Chapter 6 Molecular Biology of DNA Replication and
advertisement

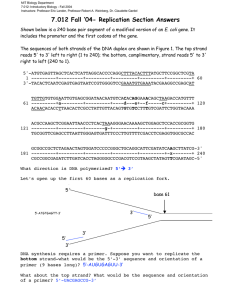
Chapter 6 Molecular Biology of DNA Replication and Recombination Jones and Bartlett Publishers © 2005 Replication of DNA by strand separation and copying of the 2 template strands using A-T and G-C base pairing (semiconservative replication) Experimental proof of semi-conservative replication of DNA Use of a thymidine analog (BUdR) provides cytological proof that DNA in chromosomes also replicates semi-conservatively Replication of a circular DNA molecule through a qstructure Replication can be uni- or bi-directional During q-form replication, both parental DNA strands remain intact. DNA replication begins at special sequences called “origins of replication”. A single DNA molecule may have one (E. coli chromosome) or many origins (a human chromosome can have more than a thousand origins). Replication of a circular DNA molecule by a “rolling circle” mechanism During rolling circle replication, one of the template DNA strands is cut to create a primer 3’-OH end. For every round of replication, the tail of the rolling circle becomes one unit longer One strand of the rolling circle grows continuously while the other is made in small pieces (Okazaki fragments). Replication of a linear eukaryotic chromosome Model of an E. coli DNA replication fork showing the many proteins that play a role there Prevention of knotting of DNA (as strands are separated) by DNA gyrase working ahead of the DNA replication fork The components that are different in RNA relative to DNA Priming of DNA synthesis with and RNA segment New DNA chains are initiated by short RNA primers Addition of a deoxynucleotide to the 3’-OH end of a primer chain A misinserted deoxynucleotide is excised by the proofreading exonuclease function of DNA polymerase One side of the fork grows continuously (leading side) while the other side grows by making small DNA pieces (lagging side) Sequence of events in the joining of adjacent precursor fragments in eukaryotes Addition of a dideoxynucleotide to the 3’-OH end of a DNA chain terminates chain elongation The chain termination method of DNA sequencing DNA sequencing machines use fluorescent dideoxynucleotides A “shotgun sequencing” approach for sequencing of a large DNA molecule Analogs of Normal deoxynucleotides useful as anti-AIDS agents Excision repair of a mismatched base pair Variations from normal 2:2 segregation (gene conversion) can result from mismatch repair Symmetric model of homologous recombination showing some of the intermediate steps and structures Note the 2 nicks in the same location in the two duplexes in step (B). This is followed by strand exchange and Holliday junction formation (C) An asymmetric model of homologous recombination Recombination is initiated by a cut in only one of the 2 recombining duplexes (A) followed by strand invasion (B) and Holliday junction formation (C) A recombining molecule showing the parental and the recombinant duplexes joined at the Holliday junction Resolution of a Holliday junction can occur by 2 alternative cleavage pathways Model for the recombinational repair of a double strand break in DNA