Proteins
advertisement

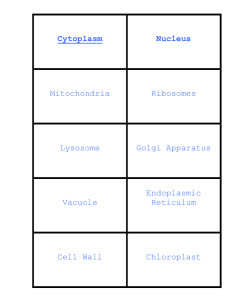
Cells and Their Housekeeping Functions – Nucleus and Other Organelles Shu-Ping Lin, Ph.D. Institute of Biomedical Engineering E-mail: splin@dragon.nchu.edu.tw Website: http://web.nchu.edu.tw/pweb/users/splin/ Chloroplasts, Cell Wall, and Vacuole Chloroplasts: green organelles that make food, found only in green plant cells Convert energy of light into chemical energy Chlorophyl: green pigment that gives leaves & stems their color Captures sunlight energy that is used to produce food called glucose (Glucose is a type of sugar) Cell wall: restrict shape change and mobility Vacuole: collect and store nutrient molecules and waste products Chloroplasts Cell wall www.worldofteaching.com From Cell to Organism Cell The basic unit of life Tissue Group of cells working together Organ Group of tissues working together Organ System Group of organs working together www.worldofteaching.com Organism Any living thing made of 1 or more cells www.worldofteaching.com 1- Nucleus 2- Chromosomes 3- Mitochondria 4- Ribosomes 5- Chloroplasts 6- Vacuoles 7- ER 8- Cell Membrane Nucleus Largest organelle within the cell, containing DNA DNA replication: necessary for cell division (so that both daughter cells have identical copies of DNA) Transcription: formation of an RNA template of a gene, essential for protein synthesis Library of genetic information Bounded by nuclear membrane, 2 lipid bilayers of double membrane are separated by a gap (20~ 40nm) with many openings or pores for trafficking small molecules and proteins Chromosome Identical sets of chromosomes (long and thin DNA molecules, store genetic information) in each eukaryote Histone: beadlike protein structure, Ex: each human cell has about 1.8 meters of DNA, but wound on the histones it has about 90 millimeters of chromatin Before cell division, DNA is replicated and tightly coiled and bound in identical pairs called chromatids Chromosomes can be seen by light microscopy. Chromatin DNA molecules are packed around and attached to beadlike protein structure (histon) Nucleosomes http://en.wikipedia.org/wiki/Chromosome Chromatin Chromosome Different levels of DNA condensation. (1) Single DNA strand. (2) Chromatin strand (DNA with histones). (3) Chromatin during interphase with centromere. (4) Condensed chromatin during prophase. (Two copies of the DNA molecule are now present) (5) Chromosome during metaphase. http://zh.wikipedia.org/zh-tw/File:Chromatin_chromosome.png Protein Synthesis-1 RNA polymerase: transcribe RNA from DNA template at average rate 30 nucleotides/sec, an enzyme consisting of 12 different polypeptides with mass of 500kDa, arrange in 10 subunits RNA polymerase II (pol II): central machine for synthesis protein of mRNA in eukaryotes Polymerase I: ribosomal RNA, polymerase III: transfer RNA 9 out of 10 subunits are identical in these 3 enzymes Begin in nucleus with binding of transcription factors to regulatory sequence (protein-coding gene) Transcription: energyconsuming process and driven by energy-released by ATP hydrolysis Protein Synthesis-2 Binding of transcription factors to regulatory sequence (protein-coding gene) on DNA ––Activate Pol II unwind DNA double helix Polymerize mRNA and proofread the resulting transcript Pol II recognize promoter region of genes if DNA interact with transcription factors Subunits 1, 5, and 9 of pol II grip DNA downstream of active center Subunits 1, 2, and 6 clamp on DNA near active center Growing mRNA strand locks this clamp, thus stabilizing transcribing complexes Complete RNA molecule (primary RNA transcript, pre mRNA): 20,000 nucleotides because of noncoding introns RNA splicing mRNA Transport out of nucleus and interact with ribosomes in cytoplasm to begin translation Spliceosome: recognize exon/intron Excised intron transcripts rapidly interface , mediate excision, and anneal degrade in nucleus to provide raw material ends of exons for new transcripts Ribosomes Small and darkly staining spherical structures: ~20 nm in diameter that are made of 50 proteins and several long RNAs intricately bound together Ribosomes are made in the nucleolus, compartment in nucleus. Once constructed, ribosomes leave nucleus through nuclear pores. Float freely in the cytoplasm to synthesize cytoplasmic proteins without any further modification or attach to the endoplasmic reticulum (ER) to manufacture membrane proteins Make proteins Translate sequence information on mRNA into polypeptides No membrane and disassemble into 2 subunits when not actively synthesizing protein Protein synthesis is extremely important, so eukaryotic cells contain million of ribosomes. Take 30 sec to synthesize a protein containing 400 amino acids, and human cell synthesize 1010 proteins in 24 hr Translation of mRNA by Ribosomes 1. Attachment of mRNA to a ribosome 2. Begin knitting together amino acids, according to template encoded mRNA 3. Translation is initiated by tRNA, acting as adapter and matching each codon on mRNA with amino acid the codon prescribes. tRNAs: amino-acid-specific adapter molecules Ribosome has binding sites for 2 different tRNA molecules so that 2 amino acids joined to the growing polypeptide chain at one time. Endoplasmic Reticulum A series of folded membranes that move materials (proteins) around in a cell – like a conveyer belt Large surface area provides template for enzyme-mediated chemical reactions Ribosomes manufacture membrane proteins or proteins Will be secreted or transported to other membranous organelles – ER Smooth ER – ribosomes not attached to ER; lipid synthesis in the cell (cell’s membranes); houses detoxifying enzymes (particular in liver); early stages of synthesis of steroid hormones – testosterone (contain large amounts of SER in testes, ovaries, and adrenal glands; SER a storage site for calcium in skeletal cells Rough ER – ribosomes attached to ER; protein synthesis, folding, and some posttranslational modifications – addition of carbohydrates (glycosylation), membranebound polypeptides threaded through membrane of RER have 3D shapes ER Information dictates polypeptide is synthesized on free or ER-bound ribosomes mRNA contain primary sequence of gene and does not depend on protein-synthesizing apparatus (ribosome or ER) Proteins are targeted to their specific cellular destinations by address labels. Targeting to RER – Signal sequence consists of about 20 hydrophobic amino acids at the N terminal end of the protein Nuclear proteins synthesized on free ribosomes, tag that directs them to nucleus including stretch or patch of basic amino acids, Ex: transmembrane proteins have multiple signal sequences. Ribosome attached mRNA with a signal-sequence coding sequence is directed to outer surface of RER Protein is processed by enzymes and folded into its correct 3D conformation. Proteins Processed in ER Protein are inserted into lumen of ER as they are being synthesized rather than as fully made polypeptides Protein to be passed into lumen of ER, the end of polypeptide must be identified by a signal-recognition particle (a receptor) and machinery must exist to pull the thread into the lumen (ATP and GTP-driven motors) Protein translocation across ER membrane, pore in hydrophobic lipid membrane presents a hydrophilic interior to accommodate newly synthesized protein. Proteins on the outside of pore recognize polypeptide and ribosome. Ratchet-like mechanism: ER simultaneously bind and pull polypeptide into lumen by utilizing energy driving from the hydrolysis of GTP and ATP Translocated protein enters ER lumen, signal peptide is cleaved by enzyme called signal peptidase. Emerging protein is sequestered by heat shock protein hsp70 and assisted with proper folding, this chaperone can prevent aggregation in ER lumen. Disulfide bonds are rate-limiting step in protein folding in ER. Extensive post-translation modification in secretory proteins is glycosylation: carbohydrate motifs; starts in ER with further processing in Golgi apparatus Golgi Apparatus Following synthesis, small vesicles transport concentrated proteins from ER to another membranous compartment – Golgi apparatus: stacked flattened membranes Lumenal and transmembrane proteins are concentrated in lipid vesicles Golgi complex modifies proteins by adding to and modifying their carbohydrate chains to form mature glycoproteins Golgi vesicles pinch off from edges of flattened sacks to transport the modified proteins their destinations in cell and to its exterior Mechanism of concentration, vesicle formation, and subsequent fusion with Golgi (Process proteins) – Sort and package proteins Pinch off coated vesicles (coatomer proteins or COPs coat vesicle) binding GTP to form G-protein (once release from ER, vesicle is protected and allow to fuse with Golgi) COPs cover naked lipid membrane and some selected proteins target vesicle to correct Golgi stack; others catalyze fusion of vesicle and Golgi membranes (simultaneous uncoating and fusion of vesicle requires hydrolysis of GTP and ATP) Protein Transport in Golgi Apparatus Similar process mediates vesicular transport between different Golgi stacks and between Golgi and plasma membrane. Proteins reside in ER or Golgi selectively retained during vesicle formation or retrieved from distal compartments. ER proteins possess short amino-acid sequence marking them for retention in ER Membrane vesicle involved in retrieval of ER proteins from Golgi apparatus Cisternal maturation model: intra-Golgi transport Cisternal Maturation Model Hypothesis: Vesicles are used only in retrograde transport, to retrieve ER components that pass into Golgi. Cisternal maturation models (changing their molecular makeup) are correct, but the rate is too slow to account for the speed at which many proteins pass through the Golgi stack Large aggregates of extracellular matrix proteins, ex: pro-collagen, requires a lot posttranslational modification and passes through Golgi without entering vesicles and the rate of passage coincides with its in cisternal maturation. Other proteins are concentrated in vesicles and pass rapidly from one stack to another. Final cis-trans directionally of cargo transport being dependent on a sort of iterative progression Small vesicles only contain ER proteins with ER-retrieval label are passing from the Golgi to the ER. Rapid vesicular transport system is superimposed on a more slowly maturing cisternal system. Lysosomes The word "lysosome" is Latin for "kill body". The purpose of the lysosome is to digest things. They might be used to digest food or break down the cell when it dies. Break down food molecules, cell wastes & worn out cell parts They are found in animal cells, while in yeast and plants the same roles are performed by lytic vacuole Cytoskeleton Organelles of eukaryotic cells are not freely suspended in cell cytoplasm, but are anchored to cytoskeleton. Cytoplasm: an aqueous medium containing many enzymes and other compounds needed by cell, also contains dynamic cytoskeletal network including microfilaments, intermediate filaments, and microtubules Cytoskeleton Give cell physical strength and rigidity and hold intracellular structures in place Facilitate and control movement within cell and locomotion of cell Actin Microfilaments Actin cytoskeleton: for cell motility (actin interact with molecular motor myosin leads to contraction of actin cytoskeleton and enables cells to move and change shape) and phagocytosis (macrophages eat microbes), adhesion of cells to other cells and to extracellular matrix (signaling receptors on cell surfaces anchor to actin either directly or through adapter proteins), conserved ancient protein of eukaryotes, and (~20%) the most abundant protein in cytoplasm of mammals Actin microfilaments: 5nm in diameter, including G-actin (globular monomer) and F-actin (filament, formed by head-to-tail polymerization of asymmetric G-actin) Create dynamic (continuously remodeling) and intricate cytoskeletal network and contractile (F-actin associate with myosin(motility engine)) TEM of crosslinked actin cytoskeleton of macrophage Formation of Actin Filament 3 actin monomers Unstable trimeric nucleus: dissociate back into monomers rapidly or survive long enough to permit subsequent binding of additional actin molecules Proteins, such as Arp2/3 complex, regulate nucleation of actin filaments. Treadmilling – Monomeric actin molecules (containing ATP) adding for elongating filament: Monomer rapid addition at “+” Growing filaments, ATP-actin hydrolysis to form likely dissociated ADP-actin within filament’s helical lattice Active filaments reach steady-state length ADP-actin released at “-” at about the same rate as new ATP-actin monomers added to “+” Treadmilling Actin filaments assemble and disassemble very rapidly regulated by actin-binding proteins (α-actinin) Bind to sides of actin filaments, and link them into bundles (Crosslinking creates rigid structures for cell to resist physical forces, Ex: fluid force exerted on endothelial cells covering lumen of blood vessels) Some actinbinding proteins “cap” the “+” ends to prevent depolymerization, and others sequester G-actin to prevent polymerization J. Baum, Nature Reviews Microbiology 4, 2006 Fibrous Actin Fibrous actin: found in muscle cell in association with regulatory proteins tropomyosin and troponin Thin filaments of muscle cells consist of 2 actin strands twisted into double helix together with troponin and tropomyosin. Thin filaments hardly change length during muscle contraction or stretching, but slide past each other to alter the length of muscle cells. Cells suspended in fluid are spherical shape (existence of uniform surface tension); but compose tissues deviate from spherical shape (forces cell-cell and cell-substratum adhesion pull cells in different directions) – Cytochalasin B lose cell shape and resemble a plastic bag Microtubules and Tubulin Microtubules: hollow tubes ~ 20 nm in diameter composed of αtubulin and β-tubulin, each contains 13 strands of protofilaments (polymerized tubulin dimers, α-tubulin and β-tubulin interact head to tail) Protofilaments form a sheet, then fold to form hollow microtubule. Functions: Transport of vesicles and other organelles such as those that pass through Golgi apparatus during protein sorting Components of cell appendages such as flagella used to propel spermatozoa: stable configuration of microtubules, beating motion is from sliding of microtubules along one another) Mitotic spindle during cell division: remarkably dynamic, grow and shrink at the same time as they move chromosomes and segregate daughter cells Microtubules associate with ATP-hydrolyzing motor proteins kinesin and dynein. Centrosome Centrosome: microtubules nucleated and anchored in a region of cell, also called microtubule-organizing center (MTOC) Polarized with “+” Grow from MTOC into cytoplasm Microtubules grow by the reversible addition of subunits catalyzed by GTP-nucleotide hydrolysis. Dynamic instability: “+” end of microtubules switch from rapidly growing state to rapidly shrinking state β-tubulin bound to GTP assembles with other dimers during polymerization GTP hydrolysis after polymerization and “cap” of GTP-bound subunits found at “+” of growing microtubules; size of cap depends on rate of new subunit addition Rate at GTP-bound tubulin formed or added to growing microtubule is reduced, size of GTP cap ↓, and GDP-tubulin is exposed Microtubule catastrophe: GDPtubulins dissociate more readily than GTP-tubulin, microtubule depolymerizes rapidly Enzyme then let GDP-tubulin converts to GTP- tubulin Capture “+” (stable) Implications: cell division, cancer, neurobiology, and cell motility