Panagrolaimus Genome Evolution: Impact of Reproductive Mode
advertisement

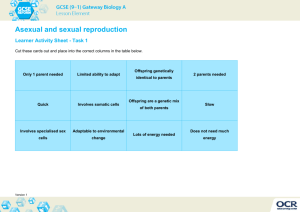
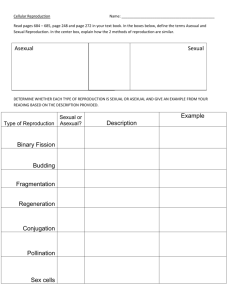
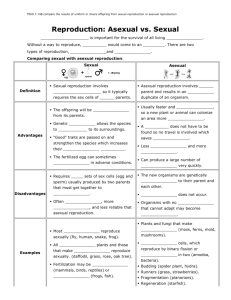
Panagrolaimus Mitochondrial Genome Evolution: Impact of Reproductive Mode Samantha C. Lewis ♦ Dr. Dee Denver Zoology Dept. ♦ Center for Genome Research and Biocomputing Background • Recombination: a force at the population, organismal and molecular levels • Gonochorists and parthenogens (Sexual species and asexual species) MUTATION MUTATION ∞ Background • Theory: asexual organisms do not persist as long as sexual ones • “Mutational Meltdown” • Can reduced selection efficiency be observed at the molecular level? Hypothesis • Inefficient selection may be observed as non-synonymous protein-coding gene changes that lead to deleterious mutation • These changes may be observed in the mitochondrial genome www.nsf.gov/news/biology/assets/interact08.jpg Methods • Sequence the mitochondrial genomes of one sexual and one asexual organism; compare Extract DNA from nematodes Genome content comparisons Amplify mtDNA Analysis in MEGA and DNAsp Sequence amplified products Perform DNA sequence alignments Work to Date Gene rRNA No sequence PS 1579 AF 36 C. elegans AF36 PS1579 sexual asexual nad2 nad2 nad2 tRNA I tRNA I tRNA I ………….. tRNA R tRNA R tRNA G tRNA G tRNA G tRNA tRNA F tRNA F tRNA F cob cob cob Genome Comparisons • Insertions; Deletions; Gene order • Ka/Ks ratio An index of selection efficiency The lower the ratio, the stronger selection is • Nucleotide diversity A measure of genetic diversity Highest in sexual species • Homopolymer repeats Theory • Insertions; Deletions; Gene order Similar to C. elegans • Ka/Ks ratio Higher in asexual Lower in sexual • Nucleotide diversity Higher in sexual Lower in asexual • Homopolymer repeats Higher in asexual Lower in sexual Results AF36 - sexual • Homopolymers cause slipped strand mispairing during DNA replication Number of Repeats 25 PS1579 - asexual 20 15 10 5 0 6T 6 77T 88T 9 9T 10T 10 11T 11 Length of Repeats (bp) • Considerably more mononucleotide runs were observed in the asexual strain Results 0.4 [Nucleotide diversity] [Ka/Ks] 0.35 0.3 0.25 0.2 • Sexual Panagrolaimus did show higher nucleotide diversity values 0.15 • Asexual Panagrolaimus did not show higher Ka/Ks values 0.1 0.05 0 Label Reproductive Mode PI-P PI-G PII-P* Panagrolaimus Parthenogens Panagrolaimus Gonochorists Other Nematode Parthenogens Theory Support Theory Support • Insertions; Deletions; Gene order Similar to C. elegans Theory Support • Insertions; Deletions; Gene order Similar to C. elegans • Nucleotide diversity Higher in sexual Lower in asexual Theory Support • Insertions; Deletions; Gene order Similar to C. elegans • Nucleotide diversity Higher in sexual Lower in asexual • Homopolymer repeats Higher in asexual Lower in sexual Theory Support • Insertions; Deletions; Gene order Similar to C. elegans • Ka/Ks ratio Higher in asexual Lower in sexual • Nucleotide diversity Higher in sexual Lower in asexual • Homopolymer repeats Higher in asexual Lower in sexual Conclusions • Panagrolaimus is a superb model to study the evolutionary causes and consequences of transitions to asexuality in animals • While some analyses were consistent with a reduced efficiency of natural selection in asexual species, others were not • Further studies with expanded data sets are needed Future Work • Confirm presence/absence of Arg tRNA in mt genome • Sequence more genes from Panagrolaimus and other nematodes – add to Ka/Ks analysis • Apply genome scale technologies to the Panagrolaimus system – high-throughput sequencing Acknowledgments • The Denver lab: Dee, Dana, Bobby, Leslie, Steph, Caroline, Peter, Larry, Ashley • Dr. Kevin Ahern • Howard Hughes Medical Institute We thank many individuals for providing Panagrolaimus strains: James Baldwin (UC-Riverside, USA) ♦ Ann Burnell (National University of Ireland Maynooth) ♦ Marie-Anne Felix (Institut Jacques Monod, France) Einhard Schierenberg (Universität Köln, Germany) Paul Sternberg (Caltech, USA) ♦ the Caenorhabditis Genetics Center (University of Minnesota, USA)