DNA Replication P. 131-137
advertisement

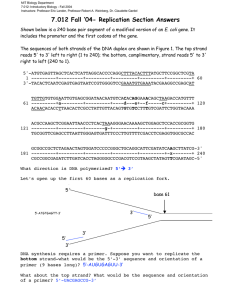
DNA Replication P. 131-137 -precise copying of DNA sequences during DNA replication(Figure) -pairing of bases in d-helical DNA double structure (Watson &Crick): -DNA templates(parental strands)---daughter strands complementary -bp template model theoretically: conservative(2 daughter strands form a new ds (duplex) or a semiconservative mechanism. DNA synthesized from pre existing duplex DNA. 4.6 DNA Polymerases Require a Primer to Initiate Replication DNA synthesized from deoxynucleoside 5_-triphosphate precursors (dNTPs) Synthesis proceeds in 5_n3_direction: - DNA polymerases initiate ; require a short, preexisting RNA or DNA strand(a primer) -adds deoxynucleotides to free hydroxyl group at 3_ end of primer - lagging strand occur in 5_n3_ direction( opposite direction from movement of replication fork) accomplishes by synthesizing a new primer every few hundred bases. - Each of primers, bp to template strand----discontinuous segments =Okazaki fragments(by: Reiji Okazaki ) RNA primer of each Okazaki fragment removed and replaced by dNTPs; - DNA ligase joins the adjacent fragments Helicase, Primase, DNA Polymerases, & Other Proteins Participate in DNA Replication Detailed understanding of eukaryotic proteins participate in DNA replication(e.g SV40 DNA, infects monkeys) These multicomponent When RNA is primer: daughter strand formed RNA at 5_ end & DNA at 3_ end. Duplex DNA Unwound, and Daughter Strands Formed at DNA Replication Fork 2 strands must be unwound to make bases available for bp with bases of dNTPs unwinding of parental DNA strands by helicases, beginning at replication origins(origins) nucleotide sequences of origins from different organisms vary, contain A_T-rich sequences. -RNA polymerase : primase forms a short RNA primer complementary to unwound template strands. -elongated by a DNA polymerase, -----forming a new daughter strand. -DNA region at which all proteins come together to carry out synthesis of daughter strands=replication fork, -Replication proceeds, growing fork & associated proteins move away from origin. -local unwinding of duplex DNA relieved by topoisomerase ---- DNA polymerases to move along and copy a duplex DNA, helicase sequentially unwind duplex & topoisomerase must remove supercoils that form -A major complication in the DNA replication fork: - two strands of parental DNA duplex antiparallel, DNA polymerases - add nucleotides to growing new strands only in the 5_n3_ direction. -Synthesis one daughter strand= leading strand--continuously from a single RNA primer in 5_n3_ direction, - problem comes in synthesis of other daughter strand= lagging strand. Replication of SV40 DNA -Replication of the SV40 DNA: hexamer of a viral protein= large T-antigen unwinds parental strands at a replication fork. -Other proteins involved in SV40 DNA replication= provided by host cell -Primers for leading & lagging daughter-strand DNA: by primase - DNA polymerase _ (Pol _)α---extends RNA primer with dNTPs.(complex of primase- Pol _ α) Primer extended into daughter-strand DNA by DNA polymerase _ (Pol _)δ, which less likely to make errors during copying of template strand than Pol _ α. Pol _ δ forms a complex with Rfc (replication factor C) and PCNA (proliferating cell nuclear antigen -------Displaces primase–Pol _ α complex following primer synthesis. PCNA( homotrimeric protein): has central hole through which daughter duplex DNA passes ----- preventing PCNA-Rfc–Pol _ δ complex from dissociating from template. -After parental DNA separated into ss templates at replication fork----bound by multiple copies of RPA (replication protein A): heterotrimeric protein -Binding of RPA maintains template in a uniform conformation optimal for copying by DNA polymerases. -Bound RPA proteins dislodged from parental strands by Pol _ δ and Pol _ α as they synthesize complementary strands bp parental strands. -Topoisomerase associates with parental DNA ahead of helicase to remove stress introduced by unwinding of parental strands. -Ribonuclease H and FEN I remove RNA at 5_ ends of Okazaki fragments--replaced by dNTPs added by DNA polymerase _ δ as extends upstream Okazaki fragment. Successive Okazaki fragments coupled by DNA ligase through standard 5_n3_ phosphoester bonds. DNA Replication Generally Occurs Bidirectionally from Each Origin - In theory, DNA replication from a single origin could involve one replication fork moves in one direction. Alternatively, two replication forks might assemble at a single origin and then move in opposite directions, --- bidirectional growth of both daughter strands. All prokaryotic and eukaryotic cells employ a bidirectional mechanism of DNA replication e.g-SV40 DNA, replication initiated by binding of two large T-antigen hexameric helicases to single SV40 origin and assembly of other proteins to form two replication forks ----- move away from the SV40 origin in opposite directions with leading- and lagging-strand synthesis occurring at both forks -left replication fork extends DNA synthesis in leftward direction; right replication fork extends DNA synthesis in rightward direction Unlike SV40 DNA, eukaryotic chromosomal DNA molecules contain multiple replication origins separated by tens to hundreds of kilobases six-subunit protein called ORC,for origin recognition complex, binds to each origin and associates with six homologous MCM proteins required to load cellular hexameric helicases. -Two opposed MCM helicases separate the parental strands at origin, with RPA proteins binding to the resulting ss DNA. - Synthesis of primers and subsequent steps in replication of cellular DNA thought to be analogous to those in SV40 DNA replication