Document 13518722
advertisement

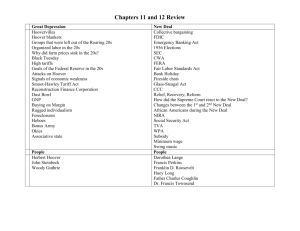
12.158 Lecture 1
Mass Spectrometry Some Fundamental Background Information for Organic Geochemists and Geobiologists
Reference:
McLafferty & Tureček
Interpretation of Mass Spectra 4th Ed
http://i-mass.com/
1
Contents
•Mass Spectrometers and their component
parts, things you need to know
•Formation of mass spectrum
•Interpretation of mass spectra
•Detection of specific compounds using ion
chromatograms
•Selected ion recording (SIR)
•Multiple Reaction Monitoring (MRM and
MRMQ)
•LC-MS
2
Components of the Mass Spectrometer (1)
•Vacuum System
•any collisions of an ion with a gas molecules in its path
destroys it --> no MS
•System to convert intact molecules to ions THE ION SOURCE
•electron impact (EI), chemical ionisation (volatiles)
• electrospray or sputtering (non-volatiles eg proteins,
DNA)
•Mass Analyser - To filter by mass (and KE)
•Ion Detector and ion current amplifier
•Display mechanisms and Data Acquisition System
3
This image has been removed due to copyright restrictions.
Image from Micromass Manual.
AUTOSPEC Mass Spectrometer
4
Example: Vacuum System Schematic for Double Focussing MS VG 70E
This image has been removed due to copyright restrictions.
Image from Micromass Manual.
5
Source Options: Micromass Autospec
This image has been removed due to copyright restrictions.
Image from Micromass Manual.
Outer: lenses and heater
This image has been removed due to copyright restrictions.
Image from Micromass Manual.
Inner: ion chamber, filament, repeller; easily cleaned/swapped 6
Components of the Mass Spectrometer (2)
•Mass Analyser - To filter by mass (and KE)
•single magnet or quadrupole --> low resolution ~ 1 dalton •double sector- magnet plus electrostatic analyser --> high resolution --> accurate mass to 1-2ppm -->
elemental composition by mass defect
•multiple sector --> hybrid such as Autospec Q or triple
quadrupole (TSQ) --> target compound analysis by
reaction chromatography
•time of flight (TOF), ion cyclotron resonance
•Ion Detector and ion current amplifier
•Display mechanisms and Data Acquisition System
7
Components of the Vacuum System
•1st Stage --> Rotary pump(s)
•monitored by Pirani guage (pump to 10-2 torr)
problems - oil level, bearings, drive, dirty oil
•2nd Stage --> Oil Diffusion pump(s)
•monitored by ion guage (pumps to 10-8 torr)
•70E and Autospec are differentially pumped to give
better vacuum in the analyser and hence better resolution
problems - oil level, cooling water, vacuum
interlocks
•Isolation valves to separate pumps, source, analyser
problem - interlocks
•2nd Stage can be turbomolecular pump(s) (pumps to 10-9 torr)
8
Components of the Ion Source
•Ion volume (or block) – Autospec accelerating
potential 6-8kV
•space to form the ions problems - dirt, high voltage
•Filament
•hot tungsten wire creates an electron beam
problems - broken, misaligned, bad contacts, low emission
•Repeller and focus places
•causes ions to exit source as collimated beam
problems - bad contacts, ion burns
•Exit slit
•causes ion beam to enter analyser with tightly defined dimensions problems - ion burns, slide mechanism
9
Components of the Analyser
•Electrostatic analyser (2 for Autospec)
•Filters ions for kinetic energy. Narrow energy
window allows magnet to gives less dispersion and
better separation
•Magnetic analyser
•Electromagnet driven by a high current
problems - current control, cooling water, interlocks
•Collision cells - fill with gas to cause fragmentation
•Collector slit
•causes ion beam to enter detector with tightly
defined dimensions
10
Components of the Mass Spectrometer (3)
•Ion Detector (must be fast response)
•Off axis detectors so need deflector plate to bend beam •Electron multiplier (HP) continuous or discrete dynode •Photomultiplier (Autospec)
•Preamplifier and amplifier --> gain and noise controls problems - slow degradation, noise
•System for Display and Acquisition
•Analogue to digital conversion noise filtering and mass
measurement
•data storage, manipulation and presentation
computer bugs, software evolution
11
Separate physical and electronic bits
•Physical bits need to be clean
•most problems come down to dirt of some kind or
other
•most problems occur in the GC or in the source •Develop skills to distinguish dirt and other
contamination from electronic faults which are much
harder to resolve
12
Inlet Systems
• Compound must be volatile for EI and
CI
• Gas Reservoir (Hot Inlet System)
• Solids Probe
• Gas Chromatograph
• Liquid Chromatograph
13
Ionization Methods
• Electron Impact
Ionization
– High energy, fragmentation = information • Chemical Ionization
– Low energy collisions with a gas such as CH4,
NH3
• Electrospray LC-MS
• Atmospheric Pressure CI LCMS
14
15
Formation of the mass spectrum
•Compound ionises in source
•Molecular ions fragments as a result of
excess energy imparted by the ionising
electrons (standard energy 70eV)
•Ions pushed from source with repeller
•Accelerated by high voltage
•Ions separated by mass analyser
•Ions counted and recorded
16
Calibration and leak checking
•Perfluorokerosene (PFK) - a mixture of
fluoroalkanes with ions to > 900 dalton
69, 100, 119, 219, 231 ……..
•Heptacosafluorotributylamine - a single compound MW 614 with 69, 119, 219 ... •Background spectrum should look like air; •17,18,28-32 and 40 Da
•Argon useful to detect leaks
17
Perfluorokerosene (PFK) - a mixture of fluoroalkanes with ions to > 900 dalton
18
m/z Rel. Abundance Formula Exact Mass ------- -------------- ------- ---------18.01
30.00
28.01
50.00
31.0
3.80
CF
30.99840
39.96
1.70
51.00
6.70
69.00 100.00
C1F3
68.99521
81.00
0.50
C2F3
80.99521
84.97
1.00
93.00
3.30
C3F3
92.99521
99.99
5.60
C2F4
99.99361
118.99
26.40
C2F5
118.99201
130.99
24.00
C3F5
130.99
19
Ionisation & Fragmentation Processes
CH3OH + e- --> CH3OH+. (m/z 32) + 2 e- ionisation
sigma cleavage
CH3OH +. --> CH2OH+ (m/z 31) + H.
sigma cleavage
CH OH +. --> CH + (m/z 15) + . OH
3
3
CH2OH +--> CHO+ (m/z 29) + H2
neutral loss
N.B. Only charged species detected
31
100
M+.
50
15
10
13C
20
20
30
Characteristic ion series
•Silicone bleed (silicon isotopes) 207, 281,
355
• Alkanes 57, 71, 85, 99, 113 ... •Alkenes 55, 69, 83, 97, 111 … •Cyclohexanes 83, 97, 111
•Terpanes 123, 149, 191, 205, 217, 231 •Monoaromatics 77, 91, 105, 119, 133 …..
21
Ionisation & Fragmentation Processes
CH3OH + e- --> CH3OH+. (m/z 32) + 2 e- ionisation
sigma cleavage
CH3OH +. --> CH2OH+ (m/z 31) + H.
sigma cleavage
CH OH +. --> CH + (m/z 15) + . OH
3
3
CH2OH + --> CHO+ (m/z 29) + H2
neutral loss
H
H
C
O
H
22
H
Appearance of the Mass Spectrum
•Average spectrum has 250 bits of information
•Molecular ion and its isotopic abundances
•Ionisation energy and structure determine degree
of fragmentation; 70 eV is standard
•Fragment abundances reflect to relative stabilities
of the fragment ions - most abundant are the most
stable
•Multiply charged and metastable ions lost in data
reduction
23
Interpretation of the mass spectrum
•Identify molecular ion
•Assess isotope abundances for:
A elements H and F
A+1 elements C, N
A+2 elements Si, S, Cl, Br
•Determine # rings and double bonds
Acyclic saturated hydrocarbons CnH2n+2
•Identify characteristic ions and low mass
series
24
Interpretation of the mass spectrum
•Identify molecular ion
–Molecular Ion corresponds to the loss of one
electron from the intact molecule
–Molecular ion is an odd electron ion
–Odd electron ions are generally present and have
an even mass unless an odd number of nitrogens 25
Identify:
1947 sats
Molecular Ion? 64
100
Isotopic features?
Ion series?
%
256
128
160
96
192
66
57
0
50
60
71
70
98
85
80
90
100
110
120
130
140
150
160
170
180
26
190
200
210
220
230
240
250
260
270
280
290
m/z
300
Nuclidic Masses and Abundances
Element
Mass
Isotope Abundance
carbon
12.00000
13.0034
98.9
1.1
hydrogen
1.007825
2.0140
99.985
0.015
nitrogen
14.00307
15.0001
99.63
0.37
oxygen
15.9949
17.9992
99.8
0.2
fluorine
18.9984
100
sulfur
31.9720
32.9715
33.9679
94.8
0.8
4.4
27
This table has been removed due to copyright restrictions.
Please see Table 2.1
©McLafferty & Tureček: Interpretation of Mass Spectra 4th Ed
28
This table has been removed due to copyright restrictions.
Please see Table 2.2
©McLafferty & Tureček: Interpretation of Mass Spectra 4th Ed
29
This table has been removed due to copyright restrictions.
Please see Table 2.3
©McLafferty & Tureček: Interpretation of Mass Spectra 4th Ed
30
GC-MS
Commonly done on low resolution
(1 dalton) quadrupole instruments in the
selected ion (mass) mode.
e.g. Hewlett-Packard MSD, Thermo,
Shimadzu
Can be done at high resolution; accurate
mass
31
What has to be right for successful GC-MS? Every single time you try!!
•GC working with correct column in good condition, right gas
flows, autosampler working, correct conditions of injection, temp
program, hot interface, no leaks, no bleed, clean injector, clean
syringe, clean wash solvents, correct vials and septa ……...
•Good vacuum, clean source, right source temp and ionising
conditions, appropriate peak resolution & tune, correct slits,
correct gain setting, low instrument noise ……..
• DS interface (noise, threshold) set correctly, correct experiment
for the job, instrument in calibration, file structure correct,
adequate storage space, no bugs before starting sequence ………
•Samples correctly ordered in sampler, blanks & external
standards included, data archived , QUAN set up correctly ….
•Interpretive skill ….
32
Familiarity with the instruction manual
•Dont even think about operating a mass spectrometer
without being familiar with the manual!
•If it is not clear, ask or read on!
•Be familiar with the emergency shutdown procedures •Be familiar with OH&S issues (eg High Voltages)
•Mistakes are inevitable and part of the scenery
tell your colleagues immediately and log it so that
they can be avoided by others
get help or do what you can to fix the problem
collegiality is critical when machines are shared
33
A7NO12A#3-1007
Identify
100 % 55
Molecular ion
69
80
Odd electron ions; A+1 or A+2?
83
60
low mass series
97
#carbons
40
111
20
rings & double bonds
125
= 412
266
0
60
80
100
120
140
160
180
200
100 % 55
220
240
280
300
m/z
280
300
m/z
x10.00
69
80
260
266
83
60
97
40
111
20
154
168 182
125
238
196
0
60
80
100
120
140
160
180
34
200
220
240
260
1947 sats
100
57
71
%
85
55
99
113
83
127
141
67
0
50
60
70
80
90
100
110
120
130
140
155
150
160
240
169
183
170
180
35
197
190
200
211
210
220
230
240
250
260
270
280
290
m/z
300
68 sats
57
100
71
85
%
55
113
127
69
99
97
183
197
141
155
67
53
0
50
60
70
169
81
80
179
90
100
110
120
130
140
150
160
170
180
36
211
190
200
210
253
225
220
230
240
250
267
260
270
282
280
290
m/z
300
68 sats
57
100
71
85
55
%
113
69
99
183
127
97
67
53
0
50
60
70
141
155
81
80
169
90
100
110
120
130
140
150
160
197
170
180
37
190
200
225
210
220
230
239
240
253
250
268
260
270
280
290
m/z
300
1947 sats
100
57
71
%
85
55
99
69
113
83
127
141
67
0
50
60
70
80
90
100
110
120
130
140
155
150
160
254
169
183
170
180
38
197
190
200
211
210
225
220
230
240
250
260
270
280
290
m/z
300
1947 sats
100
57
71
%
85
281
69
99
83
113
97
127
141
155
169
183
67
0
50
60
70
80
90
100
110
120
130
140
150
160
170
180
39
197
190
200
211
210
253
225
220
230
240
250
295
260
270
280
290
m/z
300
1947 sats
57
100
55
97
%
71
69
83
67
81
111
99
125
53
79
65
0
50
60
70
80
91
90
105
100
110
119
120
139
153
168
131
130
140
150
160
183
170
180
40
197
190
200
207
210
224
220
239
230
240
252
250
260
270
280
290
m/z
300
Standard Interpretation Procedure
• Verify masses (calibration); determine elemental
compositions & rings+unsaturations
• Mark odd electron ions and verify molecular ion
• Study appearance of spectrum, molecular stability,
labile bonds
• Identify low mass ion series
• Identify neutral fragments, logical mass losses
• Postulate structures of low mass ions
• Postulate identity; test against ref. spectrum or library;
rationalize fragmentation pattern
©McLafferty: Interpretation of Mass Spectra
41
HPLC-ESI-MS data for Intact Polar Lipids Water Column Biomass or Sediment: Extract lipids (DCM: MeOH) Density Map
Mass
TLE
Coeluting peaks
make density map
preferable
component
separation by HPLC
Retention time ESI source
Mass spectrometer
42
Detection of microbial biomass by intact polar membrane lipid analysis in the water column and surface sediments of the Black Sea
Florence Schubotz1, Stuart G. Wakeham, Julius S. Lipp, Helen F. Fredricks, Kai-
Uwe Hinrichs
Environmental Microbiology11, 2720–2734, October 2009
This image has been removed due to copyright restrictions.
43
GC-MS of fossil hydrocarbons 44
αβ-Hopane a fossil hydrocarbon 369
Structure
191
MW = 412
191
100 %
Mass spectrum
95
81
68
50
149
109
123
137 163
177
100
150
369
206
200
250
300
45
350
412
397
400
450
500
217
362
347
NIST Chemistry WebBook (http://webbook.nist.gov/chemistry)
Triterpanes - 191 Da
C30 αβ
C29 αβ
Ts
C31 αβ
SR
Tm
Gippsland
C32 αβ
C33 αβ
C29Ts
NW Shelf
Dia
30-nor
47
Middle East
D4 Int. Std. File A6MA15C:4
SIM-GCMS: 221.221
Da
Scale Factor: 4.9
AGSO Standard
221 Da
100 %
90
80
70
60
50
40
30
20
10
0
50:00
55:00
1:00:00
1:05:00
TIME (min.) 48
1:10:00
1:15:00
100 %
90
AGSO Standard
234 Da
D3 Rec. Std.
File A6MA15C:4
SIM-GCMS: 234.232
Da
Scale Factor: 5.4
80
70
60
50
40
30
20
10
0
50:00
55:00
1:00:00
1:05:00
TIME (min.) 49
1:10:00
1:15:00
80
60
0
48:00
50
40
50:00
C28 βα 20S, 24S +R
C27 βα 20R
70
52:00
30
50
C28 ααα 20S
+ Bicad T
54:00
TIME (min.) 56:00
20
58:00
C30 ααα 20R
C29 ααα 20R
C28 αββ 20R
C28 αββ 20S
+ Bicad T1
C28 ααα20R +
Bicad R
C29 ααα 20S
C29 αββ 20R
C29 αββ 20S
C27 ααα 20R
C29 βα 20R
C30 βα 20S
C27 αββ 20S
C29 βα 20S
+C27 αββ 20R
100 %
Bicad W
C27 ααα 20S
C28 βα 20R, 24S + R
90
C27 βα 20S
File A6MA15C:4
SIM-GCMS: 217.196 Da
Scale Factor: 4.9
AGSO Standard
217 Da
10
1:00:00 51
C30 ααα 20R
C29 ααα 20R
C29 αββ 20R
C29 αββ 20S
C29 ααα 20S
C28 ααα20R + Bicad R
C28 ααα 20S
+ Bicad T
C28 αββ 20R
C28 αββ 20S
+ Bicad T1
C30 βα 20S
C29 βα 20R
C27 ααα 20R
Bicad W
C27 ααα 20S
C29 βα 20S
+C27 αββ 20R
C27 αββ 20S
C28 βα 20R, 24S + R
C28 βα 20S, 24S +R
C27 βα 20R
C27 βα 20S
AGSO Standard: steranes SIM vs. MRM
SIM-GCMS: 217 Da MRM-GCMS: 400 -> 217 Da
MRM-GCMS: 386 -> 217 Da
MRM-GCMS: 372 -> 217 Da
File A6MA15C:4
SIM-GCMS: 218.203
Da
Scale Factor: 4.1
90
80
60
50
C29 αββ 20S
C29 αββ 20R
C28 αββ 20R
70
C28 αββ 20S
100 %
C27 αββ 20S
C27 αββ 20R
AGSO Standard
218 Da
40
30
20
10
0
48:00
50:00
52:00
54:00
56:00
TIME (min.) 52
58:00
1:00:00
File A6MA15C:4
SIM-GCMS: 191.179
Da
Scale Factor: 6.4
AGSO Standard
191 Da
C24Tet
100 %
90
80
C23Tri
C19Tri
70
C21Tri
C24Tr
i
50
40
C22Tri
30
C26Tri(R + S)
C20Tri
C25Tri (R + S)
60
20
10
0
26:00 28:00 30:00 32:00 34:00 36:00 38:00 40:00 42:00 44:00 46:00 48:00 50:00 TIME (min.) 53
File A6MA15C:4
SIM-GCMS: 191.179
Da
Scale Factor: 25.1
AGSO Standard
191 Da
C29H
100
%
C30H
90
80
Compounds 1, 2 and 4 are Oleanoid Triterpanes (see
Murray et al. (1997), Geochim. Cosmochim. Acta, in
press. Tx is Taraxastane
70
60
Bicad W
10
0
50:00
C29 Ts
C31H
S
C31H
R
C31H βα
20
Tm
C30H Dia
C29H βα
30
Bicad T
Ts
29,30 C28H
C29 Dia
1,2 + 28,30 C28H
25-nor C29H+4
40
30-nor C30H
C30H βα
O
50
Tx
55:00
1:00:00
γ
C32H
S
C32H
C33H
R
S C33H
R
1:05:00
TIME (min.) 54
C34HS
C34HR
1:10:00
C35HS C35HR
1:15:00 File A6MA15C:4
SIM-GCMS: 369.352
Da
Scale Factor: 10.3
100 %
AGSO Standard
369 Da
Bicad T
W = cis-cis-trans Bicadinane
T = trans-trans-trans Bicadinane
T1,R = isomers of T
90
80
70
60
50
Bicad W
40
30
Bicad T1
20
Bicad R
10
0
50:00
55:00
1:00:00
1:05:00
TIME (min.) 55
1:10:00
1:15:00
File A6MA15C:4
SIM-GCMS: 123.117
Da
Scale Factor: 100
100 %
AGSO Standard
123 Da
8 β(H)-Homodrimane
90
80
70
60
50
40
30
20
10
0
10:00
14:00
18:00
22:00
26:00
TIME (min.) 56
30:00
34:00
38:00 File A6MA15C:4
SIM-GCMS: 205.194
Da
Scale Factor: 10.3
AGSO Standard
205 Da
C31 2MeH
100 %
90
80
70
60
C31 3MeH
50
40
30
20
10
0
56:00
58:00
1:00:00
1:02:00
TIME (min.) 57
1:04:00
1:06:00
File A6MA15C:4
SIM-GCMS: 231.212
Da
Scale Factor: 2.5
100 %
AGSO Standard
231 Da
Bicad T
90
80
70
Bicad W
C31 ααα 4Me20R
60
50
40
30
20
10
0
48:00
50:00
52:00
54:00
56:00
TIME (min.) 58
58:00
1:00:00
GC-MS-MS
True GC-MS-MS requires an instrument with
at least two, independently controllable mass
selection regions.
Various configurations possible, e.g. Triple
Sector Quadruple,
Hybrid magnet/
quadrupole etc. e.g. Autospec-Q, Finnagin
MAT 95Q etc.
Very high sensitivity and selectivity very
powerful for biomarker work.
59
60
C30 ααα 20R
C29 ααα 20R
C29 αββ 20R
C29 αββ 20S
C29 ααα 20S
C28 ααα20R + Bicad R
C28 ααα 20S
+ Bicad T
C28 αββ 20R
C28 αββ 20S
+ Bicad T1
C30 βα 20S
C29 βα 20R
C27 ααα 20R
Bicad W
C27 ααα 20S
C29 βα 20S
+C27 αββ 20R
C27 αββ 20S
C28 βα 20R, 24S + R
C28 βα 20S, 24S +R
C27 βα 20R
C27 βα 20S
AGSO Standard: steranes SIM vs. MRM
SIM-GCMS: 217 Da MRM-GCMS: 400 -> 217 Da
MRM-GCMS: 386 -> 217 Da
MRM-GCMS: 372 -> 217 Da
MRM-GC-MS
Metastable Reaction Monitoring GC-MS
Monitoring of ion reactions (e.g. MolecularDaughter) by selecting metastable ions in the
first field free region.
Not true
GCMSMS but allows selective
recording of biomarker molecular weight
groups (e.g. only C30 steranes cf. all steranes
at once).
61
MRM-GC-MS (2)
Metastable Reaction Monitoring GC-MS
Higher selectivity and sensitivity.
Can be done on double focussing, high
resolution instruments such as VG 70E etc
but parent ions can only be selected at low
resolution - some interference or cross talk
between traces.
62
90
AGSO Standard
C28
389 → 234 Da
8.6
3-CD3-5α-(H)-cholestane
100 %
80
70
60
Method recovery standard
50
40
30
20
10
0
48:00
50:00
52:00
54:00
56:00
63
58:00
1:00:00 90
C27 βα 20S
100 %
AGSO Standard
C27
372 → 217 Da
18.3
50
40
30
C27 ααα 20R
60
C27 ααα 20S
C27 αββ 20R
C27 αββ 20S
70
C27βα 20R
80
20
10
0
48:00
50:00
52:00
54:00
56:00
64
58:00
1:00:00
C28 ααα 20S
50
40
C28
386 → 217 Da
5.2
C28 ααα 20R
{
60
C28 αββ 20S
C28 αββ 20R
70
C28 βα 20R
80
C28 βα 20S
90
{
100 %
AGSO Standard
30
20
10
0
48:00
50:00
52:00
54:00
65
56:00
58:00
1:00:00
90
80
GCMS Internal Standard
C29
404 → 221 Da
13.4
70
60
D4-ααα-stigmastane 20R
100 %
AGSO
Standard
50
40
30
20
10
0
48:00
50:00
52:00
54:00
56:00
66
58:00
1:00:00
70
60
50
C29 ααα 20R
80
C29 βα 20R
90
C29
400 → 217 Da
13.4
C29 ααα 20S
C29 αββ 20R
C29 αββ 20S
100 %
C29 βα 20S
AGSO Standard
40
30
20
10
0
48:00
50:00
52:00
54:00
56:00
67
58:00
1:00:00
70
60
50
40
30
20
10
0
48:00
50:00
52:00
54:00
56:00
68
58:00
1:00:00
C30 ααα 20R
80
C30 αββ 20(R+S)
C30
414 → 217 Da
0.9
C30 ααα 20S
C30 βα 20R
90
C30 βα 20S
100 %
AGSO Standard
100 %
90
0
48:00
C30
414 → 231 Da
2.2
50:00
70
60
50
40
30
52:00
54:00
56:00
69
58:00
C30 3β Me 20R
C30 4α Me 20R + Dino
20
C30 4α Me 20S
C30 4α ββ Me 20R
C30 2α Me 20R
+ 4α Me ββ20S
C30 3β Me ββ 20R + S
80
C30 2αMe 20S
C30 3βMe 20S
AGSO Standard
10
1:00:00
90
AGSO Standard
C31
426 → 205 Da
4.7
C31 2α Me
80
70
60
C31 3β Me
100 %
50
40
30
20
10
0
56:00
58:00
1:00:00
1:02:00
70
1:04:00
1:06:00
File:A6MA16 #1-986 Acq:17-MAR-1996 00:09:04 GC EI+ MRM Autospec-UltimaQ
Sample Text:AGSOSTD 1/2A File Text:Ultra-1, 50m, H2@20psi
S:6 F:2
Exp:MRM_OZOILS_STD
100 %
90
Bicad T
C30
412 → 369 Da
100
AGSO Standard
80
70
60
50
30
Bicad W
20
Bicad R
Bicad T1
40
10
0
48:00
50:00
52:00
54:00
56:00
71
58:00
1:00:00
Ts
100 %
90
80
AGSO Standard
C27
370 → 191 Da
23.2
Tm + Bic T1
70
Bic T
60
50
40
C27 17β(H)
30
20
10
0
48:00
50:00
52:00
54:00
56:00
72
58:00
1:00:00 AGSO Standard
100 %
90
80
C28
384 → 191 Da
7.6
70
29,30 C28H
60
50
28,30 C28H
40
30
20
10
0
48:00
50:00
52:00
54:00
56:00
73
58:00
1:00:00
AGSO Standard
100 %
90
80
C29
398 → 191 Da
40.2
C29
H
70
60
50
40
30
C29 Dia
C29 Dianeo
C29Ts
20
10
0
48:00
50:00
52:00
54:00
56:00
74
C29H βα
58:00
1:00:00
C30H
AGSO Standard
100 %
90
80
C30
412 → 191 Da
35.4
Compounds 1 - 6 are Oleanoid Triterpanes (see Murray
et al. (1997), Geochim. Cosmochim. Acta, in press. Tx
is Taraxastane
70
60
50
40
C30H Dia
30
Oleanoid triterpanes
20
4
10
1
O
30-nor C30H
C30H βα
6
2 3
Tx
γ
0
56:00
58:00
1:00:00
1:02:00
75
1:04:00
1:06:00
90
80
AGSO Standard
C31HS
100 %
C31
426 → 191 Da
15.9
C31HR
70
60
50
40
C31Dia S
C31 Dia R
30
20
10
0
56:00
58:00
1:00:00
1:02:00
76
1:04:00
1:06:00
Note: This trace is from run A6FE24:5 using MRM-OZOILS_50
C34HS
100%
90
AGSO Standard
C34
468 → 191 Da
80
70
C34HR
60
50
40
30
20
10
0
45:00
50:00
55:00
1:00:00
77
1:05:00
Note: This trace is from run A6FE24:5 using MRM-OZOILS_50
C35HS
100%
90
AGSO Standard
C35
482 → 191 Da
80
70
60
C35HR
50
40
30
20
10
0
45:00
50:00
55:00
1:00:00
78
1:05:00
MIT OpenCourseWare
http://ocw.mit.edu
12.158 Molecular Biogeochemistry
Fall 2011
For information about citing these materials or our Terms of Use, visit: http://ocw.mit.edu/terms.
79