Identification of Two Unknown Bacteria in Mixed Culture
advertisement
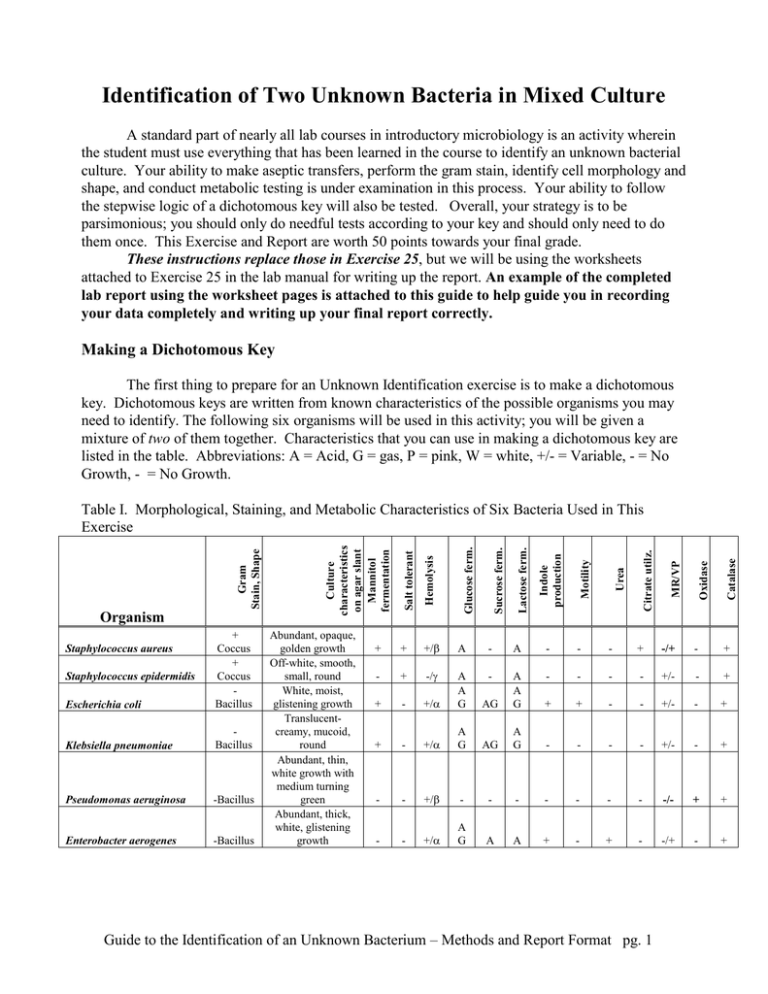
Identification of Two Unknown Bacteria in Mixed Culture A standard part of nearly all lab courses in introductory microbiology is an activity wherein the student must use everything that has been learned in the course to identify an unknown bacterial culture. Your ability to make aseptic transfers, perform the gram stain, identify cell morphology and shape, and conduct metabolic testing is under examination in this process. Your ability to follow the stepwise logic of a dichotomous key will also be tested. Overall, your strategy is to be parsimonious; you should only do needful tests according to your key and should only need to do them once. This Exercise and Report are worth 50 points towards your final grade. These instructions replace those in Exercise 25, but we will be using the worksheets attached to Exercise 25 in the lab manual for writing up the report. An example of the completed lab report using the worksheet pages is attached to this guide to help guide you in recording your data completely and writing up your final report correctly. Making a Dichotomous Key The first thing to prepare for an Unknown Identification exercise is to make a dichotomous key. Dichotomous keys are written from known characteristics of the possible organisms you may need to identify. The following six organisms will be used in this activity; you will be given a mixture of two of them together. Characteristics that you can use in making a dichotomous key are listed in the table. Abbreviations: A = Acid, G = gas, P = pink, W = white, +/- = Variable, - = No Growth, - = No Growth. -Bacillus Enterobacter aerogenes -Bacillus Citrate utilz. Lactose ferm. Sucrose ferm. Glucose ferm. +/ A - A - - - + -/+ - + - + -/ - - - - +/- - + - +/ AG A A G - + A A G + + - - +/- - + + - +/ A G AG A G - - - - +/- - + - - +/ - - - - - - - -/- + + - - +/ A G A A + - + - -/+ - + Urea Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 1 MR/VP + Motility + Indole production Catalase Pseudomonas aeruginosa Oxidase Bacillus Hemolysis Klebsiella pneumoniae Staphylococcus epidermidis Abundant, opaque, golden growth Off-white, smooth, small, round White, moist, glistening growth Translucentcreamy, mucoid, round Abundant, thin, white growth with medium turning green Abundant, thick, white, glistening growth Salt tolerant Escherichia coli + Coccus + Coccus Bacillus Staphylococcus aureus Culture characteristics on agar slant Mannitol fermentation Organism Gram Stain, Shape Table I. Morphological, Staining, and Metabolic Characteristics of Six Bacteria Used in This Exercise Dichotomous keys are charts that require decisions at branch points, much like a flow chart in computer logic: If the answer to a question is yes, then do X; if the answer is no, then do Y. Your job is to make the shortest key, with the shortest pathways, to a final identification of an organism. Ideally, any pathway in your key should have no more than four steps (including gram staining and cell morphology as separate steps) leading to an identification. To make a dichotomous key, you first need to sort all the different species in your list into two big groups based on common characteristics between them. Most microbiologists begin with gram stain results, and I encourage you to do the same. Using the Example Dichotomous Key in this Guide, you will see that you should begin your own key by writing out the names of all the possible bacteria in the test near the top of the page. I used 8 bacterial names in my Example Key; you should use the 6 organism names from Table I. I recommend using landscape (sideways) mode in laying out a key since the wider the page, the better. After listing all 6 potential species on your key sheet, you should next draw two arrows downwards that show this group being separated by the gram stain. Write gram positive at the end of one arrow and gram negative after the other. Follow each result with a list of bacteria still remaining in each group. In my Example Key, I was able to divide my list of 8 bacteria into two groups of four. The next dichotomy to further separate the gram positives from each other, and the gram negatives from each other, should be one of salt tolerance, lactose fermentation, or cell morphology (bacilli or cocci) since you will get results of these tests from the selective media (MacConkey and mannitol salt agar) you will use early in the exercise. The important thing is that you are subdividing each group at every dichotomy into two subgroups of nearly equal size. If you separate off only one species at a time at each dichotomy, your key will be far too long and will not be parsimonious. In my example key, I decided to separate the gram negatives using galactose fermentation. I decided on this test after discovering that the four gram negative bacteria differed in their fermentation reactions to galactose. Half fermented the sugar and half did not. That's the strategy you should employ at each step in the key: find a characteristic that differs in the group of organisms you need to separate. Ideally this characteristic or test divides them into roughly equal subgroups. You continue finding tests that subdivide those groups until you only have one organism left per group. You will need a completed dichotomous key by the first period of the Unknown ID activity. The instructor will not let you progress further unless you can demonstrate that you have a completed and correct dichotomous key to work from. Ultimately you will handwrite or type a copy of your dichotomous key and (if typed) paste it onto the blank page of your lab manual (pg. 210). Try, if you can, to get it all on one page. Use the Example Key on the next pages an example. Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 2 Here's the list of 8 bacteria and their characteristics that I used in making my Example Dichotomous Key of Eight Bacteria on the following page. You are not using these bacteria in Ex 25; these are just for demonstration purposes. Table II. Characteristics of 8 Example Bacteria Used to Make a Dichotomous Key Organism Characteristics Alcaligenes faecalis Gram negative bacillus, acid-fast negative, ferments galactose, catalase positive, motile, alkaline with litmus milk, not hemolytic, H2S negative Citrobacter freundii Gram negative bacillus; acid-fast negative, does not ferment galactose, catalase positive, non-motile, acid produced in litmus milk, not hemolytic, H2S negative Pseudomonas fluorescens Gram negative bacillus, acid-fast negative, does not ferment galactose, catalase positive, non-motile, alkaline in litmus milk, hemolytic, H2S negative Streptococcus pneumoniae Gram positive oval coccus found as streptococci, acid-fast negative, galactose fermentation negative, catalase negative, non-motile, acid in litmus milk, hemolytic, H2S negative Mycobacterium smegmatis Gram positive club-shaped bacilli, acid-fast positive, galactose fermentation negative, catalase-negative,, non-motile, alkaline in litmus milk, catalase positive, not hemolytic, H2S negative Mycobacterium leprae Gram positive club-shaped bacilli, acid-fast positive, galactose fermentation negative, catalase-positive, non-motile, alkaline in litmus milk not hemolytic, H2S negative Corynebacterium xerosis Gram positive club-shaped bacilli, acid-fast negative, galactose fermentation positive, non-motile, acid from litmus milk, catalase positive, not hemolytic, H2S negative Aeromonas hydrophila Gram negative bacilli, acid-fast negative, ferments galactose, catalase positive, motile, acid from litmus milk, not hemolytic, H2S positive Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 3 Be sure to list of all the bacteria that remain at each step of the process. At the start, you list all the bacteria being considered. Use the name of the actual test or molecule being detected (lipid hydrolysis), rather than using the name of the medium (e.g. mannitol fat medium) Example Dichotomous Key to Identifying Eight Bacteria Alcaligenes faecalis Citrobacter freundii Pseudomonas fluorescens Streptococcus pneumoniae Gram stain negative positive Alcaligenes faecalis Citrobacter freundii Pseudomonas fluorescens Aeromonas hydrophila Mycobacterium smegmatis Mycobacterium leprae Corynebacterium xerosis Streptococcus pneumoniae Acid-fast staining Galactose fermentation positive acid fast negative Alcaligenes faecalis Citrobacter freundii Aeromonas hydrophila Pseudomonas fluorescens H2S production positive Aeromonas hydrophila You will be working with nine bacteria, as listed in Exercise 25 of your lab book. This example key was made up using 8 bacteria Mycobacterium smegmatis Mycobacterium leprae Corynebacterium xerosis Aeromonas hydrophila negative Litmus milk acid Citrobacter Alcaligenes freundii faecalis alkaline Pseudomonas fluorescens not acid fast Mycobacterium smegmatis Mycobacterium leprae Catalase produced positive Mycobacterium leprae negative Hemolysis positive Mycobacterium Streptococcus smegmatis pneumoniae Figure 1. An Example Dichotomous Key Using Characteristics From Table II. Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 4 Corynebacterium xerosis Streptococcus pneumoniae negative Corynebacterium xerosis Getting Started on the First Day (Instructions replace those in Ex. 25 in the Lab Manual) On the first day: 1. Pick up an assigned bacterial culture from the instructor. The culture may be provided as a broth or an agar slant. You should ask whether the culture is < 24 hours old as this will determine whether or not you can perform a reliable gram stain (re-read the Gram Staining test in your lab book if you don’t understand why). You should see a mixed culture with one coccus species and one bacillus species. Record your observations on pg. 211, including the shape, arrangement, and gram stain reaction. Include the color of your gram stained cells too. 2. Write down in your lab book the number of your assigned culture. It is very important that you do not lose this number, for it is exclusively yours. 3. Next, inoculate TS agar, MacConkey agar, and mannitol salt agar plates from your mixed culture, with proper streaking technique to get isolated colonies. Incubate these at a temperature and a time period consistent with mesophiles that we've grown before during this lab. Part of your final score will be based on your ability to correctly streak a plate and get isolated colonies. 4. Next, perform a gram stain on your culture at your earliest opportunity. You may need to transfer and grow the culture on a TSA plate for the appropriate amount of time before it is ready for gram staining. You may label your original culture (usual labeling rules) and place it in the 10oC incubator (refrigerator) so you have a backup of your culture. Write the results of your gram stain at the beginning of the worksheets for Ex 25 in the appropriate space for "gram stain of original culture. 5. All plates, tubes, or other media that are to be placed in an incubator or the refrigerator must minimally have your full name, your culture number, and your lab section (morning, afternoon, evening). You should also write the date of inoculation on all media so you know when you started the test. I’ll leave it to you to keep track of what medium is in the tube! 6. As part of your report, you will be keeping a handwritten log or journal of everything you do in regards to your culture, including inoculation and incubation times and dates, reagents added, colors seen, interpretations made, etc. Start writing down what you do on the first day in a notebook that can be kept in a safe place; you cannot lose your journal. 7. Be sure you show appropriate actions in cleaning up your lab space and in working safely while in the lab. I will be looking out for correct and safe placement of the Bunsen burners and incinerators, appropriate placement of notes versus cultures, whether you keep your chair pushed in, hair tied back (if appropriate), books and packs underneath, etc. Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 5 The Second Lab Period 1. Interpret the results of your TSA, MacConkey, and mannitol salt agar plates and record the data (colony color, morphology) on pg. 211. Did you get separation on the TSA? What type of organisms should grow on MacConkey? What else can you learn from colony color on MacConkey? Write down your results. 2. Gram stain a colony (note it's morphology!) on MacConkey and write down your results on pg. 211. Designate this as Organism #1. Is the gram stain consistent with what you expected on MacConkey? What morphological information or metabolic ability has been tested on MacConkey? Use your answers to these questions by recording your data in the table on pg. 211. Include both color changes (if any) and the interpretation of this result. 3. Use your answers to these questions by recording your data in the table on pg. 211. Gram stain a colony from the mannitol salt agar, and call this Organism #2 . Is the gram stain consistent with what you expected on MSA? What morphological information or metabolic ability has been tested on MSA? Use your answers to these questions by recording your data in the table on pg. 211. Include both color changes (if any) and the interpretation of this result. 4. Subculture a colony of Organism # 1 and #2 a TS slant to grow pure cultures for next time. What would be a good temperature and time for your organisms? 5. Look at your dichotomous key and determine what remaining tests you need to perform to complete the identification. If you haven't already shown your instructor your completed and corrected dichotomous key, do so early in this period. You will be asked to defend your choices of tests based on what's in your key. You must do this before you are allowed to proceed with futher testing. 6. Are you keeping careful records of the date and everything you see and do? I hope so, for you'll need this detailed information to write up your final rationale for how you identified your bacteria. 7. Inoculate all the media that you need in order to complete the identification for next time. Find the procedures for how to initiate and interpret each test from your lab manual. Remember, you can't ask your lab partner (that is considered cheating, which comes with grade penalties). 8. Turn in your TSA streak plate for grading. Be sure it is properly labeled and includes your culture number. Third Lab Period 1. Interpret the tests you have set up and do any remaining "on-the-spot" tests like oxidase, catalase, gram staining, etc. on your fresh colonies on TS agar slants. Write down all your data in the appropriate spaces on pg. 211 and 212. Don't forget to include color changes as well as interpretation. Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 6 2. Complete the identification of your organism. Begin writing up your report on pg. 213 using the following example report as your guide. Remember to paste in or write in your dichotomous key on pg. 210 AND show the pathway you took to reach an identification (See example lab report following). How I will grade your Unknown ID Exercise: UNKNOWN REPORT GRADING RUBRIC Student: Possible Your GRADED ITEM Pts Points Dichotomous Key Flow Chart: - Is the chart parsimonious with four or less steps needed to reach the IDs? - Are all steps true dichotomies and not trichotomies? - Did you use only the characteristics for the 9 bacteria that are listed in your lab manual for this exercise? - Are all nine bacteria separated out? - Is path you took for Organisms #1 and #2 clearly marked on your key? - Did you have the key ready by the start of the third lab period? - Was the key draw onto or pasted onto blank pg. 210 in your lab manual? 10 Gram Stain and Cell Morphology, Organism assignments - Did you fully describe the shape, arrangement, and gram color and interpretation for both organisms? - Was this information consistent with the assigned organisms (teacher grades this) Streak Plate (TSA) - Graded by instructor - Did student achieve isolation of colonies, especially of both types? - Was the technique faulty or beyond the control of the student? Experimental Results ~ For every test you did, is there BOTH visual AND interpretive Information for each test result? - Did you include the media used in each test, along with the temperature and time of incubation? - Did you include any problems you encountered along the way? ~ Did you follow your key and only do the tests your key said to do? - Were results correctly interpreted? (Teacher grades this) Correct ID ~ Was the unknown correctly identified? Verified by instructor? ~ If unknown ID incorrect, was it the result of student error (technique or judgment) or a factor out of the student's control? Final Report on pg. 213 & 214 - Did you neatly write or type and paste in your final report on these pages? ~ Did you include dates, media used, and results (positive or negative) for each test you inoculated or interpreted? ~ Did you make appropriate conclusions based on your key and results? ~ Has you identified any errors in technique or judgment and suggested alternatives for future work? ~ Is report written in a professional and scientific manner? Neatness, spelling, grammar, nomenclature ~ Is report written in professional, objective manner--no personal pronouns ~ Have all scientific names been written using proper nomenclature? ~ Is grammar and spelling proper and accurate? ~ Is report prepared in a tidy fashion according to requirements? 5 5 10 5 10 5 If lab book submitted late (assign -20% per day to this report) Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 7 TOTAL 50 Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 8 Dichotomous Key to Identifying Eight Bacteria NOTES: Alcaligenes faecalis Mycobacterium smegmatis Citrobacter freundii Mycobacterium leprae Pseudomonas fluorescens Corynebacterium xerosis Streptococcus pneumoniae Aeromonas hydrophila Gram stain negative Alcaligenes faecalis Citrobacter freundii Pseudomonas fluorescens Aeromonas hydrophila Mycobacterium smegmatis Mycobacterium leprae Corynebacterium xerosis Streptococcus pneumoniae Acid-fast staining Galactose fermentation positive Alcaligenes faecalis Aeromonas hydrophila H2S production positive Aeromonas hydrophila negative Citrobacter freundii Pseudomonas fluorescens Litmus milk Citrobacter Alcaligenes freundii faecalis Organism #2 acid fast negative acid Don't forget to highlight he exact pathway you followed in your key to reach an identification of BOTH your organisms. positive alkaline Pseudomonas fluorescens not acid fast Mycobacterium smegmatis Mycobacterium leprae Catalase produced positive Mycobacterium leprae negative Corynebacterium xerosis Streptococcus pneumoniae Hemolysis positive Mycobacterium Streptococcus smegmatis pneumoniae The name of only one organism should be found at the end of each pathway. Be sure this scientific name is spelled correctly and written in proper scientific format. Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 9 negative Corynebacterium xerosis Organism #1 Name Philo Microbius Partner leave blank EXERCISE Date 4/25/14 Unknown # 25 16 Section MW 7:30-9:20 pm Source leave blank Be sure to describe the color, Laboratory Report: Worksheet Final Report: shape and and arrangement that you actually see along with your Clinical Unknown Identification interpretation Gram stain of original specimen: Two organisms were seen, a purple (gram positive) bacillus in chains (concobacillus) arrangement and a pink (gram negative) coccus in groups of 6 (hexacoccal). (describe cell shape, arrangement, and Gram reaction) Gram stains of TSA plate subcultures: Organism #1 was a purple (gram positive) bacillus in streptobacillus arrangement and Organism #2 was a purple (gram positive) coccus in staphylococcal arrangement. Note: I made a mistake in gram staining and describing the cocci on the original specimen. (describe cell shape, arrangement, and Gram reaction) Organism #1 Organism #2 Colony description Test White, round, entire margin, convex (TS agar or blood?) Gram stain Viewed on TSA Purple (gram positive) of 24 hr old colonies grown at 37oC Yellow, irregular, lobate margin, flat Viewed on TSA Purple (gram positive of 86 hr old colonies grown at 37oC Colony appearance on MacConkey No growth after 24 hrs at 37oC Dark purple with metallic sheen after 24 hrs at 37oC Colony appearance on mannitol salt agar (MSA) Pink tinged whitish colonies, round, entire, and elevated after 24 hrs at 37oC Yellow, irregular, lobate margin, convex after 24 hrs at 37oC Did not perform Did not perform Did not perform Did not perform Positive (blue, acid from on MacConkey agar, 37oC, 24 hrs) Negative XX: Not turbid, no gas, yellow in glucose PR ferm. tube. Positive AG: turbid and gas, yellow color in sucrose PR ferm. tube. Did not perform Negative turbid, gas in tube Be(no sure to describe the Special stains capsule endospore These LactoseWarning: fermentation usingresults, media X including the color change and reactions are only examples Glucose(not fermentation media correct!)using -- copy theX format of these examples. colorusing or physical SucroseInclude fermentation media X change relative to control, what this means (positive or Mannitolnegative), fermentation using media X what medium was used, and for how long and at what temp. it was incubated. 25-6 Exercise 25 Clinical Unknown Identification color, or physical characteristics that you actually see along with your Did not perform interpretation. Negative XX: Not turbid, no gas, yellow in sucrose PR ferm. tube. Positive: AG: Turbid with lots of gas using mannitol PR ferm. tube 211 Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 10 Test Organism #1 Organism #2 Did not perform Did not perform Indole production Methyl red Voges-Proskauer Citrate utilization Urea hydrolysis Motility Catalase test Coagulase Oxidase Remained deep purple (non change (negative for ) after adding methyl red to culture in BBSF broth, 24 hrs at 37oC Changed from yellow to bright blue (positive for esculitin) after adding VP reagents to culture grown in BBSF broth, 24 hrs at 37oC Remained pink, no change (negative for calbutrin) using supbacter media grown in BBSF broth, 24 hrs at 37oC Did not perform No flagella visible (negative) after staining with colortail dye; taken from culture in thioglycollate, 72 hrs, at 25oC Bubbles appeared after adding H2SO4 (positive for cats) to a sample 25 hrs old cultured in TS broth Did not perform Negative XX: Not turbid, no gas, yellow in glucose PR ferm tube. Warning: These results, Changed from deep purple to hot including the color change pink (positive) after adding methyl and reactions are only red to culture in BBSF broth, 24 o hrs at 37 C examples (not correct!) -Remained yellow copy the (negative format offorthese esculitin) after adding VP reagents examples. Include color or to culture grown in BBSF broth, o physical change relative to 24 hrs at 37 C control, what this means Change from pink toorgreen, with what (positive negative), gas (positive for calbutrin) using medium was used, and for supbacter media grown in BBSF how at what temp. broth, 24long hrs atand 37oC was incubated. Did not perform it Warning: These results, 4-6 flagella visiblethe percolor cell change including (positive) after staining with and reactions only colortail dye; taken fromare culture examples (notatcorrect!) -in thioglycollate, 72 hrs, 25oC copy the format of these No bubbles appeared after adding H2SO4 (negative forInclude cats) to acolor or examples. sample 25 hrs old cultured in TS physical change relative to broth control, what this means (positive or negative), what Did not perform medium was used, and for how long and at what temp. it Positive AX: turbid but no gas, was incubated. red color in glucose PR ferm tube. Consult your dichotomous key Consult the table compiled from class results at the end of the second session and figure 23.1 to help you identify your organisms. (made from the table of organism characteristics) to identify your organism. Final Identification Organism #1 Organism #2 212 Please change this. You should be using your dichotomous key Clostridium difficule to direct you at each step as to what test to perform (look up what media you should use and how to interpret it in your lab Yersinia pestis book) This exercise is open book, open notes. You just can't 25-6 Exercise 25consult Clinicalother Unknown Identification students in the lab. You can always ask me if you need help on something. Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 11 Final Report 1. What is the identification of your organism? Write out step-by-step the tests you did and why you did them (support this with what your key indicates should be done next). Include the outcome (positive or negative) of each test. Finish with the name of your organism. Discuss the process of identification (reasons for choosing specific tests, any problems, and other comments). Organism #1: Please change this. You can number each of your steps. Be sure to include lots of detail. It is better to go overboard and be too detailed rather than too general. I won't fault you for being too detailed. 1. On the first day, 4/20/12, I subcultured double unknown # 16 onto a TS agar plate and grew it for 48 hrs at 37. I streaked this plate with the idea of isolating the two species from each other. Next, I gram stained the broth culture we were given and discovered two species, a gram-positive coccus (staphylococcal arrangement) and a gram-negative bacillus (single bacilli arrangement). Finally, I inoculated two additional selective /differential plates and streaked for isolation: MacConkey agar and Mannitol salt agar. 2. On 4/22/12 I found that I had good isolation of two different colony morphologies. The yellow, convex colony with an entire margin and round shape on TSA I designated as Organism #1. I gram stained #1 and found gram-positive cocci in staphyococcal. arrangement. A gram-positive organism also grew on EMB agar (producing green colonies, meaning it was galactose positive), but it did not grow on MacConkey. This result is consistent with the notion that gram-positive colonies should grow on EMB, and that galactose fermenters should produce green colonies. I subcultured a colony of #1 from EMB onto a TS agar slant to grow for next time at 37oC for 48 hrs. My dichotomous key indicates that for a gram-positive coccus, I need to do the oxidase test next. 3. On 4/24/12 I performed the oxidase test on Organism #1, using colonies on my younger, TS agar slant culture. The result was positive. Since I was able to do this during class time, I was able to move to the next test in my key, which is for fermentation products from glucose (MRVP). I asked the instructor for an MRVP broth and inoculated it from the TS slant for next time. 4. On 4/28/12 I performed the MR and VP tests according to the lab manual procedure and discovered it to be MR positive but VP negative. I was surprised at the negative VP test since my key says all bacteria at this point should be VP positive. I asked permission to do the VP test again, and was given a "virtual" result of VP negative from the instructor. So, given a You should keep a log positive MR and negative VP, my organism should be Proteus vulgaris. or journal of all the things you do each day Organism #2 (in addition to test 1. On the first day, 4/20/12, I subcultured double unknown # 16 onto a TS agar plate and grew it for 48 hrsin at the 37. tables I streaked results on this plate with the idea of isolating the two species from each other. Next, I gram stained the broth culture we were given previous pages. You and discovered two species, a gram-positive coccus (staphylococcal arrangement) and a gram-negative bacillus (single write up your bacilli arrangement). Finally, I inoculated two additional selective /differential plates and streakedcan for then isolation: step-by-step process on MacConkey agar and Mannitol salt agar. pg. 213 and 214, or type 2. On 4/22/12 I saw a different type of colony (blue with entire margins) on my TSA, which presumably wasand the glue gram-them them up negative bacterium I saw on the first day. I designate this as Organism #2. I gram stained this newer, younger culture but into your lab book on surprisingly found it to be a gram-positive bacillus in single arrangement. The difference in the cell morphology of Be sure Organism #2 compared to the first day may be due to the older age of the cultures given to us. Oldthese cells pages. produce unreliable they are formatted to fit gram stain results. A gram-positive organism also grew on EMB agar (producing yellow colonies, meaning onto it was these galactose negative), pages! but it did not grow on MacConkey. This result is consistent with the notion that gram-positive colonies should grow on EMB, and that galactose non-fermenters should produce yellow colonies. My dichotomous key indicates that for a gram-positive bacillus, I need to do the urease test next. 25-6 Exercise 25 Clinical Unknown Identification 213 Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 12 Organism #2, continued 3. 4/24/12. For Organism #2, I need to do the sorbitol test next. I went to the instructor and asked for a sorbitol fermentation tube with phenol red. I inoculated this for next time. 4. On 4/28/12 I learned that Organism #2 could produce both acid and gas from sorbitol; it was positive for sorbitol fermentation. My next step in the key is to an esculiten production test. I went to the instructor and asked for mebatunin media and inoculated this broth for next time. 5. On 4/30/12 I learned that Organism #2 was positive for esculiten production. According to my key, this means this is Yersenia pestis. 214 25-6 Exercise 25 Clinical Unknown Identification Guide to the Identification of an Unknown Bacterium – Methods and Report Format pg. 13





