COMP 571 Bioinformatics: Sequence Analysis Homework Assignment #2 Date Assigned: 28 September 2010.
advertisement

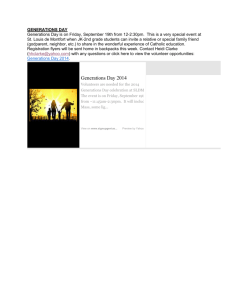
COMP 571 Bioinformatics: Sequence Analysis Homework Assignment #2 Date Assigned: 28 September 2010. Date Due: 7 October 2010. 1. Suppose that a diploid population of size 50 undergoes a change in average heterozygosity across loci from 0.50 to 0.42 in a single generation. Is it plausible to attribute this magnitude of change to random genetic drift? Justify your answer. 2. An isolated population loses half its heterozygosity in 30 generations. What is its effective population size? 3. Show that approximately 2N generations of random genetic drift are required to reduce the number of segregating genes (heterozygotes) by a factor of e (e = 2.71828...), given initial allele frequencies close to 0.5. 4. In a haploid population of constant effective size 50, what is the probability that two randomly drawn alleles shared a common ancestor exactly 100 generations ago? 5. In a population of effective population size 30, how many generations are required on the average to coalesce from 4 alleles to 3? From 3 alleles to 2? From 2 alleles to 1? 6. Consider the evolutionary history of three populations A, B, and C, shown in Figure 1. Using the coalescent model, compute the probabilities of each of the following three scenarios for alleles a, b, and c, randomly drawn from A, B, and C, respectively: (a) a and b coalesce first, and then their MRCA coalesces with c. (b) a and c coalesce first, and then their MRCA coalesces with b. (c) b and c coalesce first, and then their MRCA coalesces with a. t2 t1 A B C Figure 1: An evolutionary history of three populations A, B, and C. The effective population size is constant and equals N . The times t1 and t2 are given in numbers of generations. 1