,000 top-scoring molecules against L99A and L99A/M102Q and the 5400 top-scoring molecules against CCP were saved and rescored by the MM–GBSA protocols. AMBERDOCK is based on the amber_score() scoring module in DOCK6. The ligand structures were modified using the antechamber suite of programs to create input files that could be read by Leap to generate the parameter and topology files for AMBERDOCK. Antechamber51 has been developed to be used with the general AMBER force field for small molecules.52 Charges for the ligands were generated using three charge methods in AntechamberPEOE,53 AM1-BCC,54 and HF/6-31G* RESP.55 The protonation states of the ligands were kept the same as the previous docking run for consistency in rescoring. AMBER ff94 parameters were assigned to all the protein atoms. The standard parameters for the heme cofactor as implemented in the Amber 9 program was used for the CCP cavity.56 The protonation states of histidine residues were predicted based on their close neighbors. The GB model corresponding to igb = 5 in the AMBER 9 program was used.57 The surface area term was calculated using the LCPO model.58 A nonbonded cutoff of 18 Å was used for the calculations. Rescoring Docking Hit Lists 932 The starting structures were taken from the docked pose. The structures were subjected to 100 steps of conjugate gradient minimization, 3000 steps of MD simulation with a 1-fs time step at a temperature of 300 K, followed by 100 steps of minimization. During the minimization and MD, only the ligand and the protein residues within 5 Å of the ligand were allowed to move. To expedite the scoring process, we calculated the energy of the receptor (Rreceptor) once, and used this energy as a constant term during the subsequent energy evaluations for the rest of the ligands in the database. Binding-free-energy calculations with AMBERDOCK follows a scheme as described in Supplementary Fig. S2. Several AMBERDOCK rescoring protocols with slight variations were retrospectively tested and results are described in Supplementary Fig. S3. Protein preparation and expression T4 lysozyme mutants L99A and L99A/M102Q and CCP mutant W191G were expressed and purified as described.23,24 Binding detection of ligands to T4 lysozyme cavities by upshift of thermal denaturation temperature To detect binding, L99A and L99A/M102Q were denatured reversibly by temperature in the presence and absence of the putative ligand. Molecules that bind preferentially to the folded cavity-containing protein should stabilize it relative to the apo protein, raising its temperature of melting.22 All thermal melts were conducted in a Jasco J-715 spectropolarimeter as described.22 Each compound was screened in its neutral form. All compounds tested against L99A and L99A/M102Q were assayed in a pH 3 buffer containing 25 mM KCl, 2.9 mM phosphoric acid, and 17 mM KH2PO4 with the exception of 1-phenylsemicarbazide (3) and o-benzylhydroxylamine (14). To maintain compound neutrality, these two were assayed at pH 6.8 in a 50 mM potassium chloride and 38% (v/v) ethylene glycol buffer.22 Thermal melts were monitored by far-UV circular dichroism, except for melts in the presence of 4-(methylthio)nitrobenzene (2), 1-phenylsemicarbazide (3), and 2,6-difluorobenzylbromide (4), which absorb strongly in the far-UV region. For these three, thermal denaturation was measured by the intensity of the integrated fluorescence emission for all wavelengths above 300 nm, exciting at 283 to 292 nm, using a fluorescence detector on the Jasco instrument. Thermal melts were performed at a temperature ramp rate of 2 K/min. A least-squares fit of the two-state transition model was performed with the program EXAM59 to calculate Tm and van't Hoff ΔH values for the thermal denaturations. The ΔCp was set to 8 KJ mol− 1 K− 1 (1.94 kcal mol− 1 K− 1). Binding detection of ligands to CCP W191G Ligand binding was measured in 50 mM acetate buffer, pH 4.5. To avoid competition in ligand binding with small cations such as potassium,24 the pH of the buffer was adjusted with Bis–Tris propane. The compounds were dissolved in dimethyl sulfoxide. Binding of compounds to CCP was monitored by the red shift and increase of absorbance of the heme Soret band24 at 10 °C. crystallization buffer containing as much as 100 mM compound. In addition to soaking, drops of neat compound were added to the cover slip surrounding the drop containing the crystal. After soaking, the crystals were cryoprotected with a 50:50 Paraton-N (Hampton Research, Aliso Viejo, CA)/mineral oil mix. Crystals for CCP W191G were grown as described25 and the resulting crystals belonged to space group P212121. Crystals were soaked in 25% methyl-2,4-pentanediol with 1 to 50 mM compound for 4 h or overnight with the exception of pyrimidine2,4,6-triamine (24), which was soaked for 15 min. Diffraction data for the complexes of L99A with β-chlorophenetole (1), 4-(methylthio)nitrobenzene (2), and 2,6-difluorobenzylbromide (4) and the complex of L99A/M102Q with 3-methylbenzylazide (6) were collected using a Rigaku X-ray generator equipped with a rotating copper anode and a Raxis IV image plate. Data for the complexes of L99A/ M102Q with n-phenylglycinonitrile (10) and 2-nitrothiophene (11) and the complex of CCP with n-methylbenzylamine (18) were collected on beamline 9-1 at the Stanford Synchrotron Radiation Laboratory using an ADSC CCD detector. Data for all other complexes were collected on beamline 8.3.1 of the Advanced Light Source at Lawrence Berkeley National Laboratory using an ADSC CCD detector. All data sets were collected at 100 K. Reflections were indexed, integrated, and scaled using HKL2000.60 Parameters for ligands were generated with PRODRG.61 Complexes were refined using the CCP4 software package.62 Interactive model building was performed using Coot.63 Supporting information available A description of the PLOP side-chain rotamer search and minimization algorithm, AMBERDOCK parameters and optimization, and structures for the top 100 hits predicted by DOCK, PLOP, and AMBERDOCK for the three cavity sites is available online from the journal website. Protein Data Bank accession codes The crystallographic coordinates for the complex structures presented in this work have been deposited with the RCSB Protein Data Bank with accession codes 2RAY, 2RAZ, 2RB0, 2RB1, 2RB2, 2RBN, 2RBO, 2RBP, 2RBQ, 2RBR, 2RBS, 2RBT, 2RBU, 2RBV, 2RBW, 2RBX, 2RBY, 2RBZ, 2RC0, 2RC1, and 2RC2. Acknowledgements This work was supported by GM59957 (to B.K.S.), AI035707 (to M.P.J.) and GM56531 (to D.A.C.). M.P.J. is a consultant to Schrodinger Inc. We thank Niu Huang for advice on using PLOP; Michael Keiser and Jerome Hert for help with chemical similarity calculations; Michael Mysinger, Veena Thomas, and Michael Keiser for reading this manuscript; and MDL for providing the ACD database. Structure determination Supplementary Data Crystals for L99A and L99A/M102Q were grown as described23 and the resulting crystals belonged to space group P3221. Crystals were soaked overnight to 1 week in Supplementary data associated with this article can be found, in the online version, at doi:10.1016/ j.jmb.2008.01.049 Rescoring Docking Hit Lists 933 References 1. Powers, R. A., Morandi, F. & Shoichet, B. K. (2002). Structure-based discovery of a novel, noncovalent inhibitor of AmpC beta-lactamase. Structure, 10, 1013–1023. 2. Evers, A. & Klebe, G. (2004). Successful virtual screening for a submicromolar antagonist of the neurokinin1 receptor based on a ligand-supported homology model. J. Med. Chem. 47, 5381–5392. 3. Grzybowski, B. A., Ishchenko, A. V., Kim, C.-Y., Topalov, G., Chapman, R., Christianson, D. W. et al. (2002). Combinatorial computational method gives new picomolar ligands for a known enzyme. Proc. Natl Acad. Sci. USA, 99, 1270–1273. 4. Li, S., Gao, J., Satoh, T., Friedman, T. M., Edling, A. E., Koch, U. et al. (1997). A computer screening approach to immunoglobulin superfamily structures and interactions: discovery of small non-peptidic CD4 inhibitors as novel immunotherapeutics. Proc. Natl Acad. Sci. USA, 94, 73–78. 5. Huang, N., Nagarsekar, A., Xia, G., Hayashi, J. & MacKerell, A. D., Jr (2004). Identification of nonphosphate-containing small molecular weight inhibitors of the tyrosine kinase p56 Lck SH2 domain via in silico screening against the pY + 3 binding site. J. Med. Chem. 47, 3502–3511. 6. Song, H., Wang, R., Wang, S. & Lin, J. (2005). A lowmolecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc. Natl Acad. Sci. USA, 102, 4700–4705. 7. Lyne, P. D., Kenny, P. W., Cosgrove, D. A., Deng, C., Zabludoff, S., Wendoloski, J. J. & Ashwell, S. (2004). Identification of compounds with nanomolar binding affinity for checkpoint kinase-1 using knowledgebased virtual screening. J. Med. Chem. 47, 1962–1968. 8. Cavasotto, C. N., Oritz, M. A., Abagyan, R. A. & Piedrafita, F. J. (2006). In silico identification of novel EGFR inhibitors with antiproliferative activity against cancer cells. Bioorg. Med. Chem. Lett. 16, 1969–1974. 9. Charifson, P. S., Corkery, J. J., Murcko, M. A. & Walters, W. P. (1999). Consensus scoring: a method for obtaining improved hit rates from docking databases of three-dimensional structures into proteins. J. Med. Chem. 42, 5100–5109. 10. Bissantz, C., Folkers, G. & Rognan, D. (2000). Proteinbased virtual screening of chemical databases. 1. Evaluation of different docking/scoring combinations. J. Med. Chem. 43, 4759–4767. 11. Gohlke, H. & Klebe, G. (2001). Statistical potentials and scoring functions applied to protein–ligand binding. Curr. Opin. Struct. Biol. 11, 231–235. 12. Stahl, M. & Rarey, M. (2001). Detailed analysis of scoring functions for virtual screening. J. Med. Chem. 44, 1035–1042. 13. Wang, R. X. & Wang, S. M. (2001). How does consensus scoring work for virtual library screening? An idealized computer experiment. J. Chem. Inf. Comput. Sci. 41, 1422–1426. 14. Friesner, R. A., Murphy, R. B., Repasky, M. P., Frye, L. L., Greenwood, J. R., Halgren, T. A. et al. (2006). Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein– ligand complexes. J. Med. Chem. 49, 6177–6196. 15. Wang, J., Kang, X., Kuntz, I. D. & Kollman, P. A. (2005). Hierarchical database screenings for HIV-1 reverse transcriptase using a pharmacophore model, rigid docking, solvation docking, and MM-PB/SA. J. Med. Chem. 48, 2432–2444. 16. Kalyanaraman, C., Bernacki, K. & Jacobson, M. P. 17. 18. 19. 20. 21. 22. 23. 24. 25. 26. 27. 28. 29. 30. 31. 32. 33. (2005). Virtual screening against highly charged active sites: identifying substrates of alpha–beta barrel enzymes. Biochemistry, 44, 2059–2071. Huang, N., Kalyanaraman, C., Irwin, J. J. & Jacobson, M. P. (2006). Physics-based scoring of protein–ligand complexes: enrichment of known inhibitors in largescale virtual screening. J. Chem. Inf. Model. 46, 243–253. Perola, Emanuele (2006). Minimizing false positives in kinase virtual screens. Proteins: Struct., Funct., Bioinf. 64, 422–435. Lee, M. R. & Sun, Y. (2007). Improving docking accuracy through molecular mechanics generalized Born optimization and scoring. J. Chem. Theory Comput. 3, 1106–1119. Lyne, P. D., Lamb, M. L. & Saeh, J. C. (2006). Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM–GBSA scoring. J. Med. Chem. 49, 4805–4808. Mobley, D. L., Graves, A. P., Chodera, J. D., McReynolds, A. C., Shoichet, B. K. & Dill, K. A. (2007). Predicting absolute ligand binding free energies to a simple model site. J. Mol. Biol. 371, 1118–1134. Morton, A., Baase, W. A. & Matthews, B. W. (1995). Energetic origins of specificity of ligand binding in an interior nonpolar cavity of T4 lysozyme. Biochemistry, 34, 8564–8575. Wei, B. Q., Baase, W. A., Weaver, L. H., Matthews, B. W. & Shoichet, B. K. (2002). A model binding site for testing scoring functions in molecular docking. J. Mol. Biol. 322, 339–355. Fitzgerald, M. M., Churchill, M. J., McRee, D. E. & Goodin, D. B. (1994). Small molecule binding to an artificially created cavity at the active site of cytochrome c peroxidase. Biochemistry, 33, 3807–3818. Musah, R. A., Jensen, G. M., Bunte, S. W., Rosenfeld, R. J. & Goodin, D. B. (2002). Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein. J. Mol. Biol. 315, 845–857. DeLano, W. L. (2002). The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA. Chang, C. E. & Gilson, M. K. (2004). Free energy, entropy, and induced fit in host–guest recognition: calculations with the second-generation mining minima algorithm. J. Am. Chem. Soc. 126, 13156–13164. Brenk, R., Vetter, S. W., Boyce, S. E., Goodin, D. B. & Shoichet, B. K. (2006). Probing molecular docking in a charged model binding site. J. Mol. Biol. 357, 1449–1470. Graves, A. P., Brenk, R. & Shoichet, B. K. (2005). Decoys for docking. J. Med. Chem. 48, 3714–3728. Wei, B. Q., Weaver, L. H., Ferrari, A. M., Matthews, B. W. & Shoichet, B. K. (2004). Testing a flexiblereceptor docking algorithm in a model binding site. J. Mol. Biol. 337, 1161–1182. Deng, Y. & Roux, B. (2006). Calculation of standard binding free energies: aromatic molecules in the T4 lysozyme L99A mutant. J. Chem. Theory Comput. 2, 1255–1273. Boresch, S., Tettinger, F., Leitgeb, M. & Karplus, M. (2003). Absolute binding free energies: a quantitative approach for their calculation. J. Phys. Chem. B, 107, 9535–9551. Machicado, C., Lopez-Llano, J., Cuesta-Lopez, S., Bueno, M. & Sancho, J. (2005). Design of ligand binding to an engineered protein cavity using virtual screening and thermal up-shift evaluation. J. Comp.-Aided Mol. Des. 19, 421–443. 934 34. Rosenfeld, R., Goodsell, D., Musah, R., Garret, M., Goodin, D. & Olson, A. (2003). Automated docking of ligands to an artificial active site: augmenting crystallographic analysis with computer modeling. J. Comput.-Aided Mol. Des, 17, 525–536. 35. Elowe, N. H., Blanchard, J. E., Cechetto, J. D. & Brown, E. D. (2005). Experimental screening of dihydrofolate reductase yields a “test set” of 50,000 small molecules for a computational data-mining and docking competition. J. Biomol. Screening, 10, 653–657. 36. Jacobson, M. P., Pincus, D. L., Rapp, C. S., Day, T. J., Honig, B., Shaw, D. E. & Friesner, R. A. (2004). A hierarchical approach to all-atom protein loop prediction. Proteins, 55, 351–367. 37. Jacobson, M. P., Kaminski, G. A., Friesner, R. A. & Rapp, C. S. (2002). Force field validation using protein side chain prediction. J. Phys. Chem. B, 106, 11673–11680. 38. Lorber, D. M. & Shoichet, B. K. (1998). Flexible ligand docking using conformational ensembles. Protein Sci. 7, 938–950. 39. Irwin, J. J. & Shoichet, B. K. (2005). ZINC—a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182. 40. Morton, A. & Matthews, B. W. (1995). Specificity of ligand binding in a buried nonpolar cavity of T4 lysozyme: linkage of dynamics and structural plasticity. Biochemistry, 34, 8576–8588. 41. Mobley, D. L., Chodera, J. D. & Dill, K. A. (2007). Confine-and-release method: obtaining correct binding free energies in the presence of protein conformational change. J. Chem. Theory Comput. 3, 1231–1235. 42. Song, L., Kalyanaraman, C., Fedorov, A. A., Fedorov, E. V., Glasner, M. E., Brown, S. et al. (2007). Prediction and assignment of function for a divergent N-succinyl amino acid racemase. Nat. Chem. Biol. 3, 486–491. 43. Claussen, H., Buning, C., Rarey, M. & Lengauer, T. (2001). FlexE: efficient molecular docking considering protein structure variations. J. Mol. Biol. 308, 377–395. 44. Leach, A. R., Shoichet, B. K. & Peishoff, C. E. (2006). Prediction of protein–ligand interactions. Docking and scoring: successes and gaps. J. Med. Chem. 49, 5851–5855. 45. Li, J. B., Zhu, T. H., Cramer, C. J. & Truhlar, D. G. (1998). New class IV charge model for extracting accurate partial charges from wave functions. J. Phys. Chem. A, 102, 1820–1831. 46. Chambers, C. C., Hawkins, G. D., Cramer, C. J. & Truhlar, D. G. (1996). Model for aqueous solvation based on class IV atomic charges and first solvation shell effects. J. Phys. Chem. 100, 16385–16398. 47. Banks, J. L., Beard, H. S., Cao, Y., Cho, A. E., Damm, W., Farid, R. et al. (2005). Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 26, 1752–1780. 48. Kaminski, G. A., Friesner, R. A., Tirado-Rives, J. & Jorgensen, W. L. (2001). Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B, 105, 6474–6487. Rescoring Docking Hit Lists 49. Gallicchio, E., Zhang, L. Y. & Levy, R. M. (2002). The SGB/NP hydration free energy model based on the surface generalized born solvent reaction field and novel nonpolar hydration free energy estimators. J. Comput. Chem. 23, 517–529. 50. Zhu, K., Shirts, M. R., Friesner, R. A. & Jacobson, M. P. (2007). Multiscale optimization of a truncated Newton minimization algorithm and application to proteins and protein–ligand complexes. J. Chem. Theory Comput. 3, 640–648. 51. Wang, J., Wang, W., Kollman, P. A. & Case, D. A. (2006). Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graphics Modell. 25, 247–260. 52. Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. (2004). Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174. 53. Gasteiger, J. & Marsili, M. (1980). Iterative partial equalization of orbital electronegativity: a rapid access to atomic charges. Tetrahedron, 36, 3219–3288. 54. Jakalian, A., Jack, D. B. & Bayly, C. I. (2002). Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 23, 1623–1641. 55. Bayly, C. I., Cieplak, P., Cornell, W. D. & Kollman, P. A. (1993). A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—the RESP model. J. Phys. Chem. 97, 10269–10280. 56. Giammona, D. A. (1984). An Examination of Conformational Flexibility in Porphyrins and Bulky-Ligand Binding in Myoglobin, Ph.D. Dissertation, University of California, Davis. 57. Onufriev, A., Bashford, D. & Case, D. A. (2004). Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins: Struct., Funct., Bioinf. 55, 383–394. 58. Weiser, J., Shenkin, P. S. & Still, W. C. (1999). Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO). J. Comput. Chem. 20, 217–230. 59. Kirchhoff, W. (1993). EXAM: a two-state thermodynamic analysis program, NIST, Gaithersburg, MD. 60. Otwinowski, Z. & Minor, W. (1997). Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology (Carter, C. W. J. & Sweet, R. M., eds), Methods in Enzymology, vol. 276, pp. 307–326. Academic Press, New York. 61. Schuttelkopf, A. W. & van Aalten, D. M. F. (2004). PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60, 1355–1363. 62. Bailey, S. (1994). The CCP4 Suite—programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 50, 760–763. 63. Emsley, P. & Cowtan, K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60, 2126–2132.