Gene Ontology - A Way Forwards Ruth Lovering, Varsha Khodiyar,
advertisement

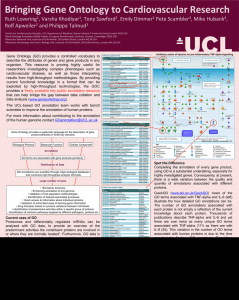
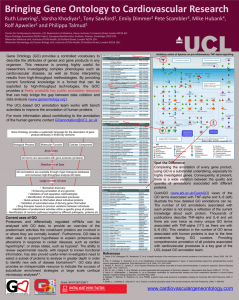
Gene Ontology - A Way Forwards Ruth Lovering, Varsha Khodiyar, Pete Scambler, Mike Hubank, Rolf Apweiler and Philippa Talmud Centre for Cardiovascular Genetics, UCL Department of Medicine, Rayne Institute 5 University Street London WC1E 6JF. Molecular Medicine Unit, Institute of Child Health, 30 Guilford Street, London WC1N 1EH. Molecular Hematology and Cancer Biology Unit, Institute of Child Health, 30 Guilford Street, London WC1N 1EH. European Bioinformatics Institute, Hinxton, Cambridge, CB10 1SD. (TNF alpha) Inhibitory action of lipoxins on pro-inflammatory TNF-alpha signalling The UCL-based GO annotation team aims to work with bench scientists to improve the annotation of human proteins. Improvements in the GO annotation of your favourite protein will lead to an improved public resource for everyone. Proteomes and differentially regulated mRNAs can be analysed with GO data, to provide an overview of the predominant activities the constituent proteins are involved in or where they are normally located1. Furthermore, often the generation of hypotheses to explain proteome-wide alterations in response to certain diseases, such as cardiac hypertrophy2, or stress states, such as hypoxia3, relies on the use of GO annotation data. The ability to review experimental results, with respect to known functional information, has also proved useful when investigators need to select a subset of proteins to analyse in greater depth in order to identify new sets of disease biomarkers4,5. GO data also provides an indispensable resource to indicate the success of subcellular enrichment strategies or large scale confocal microscopy analyses6,7. Already, drug treatments are being tailored according to molecular pathway imbalances, detected through individual-specific microarray or proteomic data. PTPN11 For more information about contributing to the annotation of the human genome contact GOannotation@UCL.ac.uk (IKBKG) Gene Ontology provides a systematic language for the description of gene product attributes in three key domains MAP3K14 (CHUK) (SFN, YWHA family) Cellular Component FOXO1 CDKN1B CCNE1 (NFKB1) (IL-6) Annotation GO terms are associated with gene products (proteins) MetaCore Map, GeneGO, www.genego.com (CCNE1, Cyclin E1) Distribution of Data GO annotations are available through major biological databases and numerous high-throughput analysis GO tools Large number of uses • Biomarker discovery • Enhancing annotation of any genome • Validation of cell separation methodologies • Identification of disease-associated processes • Quick access to information about individual proteins • Validation of automated ways of deriving gene information • Drug therapies based on process variations between individuals • Identification of predominant activities within a specific group of proteins • Identification of common pathways targeted by different pathogens, proteins etc Grant: SP/07/007/23671 www.cardiovasculargeneontology.com Spot the Difference KEY Activation Inhibition Unspecified Cytoplasm Extracellular Plasma Membrane Nucleus B Binding CR Class relation CS Complex subunit IE Influence on expression +P Phosphorylation TR Transcription regulation Z Catalysis Associated with Cardiovascular Disease Kinase Phosphatase Phospholipase Protein Transfactor Molecule Phospholipid Ligand Binding protein Receptor GPCP Protein Family Completing the annotation of every gene product, using Gene Ontology (GO), is a substantial undertaking, especially for highly investigated genes. Consequently, at present, there is a wide variation between the quality and quantity of annotations associated with different proteins. QuickGO (www.ebi.ac.uk/ego) views of the GO terms associated with TNF-alpha, IL-6 and CCNE1 (above) and the histogram, to the right, illustrate the variation in the number of unique GO terms associated with human proteins. This variation is not simply a reflection of the current knowledge about these proteins. Thousands of publications describe TNF-alpha and IL-6 and yet there are over twice as many GO terms associated with TNFalpha (68) as there are with IL-6 (28). This difference is due to the time constraints facing GO curators. At present there are only 2 projects (funding 4 curators) that prioritise the comprehensive annotation of human genes. IL-1B, IL-6, PTPN11 and TNF-alpha have been prioritised for annotation by the Cardiovascular GO Annotation Initiative, however, of these only TNF-alpha has been annotated by this project, to date. The quality of annotations also varies between proteins. Proteins annotated mostly through automated methods tend to have more general GO terms (see CCNE1). Whereas, proteins with annotations made by GO curators, based on published experimental evidence, tend to have more specific GO terms (see TNF-alpha). References 1. Pasini, E.M., Kirkegaard, M., Mortensen, P., et al. In-depth anyalysis of the membrane and cytosolic proteome of red blood cells. Blood, 2006, 108, 791-801. 2. Pan, Y., Kislinger, T., Gramolini, A. O., et al. Identification of biochemical adaptations in hyper- or hypocontractile hearts from phospholamban mutant mice by expression proteomics, Proc Natl Acad Sci U S A, 2004, 101: 2241-2246. 3. Boraldi, F., Annovi, G., Carraro, F., et al. Hypoxia influences the cellular cross-talk of human dermal fibroblasts. A proteomic approach, Biochim Biophys Acta, 2007, 1774: 1402-1413. 4. Shi, M., Jin, J., Wang, Y., et al. Mortalin: a protein associated with progression of Parkinson disease?, J Neuropathol Exp Neurol, 2008, 67: 117-124. 5. Perco, P., Wilflingseder, J., Bernthaler, A., et al. Biomarker candidates for cardiovascular disease and bone metabolism disorders in chronic kidney disease: A systems biology perspective, J Cell Mol Med, 2008. 6. Kislinger, T., Rahman, K., Radulovic, D., et al. PRISM, a generic large scale proteomic investigation strategy for mammals, Mol Cell Proteomics, 2003, 2: 96-106. 7. Barbe, L., Lundberg, E., Oksvold, P., et al. Toward a confocal subcellular atlas of the human proteome, Mol Cell Proteomics, 2008, 7: 499-508. Number of publications and GO terms associated with lipoxins/TNF-alpha signalling pathway proteins 80 6 70 Unique GO terms Publications 5 60 4 50 40 3 30 2 20 1 10 0 0 Gene Symbol Log Number of Publications Molecular Function High-throughput technologies and research into multi-factorial diseases are also highlighting how highly investigated proteins in one field of biology are relevant to processes associated with another field of biology. For example, in the central figure, several genes (IL-6, IL-8, STAT3 and TNF-alpha) are associated with the TNF-alpha pro-inflammatory signalling pathway and are also associated with cardiovascular disease. NFKBIE ERLIN1 PPAPDC2 YWHAQ YWHAZ NFKBIB PIK3R2 TNFRSF1 YWHAE AKT2 MAP3K14 AKT3 CCNE1 YWHAB YWHAG FPRL1 PIK3CA TRADD FOXO1 YWHAH PIK3CD TRAF2 CDKN1B CHUK IKBKB NFKB1 PDPK1 RIPK1 SOCS1 IKBKG JAK1 PLD1 SFN TNFRSF1 STAT3 PIK3CB NFKBIA CDK2 IL8 PRKCZ PIK3R1 RELA IL6 IL1B PTPN11 AKT1 TNF (NFKB1A) Biological Process How is GO used? Number of Unique GO Terms Gene Ontology (GO) provides a controlled vocabulary to describe the attributes of genes and gene products in any organism. This resource is proving highly useful for researchers investigating complex phenotypes such as cardiovascular disease, as well as those interpreting results from high-throughput methodologies. By providing current functional knowledge in a format that can be exploited by high-throughput technologies, the GOC provides a freely available key public annotation resource that can help bridge the gap between data collation and data analysis (www.geneontology.org).