1 2 Supplementary File Table 1. Primers and probes used in this study
advertisement
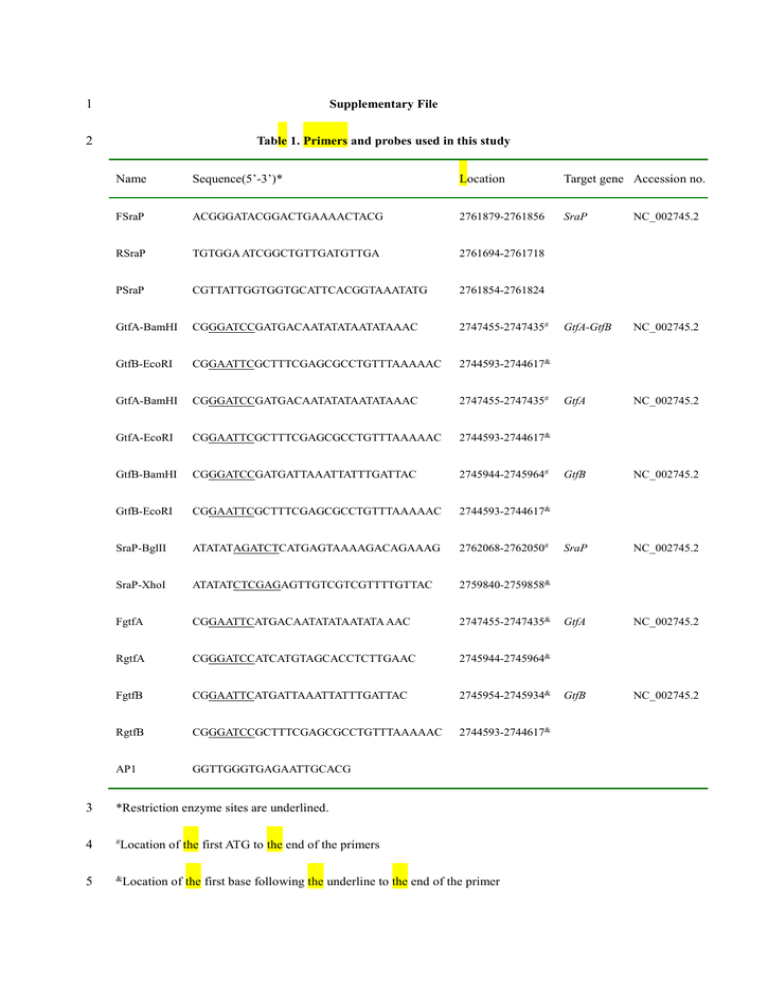
1 Supplementary File 2 Table 1. Primers and probes used in this study Name Sequence(5’-3’)* Location Target gene Accession no. FSraP ACGGGATACGGACTGAAAACTACG 2761879-2761856 SraP NC_002745.2 RSraP TGTGGA ATCGGCTGTTGATGTTGA 2761694-2761718 PSraP CGTTATTGGTGGTGCATTCACGGTAAATATG 2761854-2761824 GtfA-BamHI CGGGATCCGATGACAATATATAATATAAAC 2747455-2747435# GtfA-GtfB NC_002745.2 GtfB-EcoRI CGGAATTCGCTTTCGAGCGCCTGTTTAAAAAC 2744593-2744617& GtfA-BamHI CGGGATCCGATGACAATATATAATATAAAC 2747455-2747435# GtfA NC_002745.2 GtfA-EcoRI CGGAATTCGCTTTCGAGCGCCTGTTTAAAAAC 2744593-2744617& GtfB-BamHI CGGGATCCGATGATTAAATTATTTGATTAC 2745944-2745964# GtfB NC_002745.2 GtfB-EcoRI CGGAATTCGCTTTCGAGCGCCTGTTTAAAAAC 2744593-2744617& SraP-BglII ATATATAGATCTCATGAGTAAAAGACAGAAAG 2762068-2762050# SraP NC_002745.2 SraP-XhoI ATATATCTCGAGAGTTGTCGTCGTTTTGTTAC 2759840-2759858& FgtfA CGGAATTCATGACAATATATAATATA AAC 2747455-2747435& GtfA NC_002745.2 RgtfA CGGGATCCATCATGTAGCACCTCTTGAAC 2745944-2745964& FgtfB CGGAATTCATGATTAAATTATTTGATTAC 2745954-2745934& GtfB NC_002745.2 RgtfB CGGGATCCGCTTTCGAGCGCCTGTTTAAAAAC 2744593-2744617& AP1 GGTTGGGTGAGAATTGCACG 3 *Restriction enzyme sites are underlined. 4 # 5 & Location of the first ATG to the end of the primers Location of the first base following the underline to the end of the primer 1 Fig.1. (A) Schematic representation of SraP. The putative functional domains are delineated within the 2 individual proteins. sp, signal peptide; cw, cell wall-anchoring domain; NR, non-repeat region; R1 and 3 R2, repeat regions 1 and 2. The SraP1-743 expressed in E. coli is indicated below the full-length protein. 4 a.a., amino acids. (B) Schematic representation of the SraP operon. (C) Schematic representation of the 5 conserved domains of GtfA and GtfB SP R1 NR R2 CW SraP A SraP1-743 743aa SraP SecY2 Asp1 Asp2 Asp3 SecA2 GtfA GtfB B GtfA C GtfB 6 7 8 9 10 11 12 13 14 15 1 Fig. 2. Evolutionary relationships of the putative glycosyltransferases GtfA (A) and GtfB(B). The 2 phylogenetic analysis was performed using MEGA4. The length of the branches reflects the number of 3 nucleotide acid changes of GtfA and GtfB between different clusters, as indicated by the bar. Bootstrap 4 values are shown at the nodes. Glycosyltransferase sequences of S. staphylococcus N315 were obtained 5 from the GenBank database. 6 7 8 9 10 11 12 13 (A) 1 2 (B)