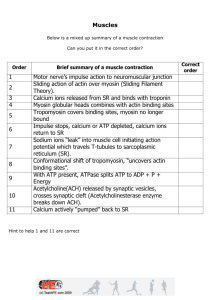
1 Muscle Contraction Proteins convert chemical energy, a scalar
advertisement

1 Muscle Contraction Proteins convert chemical energy, a scalar quantity, to motion, a vector quantity. How? I. The ATPase cycle ATP serves as the fuel for motion by undergoing hydrolysis in the presence of myosin (either whole myosin, the head (S1 region), or heavy meromyosin). The kcat for hydrolysis in this case is 0.1 s-1. This represents a fairly slow hydrolysis rate, and consequently if ATP hydrolysis in the presence of myosin were the only factor governing motion, everyone would move rather slowly. It turns out that adding actin to the myosin, forming actomyosin, increases the kcat of ATP hydrolysis to 10-20 s-1, a 1-200 fold rate increase over myosin alone. As we will see, this increase is due to an increase in the rate of Pi release from myosin. This finding explains the need for the observed cross bridges between actin and myosin. Since both myosin and actomyosin can bind and hydrolyze ATP, and actin can bind to both free myosin and to the myosin.ATP complex, the process of ATP hydrolysis in the presence of actin and myosin occurs as a cycle in which actin can bind to and dissociate from myosin at each different step. Here, M = myosin, A.M = actomyosin, T = ATP, D = ADP, and Pi = inorganic phosphate. All of the K's are given as association constants (even if the step actually involves a dissociation), so that all of the K's have the same units. ∆G°’ is the standard free energy change for the forward reaction, and kf is the rate of the forward reaction. K1=1011M-1 ∆G°': -15.4 kcal/mol kf : 106M-1s-1 K5=~1 0 200s-1 K7=~1 0 20s-1 K9=10 M-1 +1.4 0.1s-1 K11=105M-1 +7 Σ∆G°'= -7 1s-1 D T Pi M <====> M.T <====> M.D.PiR <====> M.D.PiN <====> M.D <====> M A.M <====> A.M.T <====> A.M.D.PiR <====>A.M.D.PiN <====>A. M.D <====>A. M T D Pi K4=107M-1 K6=~1 K8=~1 K10=10-1 M-1 K12=103M-1 ∆G°': -9.8 kcal/mol 0 0 -1.4 +4.2 Σ∆G°'= -7 kf : 106M-1s-1 200s-1 20s-1 100s-1 1000s-1 If we first look at just the top line of the above cycle, where myosin is hydrolyzing ATP in the absence of actin, then we see that ATP has a high affinity for myosin; K1 is a very large number and the associated free energy change is enormous. It is actually in this step (where ATP is binding to myosin) that the free energy to drive the hydrolysis is released. 2 The actual hydrolysis step has a ∆G°’ of 0, and the dissociation of both the Pi and ADP have a positive ∆G°’. However, the sum of the free energy change for the overall reaction has the expected value of -7 kcal/mol. So, the binding of ATP to myosin is rapid and releases a great deal of free energy. The hydrolysis of the terminal P of ATP goes through a SN1 reaction. The Pi that is formed is tightly bound to the myosin head and is initially in a refractory state, PiR. After a conformational change in the myosin head, the Pi enters the nonrefractory, or PiN , state. The overall rate of hydrolysis of ATP by myosin is slowed down by the slow rate of Pi dissociation from the M.D.PiN complex. Thus, in the absence of actin, Pi loss is the ratelimiting step. It occurs at a rate of 0.1 s-1 . (Since the release of Pi is the rate-limiting step, the hydrolysis of ATP by myosin alone shows burst-phase kinetics. See Problem Set 3, Question 2.) Now let us look at the bottom row of reactions: the hydrolysis of ATP by actomyosin. The free energy change associated with the overall reaction is still -7 kcal/mol as expected, but now less of the free energy change is associated with the binding of ATP to actomyosin and more from the release of Pi. The other steps have not changed appreciably. Actomyosin loses Pi rapidly (at a rate of 20s-1 ), so that the overall reaction is no longer limited by the release of Pi but by the conformational change allowing Pi R to become PiN . (Consequently, the hydrolysis of actomyosin will not show burst-phase kinetics.) In muscle, ATP hydrolysis does not occur by simply following across the bottom set of reactions. We must also consider the relative affinity of myosin for actin at each step. It is perhaps easier to consider a simplified form of the above cycle with all of the same steps but with only relative rate constants and binding affinities. A diagrammatic figure is also shown on the next page to help you follow through these steps. fast fast slow T M <====> M.T <====> M.D.PiR High Low Low very slow fast D Pi . . . <====> M D PiN <====> M D <====> M Low High A.M <====> A.M.T <====> A.M.D.PiR <====>A.M.D.PiN <====>A. M.D <====>A. M T D Pi fast fast slow fast fast Since myosin alone has a high affinity for actin, let us begin at A.M (the state seen in rigor mortis). The concentration of ATP is very high in a muscle cell, and so ATP rapidly binds to form A .M .T. However, the affinity of M.T for actin is much lower, and so the myosin will largely dissociate from actin at this point. Now, the M.T will quickly hydrolyze its ATP to give bound ADP and PiR. 3 + - power stroke conf. change ADP back to where we started, but moved one actin over towards Z line fast + D Pi ADP (rigor mortis) Start here: ADP fast productive interaction with actin fast ATP DPiN ATP fast slow rate-limiting conf. change to cock myosin head DPiR non-productive interaction with actin DPiR The M .D.PiR complex still has a low affinity for actin and so may try to form an interaction, but will not be able to form a strong interaction. The conformational change in myosin causing the conversion of PiR to Pi N is what “cocks” the myosin head for the power stroke. This is slow, about 20s-1 . Once it occurs, the M.D.PiN complex still has a low affinity for actin, BUT only when the A.M .D.PiN complex is formed can Pi release occur at a rapid rate. This drives myosin to bind actin even though the relative affinity of myosin for actin is weak. Once Pi is released, however, then a conformational change in myosin can occur which will allow for tight binding. This is the power stroke! The conformational change in myosin involves a change in the relation between the myosin head and tail, corresponding to a 5-10 nm displacement of the lever arm. Actin must be complexed to myosin for muscle movement to occur. After the power stroke, ADP is released to give AM and the cycle can begin again. If you look at V&V p. 1247 Fig. 34-65 and MBOC p.852 Fig. 16-91, you’ll notice that their models are slightly different from the one just discussed. However, the model presented in class is now thought to more accurately reflect what is occurring in the ATP cycle and is the one which you should know. So how close is this model to reality? Three experimental methods have been used to test this model of the ATPase cycle: in vitro motility, analysis of crystal structures, and mutations of the myosin lever arm. 4 II. In vitro motility A motility assay verifies that myosin heads move around on actin filaments. The experiment consists of covering a polystyrene bead with S1 myosin fragments and the adding a fluorescently labeled actin cable. The actin cable moves with respect to the beads at a rate of about 5µm/s, which is fairly fast. However, if the conditions are manipulated such that only one myosin-covered bead contacts a single actin cable, the rate of movement is about 0.2 µm/s. In a more complex version of the experiment, beads are also attached to the ends of an actin cable, and infrared laser beams are used to trap the beads, and therefore the actin cable, in place. Instead of covering the original bead with S1 fragments, a low amount of S1 is used so that only one head attaches to each bead. Similarly, a low concentration of ATP is used so that only one ATP molecule binds to each head. With this set-up, each actin cable is associated with only one myosin-conjugated bead, and the movement of the actin is measured using the actin-conjugated reporter beads. These steps demonstrate that myosin moves 5-10 nm after it interacts with actin. If the laser energy is increased, myosin tries to move the actin (relative to itself), but since the laser energy restrains the actin’s movement, the force associated with the restraint is measured. This force turns out to be 4-8 picoNewtons (pN). So are these force and displacement values consistent with the energy supplied by the hydrolysis of one molecule of ATP? A few calculations prove that they are (this is purely for your interest). First, ∆G = -12 kcal/mole = 16 x 10 -21 cal/molecule for ATP hydrolysis. At the same time, 1 pN x a 10 nm displacement = 2.5 x 10-21 calories. Since the average force observed is 5 pN, the total energy for the 10 nm displacement is 5 x 2.5 X 10-21 calories = 12.5 x 10-21 calories (or 5 x 10-13 ergs), which is indeed approximately the amount of energy supplied by the hydrolysis of a molecule of ATP. (Hydrolysis of a single ATP to ADP and Pi actually releases about 8.3 x 10-13 ergs which would make muscle contraction about 60% efficient. For converting scalar energy to movement, this is a great efficiency rate! Man-made engines are nowhere near this good.) III. Structure of myosin The crystal structure of myosin (see V&V p. 1238 Fig. 34-53) shows the head has a 25 kD domain at the N-terminus, and a 50 kD region that protrudes from and wraps around the 25 kD domain. A cleft between two parts of the 50 kD region forms the ATP-binding site, and actin binds on the other side of the cleft. The essential and the regulatory light chains are attached at the C-terminus, and a lever arm is observed at the region where the light chains are bound. The lever arm is long enough to allow for the observed 10 nm displacement. 5 The structure gives insight into why actin falls off when ATP binds to myosin. At first, the ATP cleft is open, but the Pγ end of ATP is occluded due to the presence of actin at the back of the 50 kD region. Thus, as ATP opens this side of the cleft, the actin falls off. ATP gets trapped from the other side, and hydrolysis of the Pγ occurs. In this state actin can only attach weakly to myosin and comes on and off. The hydrolyzed Pi then leaves through the side of the cleft it opened, and once it’s gone the cleft closes and actin can bind tightly again. This alternation between the closed and open cleft states should remind you of the twostate model of hemoglobin. IV. Mutation of the lever arm Since the lever arm is thought to determine the length of myosin displacement, what happens if the lever arm is shortened or lengthened by DNA mutations? It turns out that the shorter the lever arm, the slower the rate of movement. Similarly, making the arm longer by adding an extra light chain site increases the rate. The effect of changing the lever arm length is independent of ATP hydrolysis, since the rate of ATP hydrolysis is constant among all the mutants. V. Relation of ATP hydrolysis and movement rates to myosin head structure The myosin head has two loop regions, loop 1 which is part of the ATP-binding site and loop 2 which is part of the actin binding site. It has been observed that the rate of ATP hydrolysis varies among species, and it turns out that the explanation for this is the variation in the sequence of loop 2. Thus interchanging loop 2 between animals changes their overall rates of ATP hydrolysis by changing how actin interacts with myosin. Loop 1 mutations, on the other hand, affect the movement velocity since they seem to be involved in the "hand-over-hand" movement of myosin down an actin filament. Loop 1 mutations do not necessarily change the A.M-ATPase activity however. There can also be mutations in the ATP binding cleft that affect the dissociation of tightly bound actinmyosin complex by ATP. (Don’t worry about the “tc” and “ts” that were mentioned in lecture.) These features are important for insuring that ATP hydrolysis doesn’t occur without an intermediate step to prevent actomyosin dissociation, and therefore for insuring that myosin only moves forward.