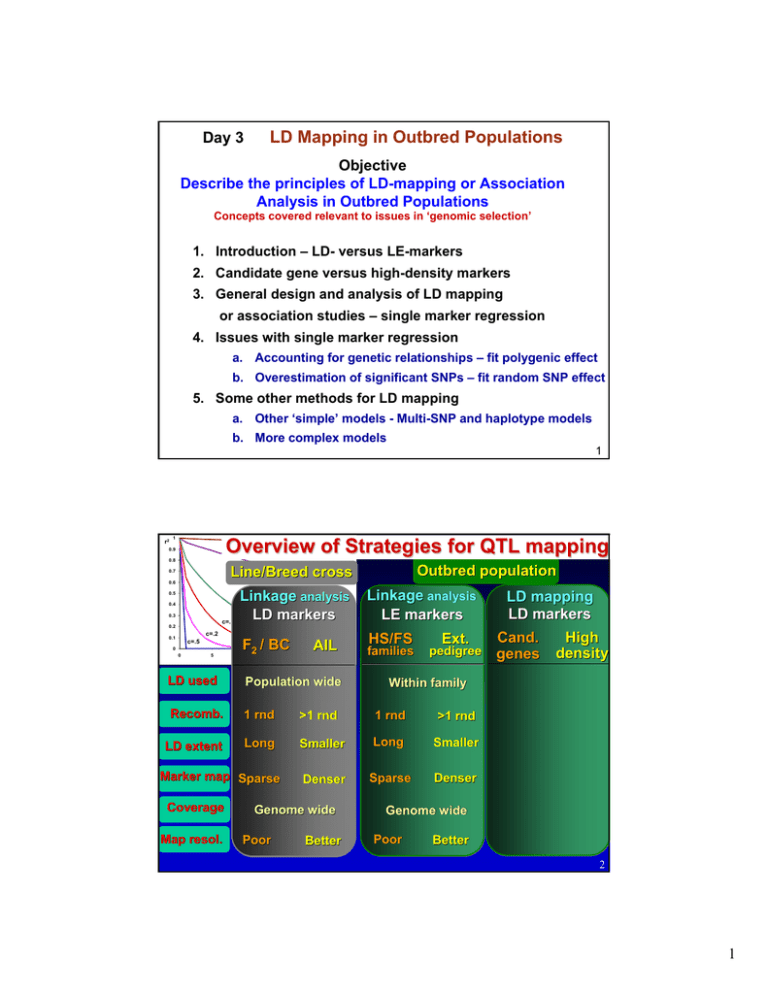
LD Mapping in Outbred Populations Day 3 Objective
advertisement
LD Mapping in Outbred Populations
Day 3
Objective
Describe the principles of LD-mapping or Association
Analysis in Outbred Populations
Concepts covered relevant to issues in ‘genomic selection’
1. Introduction – LD- versus LE-markers
2. Candidate gene versus high-density markers
3. General design and analysis of LD mapping
or association studies – single marker regression
4. Issues with single marker regression
a. Accounting for genetic relationships – fit polygenic effect
b. Overestimation of significant SNPs – fit random SNP effect
5. Some other methods for LD mapping
a. Other ‘simple’ models - Multi-SNP and haplotype models
b. More complex models
1
r2
Overview of Strategies for QTL mapping
1
c=.001
0.9
0.8
Outbred population
c=.01
Line/Breed cross
0.7
0.6
0.5
Linkage analysis
LD markers
Linkage analysis
LE markers
F2 / BC
families
c=.05
0.4
0.3
c=.1
0.2
c=.2
0.1
c=.5
0
0
5
10
15
Generation
20
AIL
HS/FS
25
Ext.
pedigree
LD used
Population wide
Recomb.
Recomb.
1 rnd
>1 rnd
1 rnd
>1 rnd
LD extent
Long
Smaller
Long
Smaller
Denser
Sparse
Denser
Marker map Sparse
Coverage
Map resol.
resol.
Genome wide
Poor
Better
LD mapping
LD markers
Cand.
Cand.
genes
High
density
Within family
Genome wide
Poor
Better
2
1
3 types of marker loci
Direct markers
LD
-markers
LD-markers
Functional mutations
- known genes
Q
q
In pop.-wide Linkage Disequilibrium
with mutation
Linkage phase
~consistent
across population
LE
-markers
LE-markers
Dekkers 2004. J.Anim.Sci
MQ
MQ
mq
MQ
mq
mq
In pop.-wide Linkage Equilibrium
with mutation
Linkage phase NOT consistent across families
Sire 2
Sire 1
Sire 3
M Q
M q
M Q
m q
m Q
m Q
Sire 4
M q
m q
3
1. Benefits of LD- over LE-Markers
Linkage phase tends to be consistent
across families and generations
MQ
MQ
mq
MQ
mq
mq
“Easier” to implement in genetic evaluation
genotype
y = marker haplotype + u + e
Estimation of effects:
does not require pedigreepedigree-based phenotypic data
Ideally, animals are unrelated
can be done in population of application vs. experimental cross
4
2
Examples of
gene tests in
commercial
breeding
Trait
Direct marker
Congenital
defects
Appearance
Milk quality
D = dairy cattle
B = beef cattle
C = poultry
P = pigs
S = sheep
Dekkers, 2004, JAS
Most tests used
commercially are
direct or LD markers
LD marker
BLAD (D)
Citrulinaemia (D,B)
DUMPS (D)
CVM (D)
Maple syrup urine (D,B)
Mannosidosis (D,B)
RYR (P)
CKIT (P)
MC1R/MSHR (P,B,D)
MGF (B)
κ-Casein (D)
β-lactoglobulin (D)
FMO3 (D)
RYR (P)
RN/PRKAG3 (P)
Meat quality
LE marker
RYR (P)
Polled (B)
RYR (P)
RN/PRKAG3 (P)
A-FABP/FABP4 (P)
H-FABP/FABP3 (P)
CAST (P, B)
>15 PICmarqTM (P)
THYR (B)
Leptin (B)
Feed intake
Disease
Reproduction
MC4R (P)
Prp (S)
F18 (P)
Booroola (S)
Inverdale(S)
Hanna (S)
Growth &
composition
Milk yield &
composition
B blood group (C)
K88 (P)
Booroola (S)
ESR (P)
PRLR (P)
RBP4 (P)
CAST (P)
IGF-2 (P)
MC4R (P)
IGF-2 (P)
Myostatin (B)
Callipyge (S)
DGAT (D)
GRH (D)
κ-Casein (D)
QTL (P)
QTL (B)
Carwell (S)
PRL (D)
QTL (D)
5
In outbred populations only some closely linked
markers will be in sufficient LD with QTL
See Day 1
r2
c=.001
1
0.9
0.8
c=.01
0.7
0.6
0.5
0.4
c=.05
0.3
c=.1
0.2
c=.5
0.1
c=.2
0
0
5
10
15
Generation
20
25
6
3
Extent of LD is driven by Ne
1.0
0.9
2
E(r2)E(r
= 1) =/ 1/(1+4N
(1 + 4N
ed)
ec)
(Sved, 1971)
0.8
Distance (Morgans)
2
Expected LD (r )
0.7
0.6
Ne=10
0.5
0.4
Ne=25
0.3
Ne=50
0.2
Ne=100
Ne=250
0.1
Ne=500
0.0
0
0.02
0.04
0.06
0.08
0.1
0.12
0.14
0.16
Distance (Morgans)
r2
c=.001
1
0.9
0.8
c=.01
0.7
2.
0.6
0.5
0.4
c=.05
0.3
c=.1
0.2
c=.5
0.1
c=.2
0
0
5
10
15
Generation
20
25
7
Candidate genes vs
high-density markers
How to find markers close enough
to QTL for population-wide LD?
Candidate gene
analysis
Find markers in genes that may
contain QTL based on
z their biological role
z location in a QTL region
z
Comparative data
z
Gene expression data
High density
genotyping
Genotype enough markers
such that each QTL will
have several markers close
enough such that at least
one marker will be in
sufficient LD with the QTL
to show an association
with phenotype.
8
4
A Revolution in Molecular Genetic Technology
2.8 million SNPs
Nature 2004
S
ingle
Single
N
ucleotide
Nucleotide
P
olymorphisms
Polymorphisms
High-through-put
SNP genotyping
AAGCCTTGATAATT
International Swine Genome
Sequencing Consortium
AAGCCTTGCTAATT
Illumina Bovine
50k Beadchip
r2
9
Overview of Strategies for QTL mapping
1
c=.001
0.9
0.8
Outbred population
c=.01
Breed/Line cross
0.7
0.6
0.5
Linkage analysis
LD markers
c=.05
0.4
0.3
c=.1
0.2
c=.2
0.1
c=.5
0
0
5
Linkage analysis
LE markers
HS/FS
LD mapping
LD markers
Extended Candidate High
genes density
F2 / BC AIL/RIL families pedigree
Generation
10
15
20
25
LD used
Population wide
Recomb.
Recomb.
1 rnd
>1 rnd
1 rnd
>1 rnd
LD extent
Long
Smaller
Long
Smaller
Denser
Sparse
Denser Few loci
Marker map Sparse
Coverage
Map resol.
resol.
Genome wide
Poor
Better
Within family
Genome wide
Poor
Better
Population wide
>>> 1 round
Small
Local
Dense
Genome
High
LDLD-LA analysis – see later 10
5
Candidate Gene Examples
Estrogen Receptor Gene
(Rothschild et al. 1991, Short et al. 1997)
Effect on Number Born Alive
ESR
genotype
AA
First parity
n=4,262
9.4
Later parities
n=4,753
10.0
AB
9.9
10.5
BB
10.2
10.7
11
MC4R mutation
and Test
(Kim et al., Mam. Gen. 2000)
a
293
C
N
C
N
295
S
I
297
I
299
D
P
N
P
L
300
I
Y
Allele 1
homozygote
sequence
S
I
I
L
I
Y
Allele 2
homozygote
sequence
11 vs 22 genotype
in 2 commercial types
Transmembrane
domains
NH2
I
II
1/1
III
IV
2/2
VI
V
VII
COO
H
1/2
542
466
Backfat Loin
(mm) depth
(mm)
Daily
Daily
Feed
Gain
Intake
(g/d)
(kg/d)
-1.3
+1.4
-26.0
-0.15
P<.05
P<.10
P<.10
P<.05
Slide courtesy Max Rothschild 12
6
3. General Design and Analysis of LD Mapping
or Association Studies – single marker regression
II.
On a ‘random’
random’ sample
of (unrelated) individuals
obtain:
I.
Phenotype for
quantitative trait
II.
Genotypes for
one or many markers
Genotype many cows
with phenotype
(or progenyprogeny-tested bulls)
for 50,000 SNPs
TRAINING DATA
Conduct statistical analysis for association between
genotype at a marker and phenotype (repeat for each marker)
Y = μ + marker genotype + e
Test for significance
13
Principle of LD marker
effect estimation
AAGCCTTGATAATT
AAGCCTTGCTAATT
Progeny tested bulls grouped by their genotype for a
particular SNP
A
A
A
C
C
C
SNP
Genotype
AA
Average
PTA protein
+20
AC
+15
CC
+10
Î SNP effect e
stimate = +5 for A
estimate
Repeat for all markers
I.
7
LD mapping / association analysis by
single marker regression analysis
y = 1n μ + Xg + e
y = vector of phenotypes
1n = vector of 1s allocating the mean to phenotype,
X = design matrix allocating records to the marker effect (0/1/2 or -1/0/1)
g = effect of the marker (= allele substitution effect)
e = vector of random deviates ~ N(0,σe2)
• Underlying assumption is that the marker will only affect the
trait if it is in LD with a QTL.
Hayes ‘07
15
Single marker regression
y = 1n μ + Xg + e
• The design vector 1n allocates phenotypes to the mean
• The design vector X allocates phenotypes to genotypes
X, Number of “2”
Animal
Phenotpe
SNP allele 1
SNP allele
Animal
1n
alleles
1
2.030502
1
1
1
1
0
2
3.542274
1
2
2
1
1
3
3.834241
1
2
3
1
1
4
4.871137
2
2
4
1
2
5
3.407128
1
2
5
1
1
6
2.335734
1
1
6
1
0
7
2.646192
1
1
7
1
0
8
3.762855
1
2
8
1
1
9
3.689349
1
2
9
1
1
10
3.685757
1
2
10
1
1
y vector
Hayes ‘07
16
8
Single marker regression
Estimate marker effect and mean as:
⎡ ∧ ⎤ ⎡1 '1
⎢ μ∧ ⎥ = ⎢ n n
⎢ g ⎥ ⎣ X'1n
⎣ ⎦
−1
1n ' X ⎤ ⎡1n ' y ⎤
X' X ⎥⎦ ⎢⎣ X' y ⎥⎦
⎡1⎤
⎢1⎥
⎢⎥
⎢1⎥
⎢⎥
⎢1⎥
⎢1⎥
[1111111111] ⎢ ⎥ = 10
⎢1⎥
⎢1⎥
⎢⎥
⎢1⎥
⎢1⎥
⎢⎥
⎢⎣1⎥⎦
⎡0⎤
⎢1⎥
⎢ ⎥
⎢1⎥
⎢ ⎥
⎢2⎥
⎢1⎥
[1111111111] ⎢ ⎥ = 8
⎢0⎥
⎢0⎥
⎢ ⎥
⎢1⎥
⎢1⎥
⎢ ⎥
⎣⎢1⎦⎥
Conduct FF-test for significance
⎡ ∧ ⎤ ⎡10 8 ⎤ −1 ⎡33.8⎤
⎢μ∧ ⎥ = ⎢
⎥ ⎢
⎥
⎢ g ⎥ ⎣ 8 10⎦ ⎣31.7⎦
⎣ ⎦
⎡ ∧ ⎤ ⎡ 0.28 − 0.22⎤ ⎡33.8⎤
⎢μ∧ ⎥ = ⎢
⎥⎢
⎥
⎢ g ⎥ ⎣− 0.22 0.28 ⎦ ⎣31.7⎦
⎣ ⎦
⎡ ∧ ⎤ ⎡2.36⎤
⎢ μ∧ ⎥ = ⎢
⎥
⎢ g ⎥ ⎣1.38 ⎦
⎣ ⎦
Hayes ‘07
17
Example results from single SNP analyses
Estimates of Marker Effects for Milk yield US Holsteins
‘Manhattan plot’
National Swine Improvement Federation Symposium, Dec. 2008 (18)
Paul VanRaden
2008
18
9
Sample results – 1 line, 1 chr, 1 trait
-lo g 1 0 (p -v a lu e )
4
1-SNP P-value
Favorable
Unfavorable
3
Allele frequency
2
1
0
1-SNP Estimates
1.0
Favorable
Unfavorable
Allele frequency
0.5
F re q .
E s tim a te /s d
1.5
0.0
Issues with LD mapping using
single marker regression
• Significance testing – e.g. F-test
• Many tests – need to control for false positives
• Could use permutation test - see before – difficult if individuals related
• False ++ because of population structure (see problem set 1)
• Simple model assumes all animals equally (un)related
(un)related = unlikely
• Presence of breeds, strains, or families all create pop.structure
• i.e. presence of extensive genetic relationships
• To try to account for this – fit breed composition (if available)
– fit breed polygenic effect with relationships
• Overestimation of significant SNPs – fit SNP effect as random
20
10
a. Impact of Genetic Relationships
• Results in underestimation of standard errors - E.g. Hassen et al.
JAS’09
Distribution of p-values
100
Freq
80
W/out polygenic effect
120
With polygenic effect
100
Excess of low p-values
Freq
120
80
60
60
40
40
20
20
0
0
• Could also give biased estimates - simple example (Hayes ’07)
07)
– a sire with high EBV has many progeny in the population.
– a rare allele at some SNP is homozygous in the sire (aa
(aa))
– Then subsub-pop. of his progeny has higher frequency of a than overall pop.
– As the sires’
sires’ EBV is high, his progeny will also have higher EBV
– If we don’
don’t account for this, the a allele will appear to have a ++ effect.
21
Extension of 1-SNP model by fitting a
polygenic effect
y = 1n ' μ + Xg + Zu + e
u = vector of polygenic effect with covariance structure u ~ N(0,Aσa2)
A = average relationship matrix built from the pedigree σa2 = genetic var.
Z = design matrix allocating animals to records.
λ=σe2/σa2
Henderson’
Henderson’s Mixed Model Equations:
⎡∧⎤
⎢ μ∧ ⎥ ⎡1n '1n
⎢ g ⎥ = ⎢ X'1
n
⎢∧⎥ ⎢
⎢u ⎥ ⎢⎣ Z'1n
⎢⎣ ⎥⎦
1n ' X
X' X
Z' X
⎤
⎥
⎥
Z' Z + A −1 λ ⎥⎦
1n ' Z
X' Z
−1
⎡1n ' y ⎤
⎢ X' y ⎥
⎥
⎢
⎢⎣ Z' y ⎥⎦
Hayes ‘07
22
11
Example
Hayes ‘07
Animal
1
2
3
4
5
6
Sire
0
0
0
1
1
1
Dam
0
0
0
2
2
3
Phenotype
10.1
2.2
2.31
6.57
6.06
6.21
SNP alleles
Pat
Mat
0
1
1
1
1
1
0
1
0
1
0
1
Simple regression model
y = 1n μ + Xg + e
⎡ ∧ ⎤ ⎡1 '1
⎢ μ∧ ⎥ = ⎢ n n
⎢ g ⎥ ⎣ X'1n
⎣ ⎦
1
2
X = 2
1
1
1
−1
1n ' X ⎤ ⎡1n ' y ⎤
X' X ⎥⎦ ⎢⎣ X' y ⎥⎦
23
The A matrix
Elements = additive genetic relationship
= the proportion of genes shared
See more later (IBD)
Pedigree
Animal
Sire
1
2
3
4
5
6
Dam
0
0
0
1
1
1
Animal 1
Animal 1
Animal 2
Animal 3
Animal 4
Animal 5
Animal 6
Half genes from mum, half from dad
0
0
0
2
2
3
1
0
0
0.5
0.5
0.5
Animals 6 is a half sib of 4 and 5
Animal 2
1
0
0.5
0.5
0
Animal 3
1
0
0
0.5
Animal 4
1
0.5
0.25
Animal 5
Animal 6
1
0.25
Hayes ‘07
1
24
12
Example
Hayes ‘07
Animal
1
2
3
4
5
6
Sire
0
0
0
1
1
1
Dam
0
0
0
2
2
3
Phenotype
10.1
2.2
2.31
6.57
6.06
6.21
SNP alleles
Pat
Mat
0
1
1
1
1
1
0
1
0
1
0
1
y = 1n ' μ + Xg + Zu + e
⎡∧⎤
⎢ μ∧ ⎥ ⎡1n '1n
⎢ g ⎥ = ⎢ X'1
n
⎢∧⎥ ⎢
⎢u ⎥ ⎢⎣ Z'1n
⎢⎣ ⎥⎦
1n ' X
X' X
Z' X
⎤
⎥
⎥
Z' Z + A −1 λ ⎥⎦
1n ' Z
X' Z
−1
⎡1n ' y ⎤
⎢ X' y ⎥
⎥
⎢
⎢⎣ Z' y ⎥⎦
λ= σe2/σa2 = (1(1-h2)/h2 = (1(1-.75)/0.75 = 0.33
Hayes ‘07
25
b. Overestimation of significant SNPs
• Least squares (fixed effect) estimates of SNP effects are
equal to the true value + estimation error: g
ˆ = g +e
gˆ
• Thus, SNPs that are significant tend to have larger
estimation errors – e.g. SNPs with small minor allele freq.
• This can be addressed by fitting SNP effects as random
e.g. assuming g ~ N(0, σg2) for some choice of σg2
Fitting g as random regresses or shrinks estimates back to 0
to account for the lack of information
If the choice of σg2 is correct (?) then the resulting estimates
are BLUP, which have property:
Where peg is the prediction error
g = gˆ + pegˆ
Note the similarity to BLUP estimation of breeding values
Differences between random / fixed are small if the amount of
data is large (Æ small errors) or if λg=σe2/σg2 is small
26
13
Fitting SNP Effects Random vs. fixed
y = 1n ' μ + Xg + Zu + e
Add λg=σe2/σg2 to the diagonal of the X’X matrix
⎡∧⎤
⎢ μ∧ ⎥ ⎡1n '1n
⎢ g ⎥ = ⎢ X'1
n
⎢∧⎥ ⎢
⎢u ⎥ ⎢⎣ Z'1 n
⎣⎢ ⎦⎥
1n ' X
⎤
⎥
X' Z
⎥
−1
Z' Z + A λ ⎥⎦
1n ' Z
X' X + Iλ g
Z' X
−1
⎡1n ' y ⎤
⎢ X' y ⎥
⎢
⎥
⎢⎣ Z' y ⎥⎦
σ g2 could be set such that Xi g explains variance equal to some value = σ M2
Î Var(Xi g ) = σ M2 , which must be solved for σ g2
Using the conditional expectation theorem:
Var(Xi g) = E{Var(Xi g | Xi = k)} + Var{ E(Xi g | Xi = k)}
2
=
∑ Pr( X
k =0
i
= k ) k 2σ g2
= {p2 + (1-p)2} σ g2
+
0
with Xi = -1 , 0 , or 1
= {p2(-1)2 + 2p(1-p)(0)2 + (1-p)2(1)2} σ g2
Î σ g2 = σ M2 /{p2 + (1-p)2}
p = freq. allele 0
27
5. Some other models for LD mapping
a. Some other ‘simple’ models
• SNP Genotype models
• Single SNP regression
u ~ Aσ
Aσa2
yi = μ + Xij gj + ui + ei
Xij= #1 alleles
(0/1/2) - estimates allele substitution effect
Or fit as class variable Æ dominance
• MultiMulti-SNP regression
yi = μ + Xijgj + Xi,j+1 gj+1 + ui + ei
j14
10011001001100110100
01111001001001011010
00100111001000010111
00111011001101101110
j15
01101000001001100010
00011001010001000111
j16
11101001001011101111
01011000001001101010
• Haplotype methods
j13
• Fixed/random haplotype effects
y = Xg + u + e g’ = [μ00 , μ01 , μ10 , μ11]
separate mean for each haplotype
28
14
Slide 28
j13
Composite likelihood
j14
Long and Langley 1999
Fan and Xiong 2002
jdekkers, 8/7/2006
jdekkers, 8/7/2006
j15
Or using any combination of markers, as implemented by Bonnen et al. (Nat. Genet 38 2006)?
Found not to be better by Hong-hua - threshold more stringent because of larger # tests.
jdekkers, 8/7/2006
j16
Mixture distribution for presumed biallelic QTL
jdekkers, 8/7/2006
b. More complex models
• IBD Mixed Models (Meuwissen & Goddard 2000)
y = ZgQ + u + e
gQ ~ N(0,GvσQ2)
Gv = IBD matrix
– see LATER
Cov. from Prob(IBD at Q | markers)
• Combined Linkage Disequilibrium and Linkage
– see LATER
Analysis Models (LD-LA)
• Whole genome analysis methods
Fit all SNPs simultaneously using genomic selection’ type
models (Xu. 2003, Meuwissen et al. 2001)
yi = μ + Σβjgij + (ui) + ei
random (Bayesian) See Module B - Genomic selection 29
r2
Overview of Strategies for QTL mapping
1
c=.001
0.9
0.8
Outbred population
c=.01
Breed/Line cross
0.7
0.6
0.5
Linkage analysis
LD markers
c=.05
0.4
0.3
c=.1
0.2
c=.2
0.1
c=.5
0
0
5
Linkage analysis
LE markers
HS/FS
LD mapping
LD markers
Extended Candidate High
genes density
F2 / BC AIL/RIL families pedigree
Generation
10
15
20
25
LD used
Population wide
Recomb.
Recomb.
1 rnd
>1 rnd
1 rnd
>1 rnd
LD extent
Long
Smaller
Long
Smaller
Denser
Sparse
Denser Few loci
Marker map Sparse
Coverage
Map resol.
resol.
Genome wide
Poor
Better
Within family
Genome wide
Poor
Better
Population wide
>>> 1 round
Small
Local
Dense
Genome
High
LDLD-LA analysis – see later 30
15
4.5
-logP
4.0
3.5
3.0
2.5
2.0
1.5
1.0
0.5
0.0
cM 0
10
20 30
40
50
60 70
80
90 100 110 120 130
Summary
and
Conclusions
• Several population designs and statistical methods are available
to map the QTL landscape
• Most studies to date have used
• line crosses
• within family linkage
Æ Broad QTL peaks
• Candidate gene analyses Æ Single sharp peak
• New technology enables genomegenome-wide LD mapping
Æ Many sharp peaks
some will stand the test of time but many will crumble . . . .
But do we really care where the peaks are – all we need is a good
predictor of breeding value / phenotype . . . . Æ Genomic Selection
31
16




