LECTURE 6: AFM IMAGING II : ARTIFACTS AND APPLICATIONS Outline :
advertisement

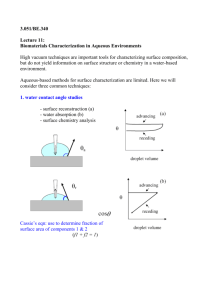
3.052 Nanomechanics of Materials and Biomaterials Tuesday 02/27/07 Prof. C. Ortiz, MIT-DMSE I LECTURE 6: AFM IMAGING II : ARTIFACTS AND APPLICATIONS Outline : LAST TIME : BASIC PRINCIPLES OF ATOMIC FORCE MICROSCOPY ................................................ 2 FACTORS AFFECTING RESOLUTION .................................................................................................... 3 Tip Deconvolution .................................................................................................................... 4 IMAGING OF CELLS ................................................................................................................................. 5 IMAGING OF DNA .................................................................................................................................... 6 HRFS COMBINED WITH AFM : SPATIALLY SPECIFIC SURFACE INTERACTIONS............................. 7 Objectives: To review artifacts present in atomic force microscopy imaging and current state of the art uses. Readings: Course Reader Document 12-15. Multimedia : Listen to "Structured Water" Podcast corresponding to journal article : Higgens, et al. Biophys. J. 2006 91, 2532. 1 3.052 Nanomechanics of Materials and Biomaterials Tuesday 02/27/07 Prof. C. Ortiz, MIT-DMSE ATOMIC FORCE MICROSCOPY : GENERAL COMPONENTS AND FUNCTIONS laser diode mirror A B C D cantilever sensor output, δ, F -Basic principles position sensitive photodetector • spring which deflects as probe tip scans sample surface δ probe tip • senses surface properties and causes cantilever to deflect ≈10°-15° -Deflection vs. Height images • measures deflection of cantilever -2D Section Profiles ERROR = actual signal - set point sample -3D Images -Normal modes of operation (Contact AC/DC, Tapping AC, Noncontact AC) computer • controls system • performs data acquisition, display, and analysis piezoelectric z y scanner • positions sample x feedback loop • controls z-sample position -Other modes of operation (LFM, CFM, Force/Volume) -DEMOs (x, y, z) with Å accuracy Advantages : 1) Unlike electron microscopes, samples do not need to be coated or stained, minimal damage, 2) Unlike electron microscopes, samples can be imaged in fluid environments (near-physiological conditions), 3) Unlike STM samples do not need to be conductive, 4) Sub-nm resolutions have been achieved on biological samples (detailed information on the molecular conformation, spatial arrangement, structural dimensions, rate dependent processes, etc.) 2 3.052 Nanomechanics of Materials and Biomaterials Tuesday 02/27/07 Prof. C. Ortiz, MIT-DMSE Hoh, et al. Biophys. J. 1998, 75, 1076. D+ΔD d -Z D L+ΔL ~ +Z L voltage applied -X +Y +X Force, F (nN) ATOMIC FORCE MICROSCOPY IMAGING : FACTORS AFFECTING RESOLUTION SPECIMEN PIEZO AMPLIFIER, SENSOR AND ADHESION FORCE DEFORMATION & CONTROL ELECTRONICS, Yang, et al. Ultramicroscopy 1993, 50, 157 MECHANICAL PARAMETERS THERMAL Physik Instruments, Nanopositioning 1998 FLUCTUATIONS y Distance, D (nm) x connecting wires polarization PROBE TIP SHARPNESS CANTILEVER THERMAL NOISE Sheng, et al. J. Microscopy 1999, 196, 1. Lindsay Scanning Tunneling Microscopy and Spectroscopy 1993, 335. kt Shao, et al. Ultramicroscopy 1996, = 66, 141. Image removed due to copyright restrictions. m cantilever Fadhesion 0 z electrodes 0 δt(max) δt(max) m Image removed due to copyright restrictions. 3-D model of sharp probe tip on a protein, from Lieber et al, 2000 (http://cnst.rice.edu) m 3 3.052 Nanomechanics of Materials and Biomaterials Tuesday 02/27/07 Prof. C. Ortiz, MIT-DMSE ATOMIC FORCE MICROSCOPY IMAGING : TIP DECONVOLUTION -Imaging very sharp vertical surfaces is influenced by the sharpness of the tip. Only a tip with sufficient sharpness can properly image a given z-gradient. Some gradients will be steeper or sharper than any tip can be expected to image without artifact. False images are generated that reflect the self-image of the tip surface, rather than the object surface. Mathematical methods of tip deconvolution can be employed for image restoration. The effectiveness of these methods will depend on the specific characteristics of the sample and the probe tip. probe tip probe tip scan direction square nanoobject 2D Height Profile θ θ z w/2 substrate x height would theoretically be accurate 2D Height Profile r* tan( θ) = x → x = wtan( θ) w w + wtan( θ) 2 r* 1 = + tan( θ) w 2 -Deep feature -Depth underestimated -Need high aspect ratio probe tip. r* = -tip broadening-width is overestimated → mention pset 4 3.052 Nanomechanics of Materials and Biomaterials Tuesday 02/27/07 Prof. C. Ortiz, MIT-DMSE AFM IMAGING OF BIOLOGICAL MACROMOLECULES: DNA Image of PtyrTlac supercoiled DNA. 750 nm scan courtesy C. Tolksdorf, Digital Instruments/Veeco, Tapping Mode image of nucleosomal DNA. Courtesy of Yuri Lyubchenko. Used with permission. Santa Barbara, USA, and R. Schneider and G. Muskhelishvili, Istitut für Genetik und Mikrobiologie, Germany. Courtesy of Veeco Instruments and G. Muskhelishvili. Used with permission. Courtesy of Zhifeng Shao. Used with permission. http://people.virginia.edu/~zs9q/zsfig/DNA.html The high resolution of the SPM is able to discern very subtle features such as these two linear dsDNA molecules overlapping each other. 155nm scan. Courtesy of W. Blaine Stine. Used with permission. AFM image of short DNA fragment with RNA polymerase molecule bound to transcription recognition site. 238nm scan size. Courtesy of Bustamante Lab, Chemistry Department, University of Oregon, Eugene OR Courtesy of Prof. Carlos Bustamante. Used with permission. Courtesy of Zhifeng Shao. Used with permission. 5 3.052 Nanomechanics of Materials and Biomaterials Tuesday 02/27/07 Prof. C. Ortiz, MIT-DMSE HRFS COMBINED WITH AFM : SPATIALLY SPECIFIC SURFACES INTERACTION INFORMATION Courtesy Elsevier, Inc., http://www.sciencedirect.com. Used with permission. (Vandiver, et al. Biomaterials 26 (2005) 271–283). 6 3.052 Nanomechanics of Materials and Biomaterials Tuesday 02/27/07 Prof. C. Ortiz, MIT-DMSE SUPPORTED LIPID BILAYERS http://faculty.virginia.edu/tamm/pages/project_support.html Courtesy of Lukas K. Tamm and the Biophysical Society. Used with permission. See Wagner, M., and L. K. Tamm. Biophysical Journal 79 (2000): 1400-1414. http://en.wikipedia.org/wiki/Image:Cell_membrane_detailed_diagram.svg DPPC Higgens, et al. Biophys. J. 2006 91, 2532. DOPC Courtesy of the Biophysical Society. Used with permission. 7 3.052 Nanomechanics of Materials and Biomaterials Tuesday 02/27/07 Prof. C. Ortiz, MIT-DMSE NANOMECHANICS OF SUPPORTED LIPID BILAYERS Higgens, et al. Biophys. J. 2006 91, 2532. Courtesy of the Biophysical Society. Used with permission. 8