UNIT 3: Molecular Genetics 6.4: DNA Replication and Repair
advertisement

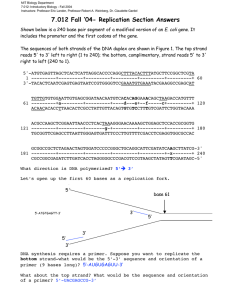
UNIT 3: Molecular Genetics Chapter 6: DNA: Hereditary Molecules of Life pg. 268 6.4: DNA Replication and Repair pg. 282 - 290 The DNA molecule is capable of replicating on its own. This is important for the reproduction of cells, ensuring there is an exact copy of the parent DNA is found in the daughter cells. DNA can be millions of nucleotides in length and must occur quickly. DNA Replication is Semiconservative Semi-conservative replication – is a mechanism of DNA replication, in which each of the two strands of a parent DNA is incorporated into two new double stranded DNA molecules. Figure 1: a) in conservative replication, the two parental strands would act as templates for replication, but then recombine afterwards. b) Disperse replication c) in semi-conservative replication the two parental strands would act as templates for replication and remain separated form each other, incorporated into new molecules. Meselson and Stahl (1958) performed an experiment to demonstrate and support the idea that replication is semiconservative. Radio-isotopes (15N) were used to tract DNA synthesis. They saturate many generation of Bacteria with 15N. This guaranteed that the sample generation would have DNA with 15N incorporated in it. They then took a sample of bacteria 15N and added it to a medium with regular nitrogen (14N) and allowed two generation to reproduce. It was assumed that the new generations would contain 14N. To separate material, a centrifuge was used, separating material by density. 15 N would be the heaviest, a hybridization of 15N and 14N intermediate, and 14 N the lightest. The results from the centrifuge indicated that the new generations of DNA were actually a hybridization of 15N and 14N, and a double strand of 14N. This supported DNA replication is semiconservative. Figure 2: Results of Meselson and Stahl’s experiment: a) position of the original parent heavy 15N DNA, b) position of hybrid DA produced after on round of DNA replication, and c) positions of DNA formed after two rounds of replication. pg. 283 DNA Replication: The Process Most detailed research has been on the prokaryotic cells (bacterium E. coli); eukaryotes cells follow a similar pattern. Since the eukaryote’s DNA in strands replication is more complicated. DNA replication has three steps: parental DNA strands separate, complementary stands assembled, and new strands are proofread. Step 1: Strand Separation Replication origin – is a specific sequence of DNA that acts as the starting point for replication. Helicase – is a replication enzyme that separates and unwinds the DNA sequence. Replication Fork – is the point of separation of the two parent DNA strands during replication. Topoisomerases – is a class of enzymes that relieve tension caused by the unwinding of parent DNA; they cleave one or two of the DNA strands, allow the strand(s) to untwist, and then rejoin the strand(s). Single-strand binding protein (SSB) – is a replication enzyme that prevents parent DNA strands from annealing to each other once they have been separated by helicase. Replication bubble – is the sequencing of DNA in both directions during replication. DNA strands must unwind from each other, the replication origins are the starting points. There’re multiple replication origins coded into the genome, to speed up the process of replication. The enzyme helicase binds to the replication origins and begins the process of unwinding DNA. To unwind the DNA the hydrogen bonds between complementary base pairs are broken. As the bonds break the gap creates a replication fork. Replication forks ring in opposite directions. The DNA molecule wants to form the double helix again, enzymes called topoisomerases are use to release the tension in the DNA strands, preventing them from rejoining. Single-stranded binding proteins (SSBs) are used to prevent the annealing of the two strands, preventing them from reattaching. These proteins are removed later and reused when necessary. The replication forks run in both directions. As the strands are separated new complementary nucleotides are added. The area created between the two replication forks and where new nucleotides are attached, is called the replication bubble. These bubbles expand in opposite direction until it merges with another bubble. The multiple replication sites allows for the DNA molecule to be replicate quickly. New DNA is created at a rate of 50 base pairs per second. If there was only one site it would take about a month to replicate. With multiple replication origins it takes 1 hour to replicate. Figure 4: Replication bubbles form. grow, and merge. Step 2: Building Complementary Strands Nucleotide triphosphate – is a building block and energy source for replicating DNA. RNA Primase – is a replication enzyme that produces RNA primer. RNA Primer – is a replication molecule that acts as a starting point for replication. DNA Polymerase III – is a prokaryotic replication enzyme that builds new DNA strands from nucleotides. Leading Strand – is the DNA strand that is copied in the direction toward the replication fork. Lagging Strand – is the DNA strand that is copied in the direction away from the replication fork. DNA Polymerase I – is a prokaryotic replication enzyme that fills in gaps in the lagging strand between Okazaki fragments; also proofreads the final strands. DNA Ligase – is an enzyme that catalyzes the formation of a phosphodiester bond between two DNA strands, as well as between Okazaki fragments. Following description applies to prokaryotic cells, eukaryotic cells have more specialized DNA polymerases, and system is quite complex. DNA polymerases are enzymes used to anneal new nucleotides to the exposed template strand. DNA polymerase reads the template and nucleotides are added to the 3' (3 prime) to the 5' (5 prime) direction, building a new strand running 5' to 3'. Nucleoside triphosphate is the energy source used by cell’s polymerase to build the complementary strand to the template strand, and the structural nucleotide. Nucleoside triphosphate consists of a three phosphate groups, deoxyribose sugar, and one of four nitrogenous bases. The nucleoside triphosphate combines to the on going new strand, releasing two phosphate groups, while the third bonds to the deoxyribose sugar group of the previous nucleotide forming a phosphodiester bond, extending the backbone while the nitrogenous base forms hydrogen bonds with the adjacent complementary nitrogenous base. Energy is supplied by the releasing of the two phosphate groups. To start the process of producing the new strand, replication bubble exposes the template strands. Since replication runs can only occur by attaching nucleotides to 3' end of an existing strand RNA primers are attached to the template strand by an enzyme called RNA Primase. These newly added RNA primers consist of 10 – 60 ribonucleotides. The replication forks which round in two directions within the bubble. At each replication fork the template strands are round in opposite direction. One replication fork is running 3' (3 prime) to the 5' (5 prime) direction, while the other is running 5' (5 prime) to 3' (3 prime) the direction. DNA polymerase III is responsible for building the new strand. The DNA polymerase III starts to build the new strand from the RNA primer in the 5' to 3' the direction, creating a leading strand proceeding towards the fork as it opens. The new strand that is built in the 3' to the 5' direction is called the lagging strand and requires a number of RNA primers to be put in place as small segments are being built. The portion that is built with a series of RNA primers and short DNA fragments that are called Okazaki fragments are approximately 1000 to 2000 nitrogen bases. The RNA primers must be removed and nucleotides must replace them, connecting Okazaki fragments together, completing the new strand. DNA polymerase I are responsible for this process of filling the gaps between Okazaki fragments and DNA ligase is the enzyme used to catalyze this reaction. Figure 6: DNA Synthesis process, pg. 287 Step 3: Dealing with Errors during DNA Replication DNA Polymerase II – is a prokaryotic replication enzyme that repairs damage to DNA, including damage that occurs between replication events. The newly built DNA strands need to be proofread for errors and corrected. DNA polymerase II is an enzyme responsible for this process. The average for errors is approximately one in every one million base pairs. These repair complexes, composed of proteins and enzymes, move along the strands looking for errors which distort the DNA molecule, and make the correction by removing the mismatched portion and adding the correct nucleotides. DNA Replication in Prokaryotes and Eukaryotes Prokaryote and Eukaryotes replication is similar but there are some differences. Prokaryotes Eukaryotes Small Genome Smaller Ringed Shaped One Replication Origin & Bubble DNA Polymerases less varied Large Genome Many Long and Linear Strands Many Replication Origins & Bubbles DNA Polymerases are more varied