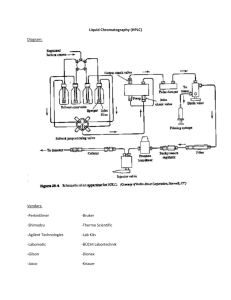
Supplementary materials for manuscript :
advertisement

Supplementary materials for manuscript : Metabolomic analysis reveals distinct profiles in the plasma and urine of rats fed a high protein diet Chunlong Mu1, Yuxiang Yang1, Zhen Luo, Weiyun Zhu* Laboratory of Gastrointestinal Microbiology, College of Animal Science and Technology, Nanjing Agricultural University, Nanjing 210095, China Corresponding Author * Corresponding Author: Weiyun Zhu, Phone: +86 25 84395523. Fax: +86 25 84395314. E-mail: zhuweiyun@njau.edu.cn Table 1. Composition and nutrient concentrations of the experimental diets Items Ingredients (%, as-fed basis) Casein Sucrose wheat starch Amylodextrin Cellulose Colza oil DL-Methionine Salt Calcium hydrophosphate Calcium phosphate choline chloride Vitamin-mineral mix* Total Chemical composition (g/100g DM) Protein Available carbohydrates Acid hydrolyzed fat Total digestible fiber Ash Protein/ CHO Energy (kcal/g) ME (kcal/100 g DM) Macronutrients on energy basis (% of ME) Protein NFE (carbohydrate) Acid hydrolyzed fat * AIN-93G mineral and vitamin mixes. MPMC diet HPLC diet 20 10 40.4 13.2 5 7 0.3 0.2 1 1.3 0.1 1.5 100 54 10 6.6 13.2 5 7 0.1 0.2 1 1.3 0.1 1.5 100 20.96 55.54 7.18 4.7 2.53 0.38 3.54 328.8 45.04 29.92 7.7 4.7 3.54 1.5 3.35 327.8 22 59 19 48 32 20 Table 2. Body weight, food intake and plasma biochemical parameters in MPMC and HPLC rats MPMC HPLC P value Body weight 365.11±11.07 264.97±8.92 <0.001 Body weight gain (g) 136.3 ± 9.07 42.42 ± 6.72 <0.001 Liver weight (g) 15.63 ± 0.7 13.41 ± 0.63 0.014 4.25±0.13 5.03±0.12 <0.001 0.62 ± 0.03 0.6 ± 0.03 0.659 0.17±0.01 0.23±0.01 <0.001 4.737 ± 0.25 2.54 ± 0.2 <0.001 1.29±0.07 0.95±0.08 <0.001 5.21 ± 0.29 2.41 ± 0.14 <0.001 0.72±0.05 0.45±0.04 <0.001 Food intake (g/d) 17.45 ± 0.47 13.87 ± 0.38 <0.001 Energy intake (kcal/d) 61.76 ± 1.66 46.48 ± 1.29 <0.001 Total protein (g/L) 44.27 ± 2.81 45.54 ± 1.41 0.631 Albumin (g/L) 21.84 ± 1.37 23.42 ± 0.66 0.579 Globulin (g/L) 22.93 ± 1.54 22.12 ± 0.9 0.315 Glucose (mmol/L) 14.62 ± 1.33 10.48 ± 0.71 0.023 Triglyceride (mmol/L) 2.31 ± 0.26 0.73 ± 0.07 <0.001 Total cholesterol (mmol/L) 1.17 ± 0.1 1.3 ± 0.06 0.247 HDLC (mmol/L) 0.76 ± 0.05 0.95 ± 0.03 0.003 LDLC (mmol/L) 0.15 ± 0.02 0.16 ± 0.01 0.436 Urea (mmol/L) 5.43 ± 0.39 11.1 ± 1.05 <0.001 Uric acid (μmol/L) 207.7 ± 17.53 173.4 ± 16.92 0.247 Creatinine (μmol/L) 23.5 ± 1.35 25.4 ± 2.08 0.631 Glutamic-pyruvic transaminase (U/L) 22.55 ± 2.76 32.23 ± 7.25 0.684 Glutamic-oxalacetic transaminase (U/L) 20.55 ± 1.72 25.35 ± 2.69 0.280 Insulin (μU/mL) 53.74 ± 8.99 19.98 ± 2.26 0.008 Glucagon (pg/mL) 65.03 ± 8.53 74.91 ± 9.28 0.4 Insulin/Glugacon 0.85 ± 0.13 0.3 ± 0.04 <0.001 thyroxine (ng/mL) 19.36 ± 3.28 22.24 ± 4.06 0.73 triiodothyronine (ng/mL) 0.32 ± 0.03 0.28 ± 0.02 0.34 Growth hormone (ng/mL) 0.91 ± 0.09 1.02 ± 0.08 0.315 Gastrin (pg/mL) 20.31 ± 3.41 24.53 ± 2.78 0.356 HOMA-IR 37.007±7.27 9.514±1.615 <0.001 IAI -6.47±0.27 -5.24±0.187 <0.001 Expressed as %body weight Spleen weight (g) Expressed as %body weight Mesenteric adipose (g) Expressed as %body weight Epididymal adipose (g) Expressed as %body weight Values are expressed as mean ± SEM, n = 10 for each group. Table 3. Quantitative concentrations of selected urinary metabolites in MPMC and HPLC groups MPMC Glucose (mmol/L) Uric acid (mg/L) Creatinine (mmol/L) 0.64±0.064 295.93±33.79 5.32±0.516 HPLC 0.276±0.055 138.03±17.1 1.8±0.285 Data were expressed as mean ± SEM, n = 10 each group. P-value <0.001 <0.001 <0.001 Table 4. Parameter description of plasma metabolite pair-wise correlation network in MPMC (M) and HPLC (H) group. Degree 1 Alanine β-Amino Isobutyrate Glutamine Glycine Isoleucine Lysine Methionine Phenylalanine Proline Serine Threonine Tryptophan Tyrosine Valine Galactose Lyxose Mannonate-γ-Lactone Threonic Acid 2-Ketoglutarate Citrate Fumarate Lactate Malate Oxalate Pyruvate 11-Cis-Octadecenoate β-Hydroxybutyrate Cis-4,7,10,13,16,19-Docosahexaenoate Glycerol Linoleate Palmitate Uric Acid 1 M 3 2 5 1 1 0 3 3 2 5 2 3 4 0 3 1 3 2 0 4 2 5 2 5 1 5 2 5 4 4 4 6 H 13 5 1 9 5 7 8 10 7 11 9 1 3 8 6 5 6 3 7 5 10 8 5 1 1 5 2 1 2 4 5 4 Betweenness Centrality 2 M H 8.00 162.59 15.00 0.00 6.03 0.00 0.00 44.18 0.00 37.00 0.00 1.24 18.00 25.34 28.00 176.40 8.00 0.83 20.00 60.73 14.00 5.36 8.33 0.00 42.08 37.00 0.00 3.63 0.00 86.80 0.00 40.84 0.00 86.80 0.00 75.47 0.00 15.21 0.67 64.53 0.00 176.40 12.07 369.47 0.00 37.00 39.35 0.00 0.00 0.00 14.40 9.12 0.00 6.53 25.98 0.00 2.63 0.00 1.80 9.00 1.80 3.24 14.60 37.00 Eigenvector Centrality 3 M H 0.12 1.00 0.06 0.46 0.81 0.05 0.18 0.58 0.02 0.39 0.00 0.65 0.13 0.75 0.15 0.74 0.03 0.64 0.17 0.97 0.06 0.76 0.17 0.03 0.28 0.19 0.00 0.68 0.41 0.13 0.01 0.33 0.41 0.13 0.09 0.05 0.00 0.55 0.75 0.07 0.04 0.74 0.80 0.31 0.04 0.34 0.54 0.00 0.13 0.00 0.67 0.30 0.04 0.02 0.67 0.05 0.66 0.01 0.57 0.16 0.57 0.25 1.00 0.07 The degree of a vertex of a graph is the number of edges incident to the vertex Betweenness centrality quantifies the number of times a node acts as a bridge along the shortest path between two other nodes 3 Eigenvector Centrality assigns relative scores to all nodes in the network based on the concept that connections to high-scoring nodes contribute more to the score of the node in question than equal connections to low-scoring nodes. 2 Table 5. Parameter description of plasma-urine metabolite pair-wise correlation network in MPMC or HPLCgroup. Betweenness Eigenvector Label Degree1 Centrality2 Centrality3 MPMC group Plasma metabolite Valine Pyruvate Alpha-Tocopherol Adipate Cis-4,7,10,13,16,19-Docosahexaenoate Urine metabolite Serine D-Fructose D-Turanose Raffinose Ribitol 9 6 5 4 3 52.50 27.00 22.50 6.00 9.00 1.00 0.38 0.55 0.11 0.21 2 2 3 2 2 7.00 7.00 13.00 7.00 5.00 0.48 0.48 0.26 0.48 0.22 HPLC group Plasma metabolite 2-oxoisocaproate 4 9.00 β-Hydroxybutyrate 4 12.50 Citrate 3 5.50 Tyrosine 3 3.00 Palmitate 2 6.00 Stearate 2 4.00 Urine metabolite L-Norleucine 4 13.67 Erythrose 2 4.00 Pentitol 2 6.00 Raffinose 2 4.00 3,4-Dihydroxybutanoate 3 7.67 1 The degree of a vertex of a graph is the number of edges incident to the vertex 2 Betweenness centrality quantifies the number of times a node acts as a bridge along the shortest path between two other nodes 3 Eigenvector Centrality assigns relative scores to all nodes in the network based on the concept that connections to high-scoring nodes contribute more to the score of the node in question than equal connections to low-scoring nodes. 0.32 1.00 0.87 0.47 0.14 0.12 0.93 0.12 0.14 0.20 0.78 (x100,000,000) a 1.1 MPMC 11-cis-octadecenoic acid 1.0 Citric acid Urea Lactic acid Threonine Ornithine 0.9 β-hydroxybutyric 0.8 acid Pyruvic acid Malic acid HPLC Glucose Glutamate Cholesterol Mannose Adipic acid Oxalic acid Palmitic acid 0.6 0.5 Stearic acid Palmitoleic acid Glutamine Proline Serine 0.7 Frucose Arachidonic acid Pyroglutamic acid 0.4 Valine 0.3 2-ketoglutaric acid 0.2 0.1 0.0 -0.1 5.0 6.0 7.0 8.0 9.0 10.0 11.0 12.0 13.0 14.0 15.0 16.0 17.0 18.0 (x100,000,000) b 3.25 MPMC 2.75 HPLC Maltose 3.00 Lactate D-glucose Succinic acid Pseudo uridine Aconitic acid 2.50 2.25 Sorbitol Glycolic acid Raffinose Citric acid Creatinine 2.00 Hippuric acid Pyruvic acid 1.75 Glucoronic acid Urea 1.50 Stearic acid 1.25 2-ketoglutaric acid 1.00 Erythrose Palmitic acid Taurine 0.75 0.50 0.25 0.00 -0.25 5.0 6.0 7.0 8.0 9.0 10.0 11.0 12.0 13.0 14.0 15.0 16.0 17.0 18.0 Retention Time (min) Supplementary Fig 1. GC/MS chromatogram overlay of MPMC and HPLC group rats depicting the plasma (a) and urinary (b) metabolite peaks. a b Fig.2 (a) The positive correlation between energy-metabolism relatives and some amino acids in plasma of HPLC rats. (b) Positive correlation between plasma amino acids. The black line indicated the unique pair-wise correlation in MPMC rats, blue line indicated the shared correlation both in MPMC and HPLC rats, and red line indicated the unque pair-wise correlation in HPLC rats. Fig.3 Plasma-urinary metabolite pair-wise correlation network using data from MPMC and HPLC group. Correlations with a p value less than 0.01 and coefficient more than 0.8 were used to construct the network. Edge color in red indicated positive correlation and blue indicated negative correlation. The node color of certain metabolite was the same as its source metabolite property. Red circle shape indicated plasma metabolites while black circle indicated urinary metabolites.