22 Statistical Modeling of DNA Sequences and Patterns TOC
advertisement

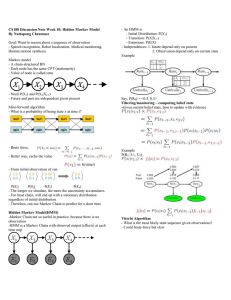
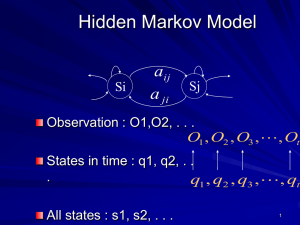
22 Statistical Modeling of DNA Sequences and Patterns TOC Table 1 Conditional Nucleotide Probabilities for the 25-nt Example Sequence ↓ Si–1 Si → A C T G A C T G 25/144 50/168 25/168 25/120 25/144 25/168 100/168 25/120 75/144 75/168 0 25/120 25/144 25/168 50/168 25/120 Table 2 Patterns and Probabilities Deduced Using IID and Markov Models Probability Pattern ATTTA TGTTTTG TTTTGGGG CTTTTACCAAT TCTTTATCTTTGCG CTGAACATTGATGCA IID Model Markov Model 2.57 × 10– 3 1.06 × 10– 4 1.40 × 10– 5 5.181 × 10– 7 6.545 × 10– 9 1.238 × 10– 9 2.0 × 10– 3 1.64 × 10– 4 4.153 × 10– 5 7.596 × 10– 7 1.968 × 10– 9 3.19 × 10– 9 Table 3 Weight Matrix for TATAA Box T C A G 6 14 8 32 49 6 4 1 1 0 58 1 56 0 4 0 6 3 51 0 22 0 38 0 6 1 53 0 20 2 30 8 Fig. 1. The set of possibe transitions in a 2nd. Order Markov chain. Note that only 8 nonzero probabilities need to be estimated for this model, as not all of the 16 transitions are possible. Fig. 2. A profile HMM utilizes the insert (diamond) and delete (circle) states. The delete states are silent and are not associated with the emissions of any symbols. The parameters of a profile HMM are learned from a multiple sequence alignment. TOC