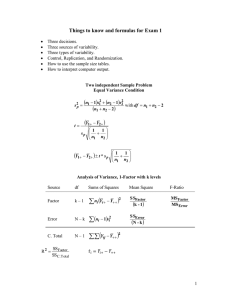
Simple Linear Regression - Graphing and Testing Model Assumptions - NBA Players Weights and Heights
advertisement

Checking Regression Model
Assumptions
NBA 2013/14 Player Heights and
Weights
Data Description / Model
• Heights (X) and Weights (Y) for 505 NBA Players in
2013/14 Season.
• Other Variables included in the Dataset: Age, Position
• Simple Linear Regression Model: Y = b0 + b1X + e
• Model Assumptions:
e ~ N(0,s2)
Errors are independent
Error variance (s2) is constant
Relationship between Y and X is linear
No important (available) predictors have been ommitted
Weight (Y) vs Height (X) - 2013/2014 NBA Players
300
275
Weight (lbs)
250
225
200
175
150
65
70
75
80
Height (inches)
85
90
Regression Model
Regression Statistics
Multiple R
0.821
R Square
0.674
Adjusted R Square
0.673
Standard Error
15.237
Observations
505
^
^
^
Y b0 b1 X b 0 b 1 X 279.869 6.331X
^
ANOVA
df
Regression
Residual
Total
1
503
504
SS
240985
116782
357767
MS
240985
232
F Significance F
1038
0.0000
s{b1} s b 1 0.197
cdf-based: t 0.975;503 = upper-tail based: t 0.025;503 1.965
^
Intercept
Height
Coefficients
Standard Error t Stat
P-value Lower 95%Upper 95%
-279.869
15.551 -17.997
0.0000 -310.423 -249.316
6.331
0.197
32.217
0.0000
5.945
6.717
b
b
6.331
H 0 : b1 0 H A : b1 0 TS : t 1 ^1
32.217
s{b1}
0.197
s b1
*
95% Confidence Interval for b1 : 6.331 1.965(0.197)
n
Total (Corrected)Sum of Squares: SSTO Yi Y
i 1
2
5.945 , 6.717
357767
2
^
Re gression Sum of Squares: SSR SSReg Y i Y 240985 df Reg 1
i 1
n
2
^
Error Sum of Squares: SSE SSRes Yi Y i 116782 df Err 505 2 503
i 1
240985 1 1038
MSR MSReg
H 0 : b1 0 H A : b1 0 TS : F *
MSE MSRes 116782 503
n
SSR
SSRes 240985
0.674
SSTO SSTO 357767
116782
s 2 MSE MSRes
232
503
r2
s 232 15.24
Checking Normality of Errors
• Graphically
Histogram – Should be mound shaped around 0
Normal Probability Plot – Residuals versus expected values under
normality should follow a straight line.
•
•
•
•
•
Rank residuals from smallest (large negative) to highest (k = 1,…,n)
Compute the percentile for the ranked residual: p=(k-0.375)/(n+0.25)
Obtain the Z-score corresponding to the percentiles: z(p)
Expected Residual = √MSE*z(p)
Plot Ordered residuals versus Expected Residuals
• Numerical Tests:
Correlation Test: Obtain correlation between ordered residuals and
z(p). Critical Values for n up to 100 are provided by Looney and Gulledge
(1985)).
Shapiro-Wilk Test: Similar to Correlation Test, with more complex
calculations. Printed directly by statistical software packages
Normal Probability Plot / Correlation Test
Extreme and Middle Residuals
rank
1
2
3
4
5
…
251
252
253
254
255
…
501
502
503
504
505
percentile
0.0012
0.0032
0.0052
0.0072
0.0092
…
0.4960
0.4980
0.5000
0.5020
0.5040
…
0.9908
0.9928
0.9948
0.9968
0.9988
z(p)*s
-46.115
-41.519
-39.045
-37.306
-35.949
…
-0.151
-0.076
0.000
0.076
0.151
…
35.949
37.306
39.045
41.519
46.115
The correlation
between the Residuals
and their expected
values under normality
is 0.9972.
Normal Probability Plot of Residuals
80
60
40
20
Residual
e
-45.583
-44.921
-39.929
-36.921
-36.590
…
-0.260
-0.260
-0.260
-0.260
0.063
…
40.748
42.079
44.417
49.740
56.079
0
-60
-40
-20
0
20
40
-20
-40
-60
Expected Value Under Normality
Based on the Shapiro-Wilk test in R, the P-value for H0: Errors are normal
is P = .0859 (Do not reject Normality)
60
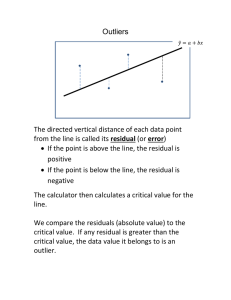
Checking the Constant Variance Assumption
• Plot Residuals versus X or Predicted Values
Random Cloud around 0 Linear Relation
Funnel Shape Non-constant Variance
Outliers fall far above (positive) or below (negative) the
general cloud pattern
Plot absolute Residuals, squared residuals, or square
root of absolute residuals
Positive Association Non-constant Variance
• Numerical Tests
Brown-Forsyth Test – 2 Sample t-test of absolute
deviations from group medians
Breusch-Pagan Test – Regresses squared residuals on
model predictors (X variables)
Residuals vs Fitted Values
60
40
Residuals
20
0
-20
-40
-60
150
165
180
195
210
225
Fitted Values
240
255
270
285
300
Absolute Residuals vs Fitted Values
60
50
Absolute Residuals
40
30
20
10
0
140
160
180
200
220
Fitted Values
240
260
280
Equal (Homogeneous) Variance - I
Brown-Forsythe Test:
H 0 : Equal Variance Among Errors s 2 e i s 2 i
H A : Unequal Variance Among Errors (Increasing or Decreasing in X )
1) Split Dataset into 2 groups based on levels of X (or fitted values) with sample sizes: n1 , n2
2) Compute the median residual in each group: e1 , e2
3) Compute absolute deviation from group median for each residual:
dij eij e j
i 1,..., n j
j 1, 2
4) Compute the mean and variance for each group of dij : d 1 , s12
5)
2
2
n
1
s
n
1
s
1
1
2
2
Compute the pooled variance: s 2
Test Statistic: t BF
n1 n2 2
d1 d 2
1 1
s
n1 n2
H0
~ t n n
Reject H 0 if t BF t 1 2 ; n 2
1
2
2
d 2 , s22
Equal (Homogeneous) Variance - II
Breusch-Pagan (aka Cook-Weisberg) Test:
H 0 : Equal Variance Among Errors s 2 e i s 2 i
H A : Unequal Variance Among Errors s i2 s 2 h 1 X i1 ... p X ip
n
1) Let SSE ei2 from original regression
i 1
2) Fit Regression of ei2 on X i1 ,...X ip and obtain SS Reg *
Test Statistic: X
2
BP
2
Reject H 0 if X BP
SS Reg * 2
2
e
n
i
i 1
2 1 ; p
n
2
H0
~
p2
p = # of predictors
Brown-Forsyth and Breusch-Pagan Tests
Brown-Forsyth Test:
Group 1: Heights ≤ 79”, Group 2: Heights ≥ 80”
H0: Equal Variances Among Errors (Reject H0)
Brown-Forsyth Test
Group
Heights(Grp)
1 69-79
2 80-87
MeanDiff
PooledVar
PooledSD
sqrt(1/n1+1/n2)
s{d1bar-d2bar}
t*(BF)
t(.975,505-2)
P-value
n(Grp)
Med(e|grp) Mean(d|Grp) Var(d|Grp)
252
-1.2673
10.8039
70.4186
253
0.7482
12.9193
108.7256
-2.1155
89.6102
9.4663
0.0890
0.8425
-2.5110
1.9647
0.0247
Breusch-Pagan Test:
H0: Equal Variances Among Errors
(Reject H0)
Regression of Weight on Height
ANOVA
df
SS
Regression
1 240984.7782
Residual
503 116782.3109
Total
504 357767.0891
Regression of e^2 on Height
ANOVA
df
SS
Regression
1 963633.2703
Residual
503 67658845.93
Total
504 68622479.2
SSE(Model1)
n
SS(Reg*)
X2(BP):Num
X2(BP):Denom
X2(BP)
Chisq(.95,1)
P-value
116782.311
505
963633.270
481816.635
53477.534
9.010
3.841
0.003
Linearity of Regression
F -Test for Lack-of-Fit (n j observations at c distinct levels of "X")
H 0 : E Yi b 0 b1 X i
H A : E Yi i b 0 b1 X i
Compute fitted value Y j and sample mean Y j for each distinct X level
c
nj
Lack-of-Fit: SS LF Y j Y j
j 1 i 1
c
nj
Pure Error: SS PE Yij Y j
j 1 i 1
2
2
df LF c 2
df PE n c
SS ( LF ) c 2 MS ( LF )
~
MS
(
PE
)
SS
(
PE
)
n
c
H0
Test Statistic: FLOF
Reject H 0 if FLOF F 1 ; c 2, n c
Fc 2,n c
Linearity of Regression
Full Model H A : E Yij j
c
nj
SSE ( F ) Yij Y j
j 1 i 1
2
^
j Y j
SS PE df F n c
c means are estimated
Reduced Model H 0 : E Yij b 0 b1 X j j Y j b0 b1 X j
^
SSE ( R ) Yij Y j
Y
c
nj
j 1 i 1
nj
c
ij
j 1 i 1
c
nj
Y j
2
c
nj
Yij Y j
j 1 i 1
Y
Y
Yij Y j
j 1 i 1
nj
c
SSE df R n 2
Yij Y j
j 1 i 1
2
2
c
nj
j
Y
Y j
j 1 i 1
2
c
nj
j 1 i 1
j
c
2
Y j
nj
j
2 means are estimated
Y j
j 1 i 1
2
c
j 1
2
2 Y j Y j
0
2
c
nj
2 Yij Y j Y j Y j
j 1 i 1
Y Y
nj
i 1
ij
j
SSE SS PE SS LF
FLOF
SSE SS PE
n
2
n
c
SS PE
nc
SSE R SSE F
df R df F
SSE F
df F
Reject H 0 if FLOF F 1 ; c 2, n c
Computing Strategy:
nj
1) For each group (j ): Compute: Y j
nj
Yij Y j
s 2j i 1
n j 1
0
Y
i 1
ij
nj
2
nj 1
otherwise
^
Y j b0 b1 X j
nj
c
nj
c
2
^
^
2) SS LF Y j Y j n j Y j Y j
i 1 j 1
j 1
3) SS PE Yij Y j
i 1 j 1
c
n 1 s
2
c
j 1
j
2
j
2
SS LF
c 2 MS ( LF )
SS PE MS ( PE )
nc
H0
~
Fc 2,n c
Height and Weight Data – n=505, c=18 Groups
Height
n
69
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
Sum
Source
df
LackFit
PureError
Mean
SD
Y-hat
SSLF
SSPE
SSE
2
182.50
3.54
156.95 1305.39
12.50 1317.89
4
175.75
15.52
169.61
150.62
722.75
873.37
13
181.00
13.00
175.94
332.27 2028.00 2360.27
16
186.13
12.09
182.28
237.15 2191.75 2428.90
21
183.33
9.26
188.61
583.79 1716.67 2300.45
41
193.71
11.58
194.94
61.96 5360.49 5422.44
32
200.84
11.96
201.27
5.74 4434.22 4439.96
31
204.13
10.70
207.60
373.06 3433.48 3806.55
43
211.00
12.83
213.93
368.86 6912.00 7280.86
49
221.35
18.70
220.26
57.94 16781.10 16839.04
46
227.33
15.13
226.59
24.90 10300.11 10325.01
67
232.49
19.63
232.92
12.30 25430.75 25443.05
53
241.49
14.79
239.25
265.64 11369.25 11634.88
44
245.66
17.55
245.58
0.26 13241.89 13242.14
34
254.62
14.70
251.91
248.66 7128.03 7376.69
7
247.86
10.75
258.24
755.21
692.86 1448.07
1
278.00
0.00
264.57
180.24
0.00
180.24
1
263.00
0.00
270.91
62.50
0.00
62.50
505 #N/A
#N/A
#N/A
5026.479 111755.8 116782.3
SS
16
5026.5
487 111755.8
MS
F(LOF)
F(.95)
P-value
314.2
1.369
1.664
0.1521
229.5
Do not reject
H0: j = b0 + b1Xj
Box-Cox Transformations
• Automatically selects a transformation from power family
with goal of obtaining: normality, linearity, and constant
variance (not always successful, but widely used)
• Goal: Fit model: Y’ = b0 + b1X + e for various power
transformations on Y, and selecting transformation
producing minimum SSE (maximum likelihood)
• Procedure: over a range of l from, say -2 to +2, obtain Wi
and regress Wi on X (assuming all Yi > 0, although adding
constant won’t affect shape or spread of Y distribution)
l
K
Y
1 i 1 l 0
Wi
K 2 ln Yi l 0
1n
K 2 Yi
i 1
n
K1
1
l K 2l 1
Box-Cox Transformation – Obtained in R
Maximum occurs near l = 0 (Interval Contains 0) – Try taking logs of Weight
Results of Tests (Using R Functions) on ln(WT)
> nba.mod2 <- lm(log(Weight) ~ Height)
> summary(nba.mod2)
Call:
lm(formula = log(Weight) ~ Height)
Coefficients:
Est Std. Error t value Pr(>|t|)
(Intercept) 3.0781 0.0696
44.20 <2e-16
Height
0.0292 0.0009
33.22 <2e-16
Residual standard error: 0.06823 on 503
degrees of freedom
Multiple R-squared: 0.6869,
Adjusted
R-squared: 0.6863
F-statistic: 1104 on 1 and 503 DF, pvalue: < 2.2e-16
Normality of Errors (Shapiro-Wilk Test)
> shapiro.test(e2)
Shapiro-Wilk normality test
data: e2
W = 0.9976, p-value = 0.679
Constant Error Variance (Breusch-Pagan Test)
> bptest(log(Weight) ~ Height,studentize=FALSE)
Breusch-Pagan test
data: log(Weight) ~ Height
BP = 0.4711, df = 1, p-value = 0.4925
Linearity of Regression (Lack of Fit Test)
nba.mod3 <- lm(log(Weight) ~
Model 1: log(Weight) ~ Height
factor(Height))
Model 2: log(Weight) ~ factor(Height)
> anova(nba.mod2,nba.mod3)
Res.Df RSS Df Sum of Sq
F
Pr(>F)
Analysis of Variance Table
1 503 2.3414
2 487 2.2478 16 0.093642 1.268 0.2131
Model fits well
on all
assumptions