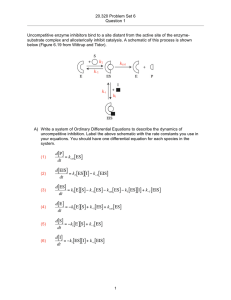
MULTICELLULAR MATHEMATICAL MODELS OF SOMITOGENESIS
by
Mark Benjamin Campanelli
A dissertation submitted in partial fulfillment
of the requirements for the degree
of
Doctor of Philosophy
in
Mathematics
MONTANA STATE UNIVERSITY
Bozeman, Montana
August, 2009
c Copyright
by
Mark Benjamin Campanelli
2009
All Rights Reserved
ii
APPROVAL
of a dissertation submitted by
Mark Benjamin Campanelli
This dissertation has been read by each member of the dissertation committee and
has been found to be satisfactory regarding content, English usage, format, citations,
bibliographic style, and consistency, and is ready for submission to the Division of
Graduate Education.
Dr. Tomàš Gedeon
Approved for the Department of Mathematical Sciences
Dr. Kenneth Bowers
Approved for the Division of Graduate Education
Dr. Carl A. Fox
iii
STATEMENT OF PERMISSION TO USE
In presenting this dissertation in partial fulfillment of the requirements for a doctoral degree at Montana State University, I agree that the Library shall make it
available to borrowers under rules of the Library. I further agree that copying of this
dissertation is allowable only for scholarly purposes, consistent with “fair use” as prescribed in the U.S. Copyright Law. Requests for extensive copying or reproduction of
this dissertation should be referred to ProQuest Information and Learning, 300 North
Zeeb Road, Ann Arbor, Michigan 48106, to whom I have granted “the exclusive right
to reproduce and distribute my dissertation in and from microform along with the
non-exclusive right to reproduce and distribute my abstract in any format in whole
or in part.”
Mark Benjamin Campanelli
August, 2009
iv
DEDICATION
I dedicate this dissertation to my family:
To my wife Amber, for her enduring patience.
To my daughter Ella, whose future I am trying to improve.
v
ACKNOWLEDGEMENTS
I would like to thank my advisor, Dr. Tomàš Gedeon, for all of his help and
guidance during this research project. I would also like to thank Jesse Berwald
for his good cheer and generosity concerning all things computational. Lastly, I
would like to thank Dr. Konstantin Mischaikow’s group at Rutgers University,
for computational time on the conley2 computer cluster.
vi
TABLE OF CONTENTS
1. INTRODUCTION ........................................................................................1
Biological Pattern Formation .........................................................................1
Developmental Biology and Somitogenesis ......................................................2
Mathematical Insights into Somitogenesis .......................................................5
Purpose and Scope of the Present Work .........................................................9
2. SURVEY OF EXISTING MATHEMATICAL MODELS ............................... 12
Early Models: Pattern Formation and Morphogenesis.................................... 12
Tissue-Based Reaction-Diffusion Models ....................................................... 14
Cell-Based Models....................................................................................... 16
Phase Oscillators..................................................................................... 16
Ordinary Differential Equation (ODE) Models .......................................... 17
Delay Differential Equation (DDE) Models ............................................... 19
Modeling Scopes and Multiple Scales ........................................................... 22
3. A MULTI-STABLE PHASE OSCILLATOR MODEL OF SOMITOGENESIS 25
Model Description....................................................................................... 25
Comparison to Existing Phase Oscillator Models........................................... 33
Lewis’s Phase Oscillator Model ................................................................ 33
Jaeger and Goodwin’s Cellular Oscillator Model........................................ 38
Discussion .................................................................................................. 39
4. A DELAY DIFFERENTIAL EQUATION MODEL OF POSTERIOR CLOCKWAVE FORMATION ................................................................................. 41
The Biological Components of the Clock ...................................................... 43
The Clock............................................................................................... 44
The Control Protein ................................................................................ 46
The Coordinating Signal .......................................................................... 47
Modeling Posterior Clock-Wave Formation: Uncoupled Cells ......................... 49
PSM Growth........................................................................................... 49
Model Variables ...................................................................................... 49
The Control Protein ................................................................................ 50
The Intracellular Clock ............................................................................ 51
Clock-Gene Regulation by a Single Repressive Transcription Factor............ 55
Modeling Posterior Clock-Wave Formation: Coupled Cells............................. 59
Intercellular Signaling .............................................................................. 59
vii
TABLE OF CONTENTS – CONTINUED
Clock-Gene Regulation by Both Repressive and Activating Transcription
Factors........................................................................................... 59
Interim Model Summary.............................................................................. 64
The Fast Dimerization Approximation.......................................................... 65
Algebraic Solution of the Fast Dimerization .............................................. 73
An Iterative Numerical Scheme for Computing the Fast Dimerization......... 75
Model Summary ......................................................................................... 78
5. MODEL VALIDATION: AN APPLICATION TO ZEBRAFISH SOMITOGENESIS ........................................................................................................ 80
Computational Considerations for Validation ................................................ 81
Clock-Wave Formation in Zebrafish .............................................................. 83
Assignment of Model Components............................................................ 84
Model Validation Criteria ........................................................................ 85
Parameter Value and Range Selection....................................................... 87
Experimentally Determined Parameter Values ....................................... 88
Parameters Estimated from a Range of Values....................................... 91
Parameters for Model Scenarios I–IV .................................................... 93
Parameter Estimation and Model Selection................................................... 94
Stage One Validation............................................................................... 95
Parameter Sensitivities......................................................................... 98
Stage Two Validation ............................................................................ 102
Model Robustness.............................................................................. 103
Reproduction of Experiments..................................................................... 106
The Mechanism of Gradient Controlled Oscillation Rate.............................. 112
Comparison to Existing Zebrafish Models ............................................... 113
Applicability of the PCW Model ............................................................ 113
Further Analyses and Future Directions .................................................. 115
6. CONCLUSION ......................................................................................... 117
Future Directions ...................................................................................... 118
REFERENCES CITED.................................................................................. 120
APPENDICES .............................................................................................. 132
APPENDIX A: Impossibility of Nontrivial Periodic Solutions in Lewis’s Uncoupled DDE Model without Delays .......................................................... 133
APPENDIX B: Competitive Dimerization of Three Proteins ..................... 135
viii
TABLE OF CONTENTS – CONTINUED
APPENDIX C: Matlab Codes ................................................................. 140
ix
LIST OF FIGURES
Figure
Page
1
Formed somites in a zebrafish embryo. ....................................................3
2
Transverse schematic of the somitic mesoderm. .......................................3
3
Formed and forming somites in a zebrafish embryo. .................................4
4
Multiple gene expression during zebrafish somitogenesis. ..........................6
5
Maturity/susceptibility plots. ............................................................... 29
6
Phase portrait snapshots of the multi-stable phase oscillator. ................. 30
7
Computed solutions of the multi-stable phase oscillator model................ 32
8
Long-term computed solution behavior of the multi-stable phase oscillator model. ........................................................................................ 32
9
Reproduction of the spatiotemporal somitogenesis pattern. .................... 34
10
Reproduction of the long-term somitogenesis pattern. ............................ 35
11
Asymptotic phase solutions for Lewis’s phase oscillator model. ............... 36
12
Asymptotic somitogenesis pattern for Lewis’s phase oscillator model. ..... 38
13
Total control protein gradients. ............................................................ 52
14
Binding site configurations. .................................................................. 58
15
Model selection. .................................................................................. 96
16
Parameter sensitivities for model scenario III, part 1 ........................... 100
17
Parameter sensitivities for model scenario III, part 2 ........................... 101
18
Model III simulated clock-wave, no noise. ........................................... 103
19
Model IV simulated clock-wave, no noise............................................. 104
20
Model III simulated clock-wave, with noise. ........................................ 105
21
Model III simulated clock-wave, no noise with longer gradient half-life. . 107
22
Model III simulated clock-wave, no noise with shorter gradient half-life. 108
23
Model III simulated clock-wave, no noise with exponential gradient. ..... 109
24
Model III simulated knockdown experiment, with noise. ...................... 110
x
LIST OF FIGURES – CONTINUED
Figure
25
Page
Model III simulated clock-wave in a rectangular lattice of cells, with
noise................................................................................................. 111
xi
ABSTRACT
Somitogenesis is an important pattern formation process in the developmental
biology of vertebrates. The phenomenon has received wide attention from experimental, theoretical, and computational biologists. Numerous mathematical models
of the process have been proposed, with the clock and wavefront mechanism rising to
prominence over the last ten years.
This work presents two multicellular mathematical models of somitogenesis. The
first is a phenomenological phase oscillator model that reproduces both the clock
and wavefront aspects of somitogenesis, but lacks a biological basis. The second is
a biologically informed delay differential equation model of the clock-wave that is
produced by coordinated oscillatory gene expression across many cells.
Careful and efficient model construction, parameter estimation, and model validation identify important nonlinear mechanisms in the genetic control circuit of the
somitogenesis clock. In particular, a graded control protein combined with differential
decay of clock protein monomers and dimers is found to be a key mechanism for slowing oscillations and generating experimentally observed waves of gene expression. This
represents a mode of combinatorial control that has not been previously examined in
somitogenesis, and warrants further experimental and theoretical investigation.
1
INTRODUCTION
Biological Pattern Formation
“How does the leopard get its spots and the zebra its stripes?” Many children
(and adults!) have wondered about such questions. The living world is replete with
patterns such as spots and stripes. Other examples of patterns include the trichome
distribution on plant surfaces, the efficient branching of tree roots, tree limbs, and the
airway passages of the lung, and certain multi-organism behaviors of insects. Nature
has devised some wonderfully useful patterns, but can we achieve some understanding
how these patterns form? The answer, of course, is yes, and the tools of science and
mathematics can be employed to do so.
Organisms may form patterns at different stages of the life cycle, from earliest
development until death. For single organisms, pattern formation typically involves
some type of cellular differentiation. This differentiation can range, for example,
from a simple color change between otherwise indistinguishable epithelial cells, to
a substantial divergence of cell type and function (e.g., beta vs. acinar cells in the
pancreas).
Such differentiation is often visible at the macroscopic level, although many important biological patterns, such as the human backbone and ribcage, initially form
in utero on a microscopic scale and are not directly observable, even in the adult
organism. Furthermore, modern molecular biology has revealed a host of genetically
orchestrated biochemical activities at the sub-cellular level that are involved in pattern
formation. Such activities include epigenetic1 intracellular and intercellular signaling
1
Epigenesis is the process by which genetic information, as modified by environmental influences,
is translated into the substance and behavior of an organism.
2
and feedback mechanisms involving, for example, regulation of protein production
and elimination.
As mentioned above, an important pattern that has evolved in higher organisms is
the repeated vertebrae and related structures of the spinal column in vertebrates (the
Verbrata subphylum of the Chordata phylum of the Animalia kingdom). Backbone
development begins early in the developmental sequence of vertebrates, during embryogenesis. This robust process occurs under a variety of conditions, for example, in
a cold-blooded zebrafish embryo in a lake or pond, in a warm-blooded chick developing
inside an egg, or in a human fetus in the womb.
Developmental Biology and Somitogenesis
Spinal column formation in vertebrates proceeds through several stages during
embryogenesis. The invention of the microscope enabled the discovery of a key early
event, called somitogenesis. Somitogenesis is the process of somite formation, which
occurs in the mesoderm, a tissue that forms just after gastrulation in the developing
embryo, see Figure 1. Gastrulation leads to three relatively flat layers of tissue,
called germ layers. The mesoderm is the middle germ layer, lying above the bottom
endoderm layer and below the top ectoderm layer, see Figure 2. Initially each germ
layer is structurally amorphous, yet the cells in each layer are already destined for
different tissues in the growing organism [1].
Somitogenesis is a fundamental stage of cell differentiation in the mesoderm.
Somites are transient, repeated blocks of epithelialized cells2 that eventually differentiate further into vertebrae, ribs, musculature, and dorsal dermis. Somites arise
sequentially and in pairs from the mesoderm, in an anterior (head) to posterior (tail)
2
Epithelialized cells are surrounded by a well-defined layer of border cells, called epithelial cells.
3
Figure 1: Side-view micrograph of recently formed somites in a zebrafish embryo. The
anterior (head) is to the left, posterior (tail) is to the right. Taken from [2, Figure 2a]
under the Creative Commons Attribution License.
Figure 2: Transverse schematic of the somitic mesoderm of a chick embryo. The
(medial) midline of the embryo occurs to the left, and one of two (lateral) sides of the
embryo is depicted to the right. Somites and other structures have already formed
in the somitic mesoderm, which lies between the top (dorsal) ectoderm and bottom
(ventral) endoderm. Taken from the public domain via the Wikipedia Commons
(http://en.wikipedia.org/wiki/File:Gray19 with color.png).
4
Figure 3: Top-view micrograph of formed and forming somites in a zebrafish embryo.
The mesoderm runs along most of the length of the embryo. Formed somites have dark
bands of stable gene expression, numbered 1–10. Two to three bands of oscillatory
gene expression (labeled (11)–(12)) move in waves from posterior (right) to anterior
(left) across the presomitic mesoderm in the posterior-most part of the embryo. Expression in the tailbud (labeled (13)) oscillates steadily as the tail elongates, and is
the source of new waves of expression that narrow as they travel from right to left.
Taken from [4, Figure 1a] under the Creative Commons Attribution License.
fashion, see Figure 3. Each pair forms on either side of the notochord, the spinal
precursor which forms along the embryonic midline. Amorphous mesoderm without
formed somites is called presomitic mesoderm (PSM), while after somite formation
the tissue is called somitic mesoderm [3], see Figure 2. This transition from presomitic
to somitic mesoderm is first marked by a pre-pattern of bands of gene expression that
help demarcate nascent somite borders, see Figures 3 and 4.
Different species of vertebrates have different numbers of vertebrae, and the PSM
elongates during somitogenesis to accommodate the length of the particular organism’s trunk, e.g., a mouse as opposed to a snake. At the posterior end of the early
embryo is the tailbud, a proliferative zone where immature cells are continually
added to the posterior PSM. Cell division and rearrangement diminish considerably in the PSM, and cells’ positions relative to each other do not change considerably. A cell’s relative position within the PSM does change, however, as the tailbud
grows away posteriorly and the oldest cells in the anterior PSM segment in groups
5
to form somites. Somitogenesis stops when the anterior formation of somites has
progressed posteriorly across the entire PSM, reaching the arresting growth in the
tailbud. [3, 5, 6, 7, 8, 9, 10, 11]
The morphological changes of somitogenesis can be more easily viewed in vivo
in certain species. In particular, zebrafish (Danio rerio) has an exposed, translucent
embryo, allowing both easy access and visualization with a light microscope, see
Figure 1. In the last several decades, modern microscopy, molecular biology, and
bioinformatics have enabled the determination of many of the underlying genetic
mechanisms of somitogenesis (compare the resolutions of Figures 3 and 4). Importantly, such investigations of the spatiotemporal dynamics of gene expression during
somitogenesis can be complemented by quantitative mathematical modeling.
In a larger context, an ever increasing portion of the biological sciences is now
conducted with a quantitative mathematical modeling component traditionally reserved for engineering and the “exact sciences” such physics or chemistry. This has
led to cross-disciplinary fields in addition to mathematical biology, such as systems
biology and computational biology. Many investigations, including the present one,
draw upon all three disciplines.
Mathematical Insights into Somitogenesis
Can mathematics help uncover the biological mechanisms of pattern formation
during somitogenesis?
The spatiotemporal dynamics of somitogenesis have long been recognized as a
good candidate for mathematical modeling. Important features of these dynamics
are shared by many organisms, and consist of the following (recall Figure 3):
1. Steady, clock-like oscillatory gene expression in the growing tail of the embryo.
6
Figure 4: High resolution confocal micrographs of gene expression during zebrafish
somitogenesis. The anterior direction is up. Individual cells are colored blue. The
mRNA expression of three genes her1, her7, and DeltaC in the PSM are shown in
green in the top three panels, respectively. Nuclear and cytosolic localization of her1
mRNA transcripts may be clearly seen in the bottom panel with higher resolution.
myoD mRNA appears in red, and demarcates the neural tube and most recently
formed somites. Taken from [4, Figure 3] under the Creative Commons Attribution
License.
7
2. Waves of expression that emanate from the oscillations in the tail and sweep
anteriorly across the PSM.
3. A posteriorly traveling determination wavefront that follows behind the growth
of the tail and periodically arrests oscillations in the anteriorly traveling waves
of expression into fixed bands of expression.
In the 1970s, even before the genetics behind somitogenesis started to be uncovered, investigators such as Cooke and Zeeman [12] adapted the catastrophe theory of
Thom [13] to propose a theoretical framework for the periodic formation of somites
as a spatiotemporal sequence of “catastrophes”. The model mechanism was termed
the clock and wavefront. In more modern mathematical parlance, somite formation
was proposed to be a periodic sequence of bifurcations of a dynamical system that
switched cohorts of cells from an undifferentiated state in the PSM to a differentiated
state in somites.
One shortcoming of the original clock and wavefront model of Cooke and Zeeman
was its abstract disconnection from any experimentally observed biological mechanism. Interestingly, the model anticipated the future discovery (in the mid-1990s) of
oscillatory gene expression in the PSM [14]. The 1980s saw other phenomenological
models applied to pattern formation, including somitogenesis. Most prominent among
these were the family of reaction-diffusion models of Meinhardt [15] that treated tissues as a spatial continuum and thus employed partial differential equations (PDEs).
In the case of somitogenesis, these models also lacked a direct connection to any
experimentally observed biological mechanism.
Extensive scientific work in the first part of the 20th century led to the emergence
early in the second half of the century of the so-called central dogma of molecular
biology. The central dogma states that genetic information stored in a cell’s DNA is
8
expressed as various proteins through intermediary, information carrying molecules
called messenger RNA (mRNA). Further advances in the second half of the last
century ultimately led to the Human Genome Project and the complete mapping
of human DNA, as well as the DNA of many other model organisms. One of the
resulting challenges of the 21st century is to understand the epigenome, that is, how
genetic information in the DNA is ultimately expressed in individuals given the genetic
control mechanisms and their past and present interaction with the environment.
Soon after the appearance of the central dogma, many of the resulting, qualitatively identified genetic control circuits were translated into quantitative mathematical models at the cellular level. For example, see [16, 17, 18, 19, 20], which discuss
both steady state and oscillatory behaviors in cellular control systems with feedback.
In many cases, this process involved extending existing compartmental models of
chemical reactions, both organic and inorganic, to mRNA and protein in cells. Examples include mass action kinetics, Michaelis-Menten enzyme kinetics [21], and the
extension of Hill’s equation for cooperative binding of oxygen to hemoglobin [22] to the
activation or repression of mRNA transcription from DNA by protein transcription
factors. Consideration of statistical thermodynamics lead to the approach of Shea
and Ackers [23] for modeling the control of gene expression in prokaryotes.
A large class of cell-based mathematical models of somitogenesis follows in the
footsteps of these initial modeling efforts [24]. Individual eukaryotic cells can be
represented by systems of ordinary or delay differential equations (ODEs or DDEs),
with state variables in the newest models representing experimentally observed mRNA
and proteins [2, 25, 26, 27, 28, 29, 30]. Multicellular tissues and interactions between
cells can be modeled by considering each cell as a subsystem in a larger coupled system
of differential equations. Cells undergoing somitogenesis are essentially fixed relative
to each other in the PSM, simplifying the cell-based approach by obviating the need
9
to track cell migrations. Furthermore, non-diffusive, contact-dependent intercellular
signaling mechanisms can be easily handled.
The existence of fast and powerful numerical solvers allows simulation of the resulting models, even for large systems of equations. Such simulations can be used to
validate the model against experiment, so that the model can ultimately guide the
formulation and verification of scientific hypotheses about the underlying biological
mechanisms.
There are, of course, challenges in mathematically modeling biological systems [31]. Like most biological systems, developmental systems usually display multiple time and/or spatial scales [24, 32, 33], and a careful accounting of such scales is
necessary to attain the proper balance between simplicity and accuracy. Stochastic
effects are often assumed to be “averaged out” in deterministic models, but this
assumption may not be strictly valid at the cellular level and may depend on the
chosen time or spatial scale [34]. Furthermore, experimental data used for model
construction and validation may be highly qualitative and the complex biological
interactions involved may be only partially known. Finally, many model parameters
may be too hard and/or costly to measure, and therefore must be estimated. In spite
of such challenges, mathematical models can provide considerable insight into the
mechanisms of biological systems, including the process of somite formation.
Purpose and Scope of the Present Work
Given the above setting for somitogenesis modeling, this dissertation presents two
multicellular, deterministic mathematical models of somitogenesis pattern formation.
These models are carefully constructed considering both existing mathematical models and the substantial body of experimental research into somitogenesis.
10
The first model represents cells in the PSM as simple, uncoupled phase oscillators.
Although this first model has limited foundation in biological experiment, it successfully captures the essential spatiotemporal dynamics of somitogenesis. As such, the
model acts as a “proof of concept” of the clock and wavefront mechanism and offers
modest improvements over similar existing models such as [35, 36].
The second model is a minimal, biologically-based model of a central feature of
somitogenesis. As a partial implementation of the clock and wavefront mechanism,
it reproduces the coordinated oscillatory gene expression that initiates the posteriormost waves of gene expression. The model is minimal in the sense that, given the
experimental data, minimal biological circuitry is used to implement the clock mechanism. Following the work of Lewis [27] and Cinquin [30], the cell-based model
employs a system of delay differential equations. The ability of the model to replicate
somitogenesis experiments in zebrafish is examined, and the robustness of this model
is examined with respect to both the estimated model parameters and heterogeneity
in parameter values across the entire cell population.
There are several important implications of this modeling work on the underlying
molecular biology of somitogenesis. First, a mathematical model is carefully constructed with regard to the important biological factors. Second, through extensive
model simulation and computational analyses, several potential biological mechanisms
are considered and eliminated, leaving only one minimal mechanism that successfully
reproduces experimental observations of somitogenesis. In particular, both the number of binding sites for self-inhibiting transcription factors and differential decay rates
of protein monomers and dimers are shown to be essential elements of the modeled
system.
A key technical feat of this work is the computational component of the model
simulation, analysis, and optimization. This involved novel use of existing computa-
11
tional tools, as well as significant algorithm development, computationally assisted
analysis and optimization, and coding in Matlab and, to a lesser extent, Mathematica. Parallel computing was employed for more efficient parameter estimation
using Monte Carlo simulations. Statistical analysis of the resulting large datasets
was necessary for model selection and also generated useful parameter sensitivity
information. Object-oriented programming tools were employed for accurate and
reproducible randomization of large parameter sets used in testing model robustness
to perturbation.
Before presenting the new models, a more thorough review of the existing mathematical models of somitogenesis will be presented in the next chapter. This will
provide the necessary background and perspective for proper consideration of the
new models presented subsequently.
12
SURVEY OF EXISTING MATHEMATICAL MODELS
Mathematical modeling of somitogenesis stretches back some thirty years. Not
surprisingly, the mathematical models have evolved alongside the growth in scientific
understanding of somitogenesis. Theoretical understanding has progressed in the
past half-century by advances in quantitative experimental molecular biology complemented more recently by mathematical models and computational tools. This
chapter presents a brief history and review of the existing mathematical models of
somitogenesis, providing perspective and a foundation for the new models presented
in Chapters 3 and 4.
Early Models: Pattern Formation and Morphogenesis
Mathematical models for pattern formation predate applications to somitogenesis.
In 1952, the pioneering work by Turing [37] showed that a system of reaction-diffusion
(RD) equations with two chemical components could produce spatial patterns. Counterintuitively, the addition of diffusion to a spatially uniform and temporally stable
system was shown to be capable of destabilizing the uniform distribution into a transient pattern (i.e., the Turing instability). Later RD models, such as the “local activation, long range inhibition” model of Gierer and Meinhardt [38], added nonlinearities
into the reaction terms which stabilized these patterns [39]. These models suggested
mechanisms of morphogenesis in developmental biology, where initially homogeneous
stem cell populations differentiate spontaneously into tissues with structure and pattern.
Mathematical interest in morphogenesis continued through the second half of the
20th century. Of special note is Wolpert’s proposed mechanism, in the late 1960’s,
13
of so-called morphogen gradients, whereby a spatial gradient of a biomolecule confers
positional information to developing tissues [40]. The existence of such gradients
was initially speculative, yet experimental evidence for them has since been found in
several systems (e.g., [41, 42, 43]). The theoretical framework of morphogen gradients
remains influential to the present day [44], and has been extended to include temporal
aspects. Contemporaneously with Wolpert’s early work, morphogenesis theories by
Thom [13] were inspired by the emergence of catastrophe theory, which lead, notably, to one of the earliest and most enduring theories of somitogenesis by Cook and
Zeeman.
In 1976, Cooke and Zeeman [12] postulated that somitogenesis could be explained
by a clock and wavefront mechanism. In this model, the susceptibility of cells in the
presomitic mesoderm (PSM) to form somites continually oscillates between susceptible and insusceptible (the clock), while a determination wavefront sweeps posteriorly
across the PSM. The passing wavefront triggers cells to form somites, but does so
only when cells are susceptible, i.e., when their clocks are in the correct phase of
oscillation. Since adjacent cells are in phase, cohorts of cells are recruited in succession to form somites. Mathematically, the theory supposes a series of bifurcations (or
“catastrophes”) that underlie the sequential formation somites.
Cooke and Zeeman proposed their model with minimal biochemical evidence for
either the clock or the wavefront. Because early heat shock experiments on developing
embryos caused a periodic disruption in somite formation whose timing agreed with
the known timing of the cell cycle, the clock was initially thought to be closely linked
to the cell cycle [45]. This lead to a line of mathematical models based upon the
apparent cell cycle connection [46, 47, 48, 49, 50].
Mounting experimental evidence has since dispelled the cell cycle as the fundamental oscillator in the clock and wavefront mechanism. In 1997, Palmeirim and
14
coworkers [14] discovered a gene with oscillatory expression in the PSM of the chick
embryo, providing an alternative candidate for the clock [51]. Experimental work has
since identified multiple oscillatory genes in each of several model organisms, including
mouse and zebrafish [3]. It should be noted that gene expression does not oscillate
synchronously throughout all the cells of the PSM. Instead, the oscillatory expression in cells is coordinated so that an anteriorly traveling clock-wave is oppositely
directed to the posterior movement of the determination wavefront. Interestingly,
the discovery of oscillatory gene expression in the PSM forged a closer mathematical
connection between somitogenesis and other biological rhythms, such as those studied
extensively by Goodwin [16, 18], Winfree [52], and Goldbeter [53].
With several variants proposed along the way (e.g., [54, 55]), the clock and wavefront mechanism has become a prominent model of somitogenesis. For additional
reviews and comparisons, see [3, 6, 7, 46, 56, 57, 58, 59, 60]. More recent mathematical
models of the clock and wavefront mechanism are discussed below.
Tissue-Based Reaction-Diffusion Models
In the 1980’s, a mathematically precise model of somite formation emerged from
the earlier pattern formation work by Gierer and Meinhardt [38]. The so-called Meinhardt models of somite formation [15, 61] rose to prominence despite the fact that
they were phenomenological in nature and lacked any direct link to experimentally
observed biological mechanisms. These models were extensions of the following nonlinear RD system of partial differential equations:
∂a
ρ a2
∂2a
=
− µ a + Da 2 ,
∂t
h
∂ x
∂h
∂2h
= ρ a2 − ν h + Dh 2 ,
∂t
∂ x
15
where a(t, x) is the concentration of a supposed activator molecule that may diffuse
through the one-dimensional tissue with diffusion constant Da , h(t, x) is the concentration of a supposed inhibitor molecule with diffusion constant Dh , and Greek letters
represent positive system parameters. The activator a auto-catalyzes but is inhibited
2
by h (note the ρ ah production term). The basic conditions for pattern formation in
this system are that the activator diffuses more slowly than the inhibitor (Da Dh )
and the activator decays more quickly than the inhibitor (µ ν) [39, 62]. This
situation is termed “local self-enhancement and long-ranging inhibition” [62]. One of
the nicest features of the model variant used for describing somitogenesis is that it
naturally forms the observed somite polarity [62].
The Meinhardt models are still employed today and can reproduce a wide range
of developmental patterns [62]. However, newer RD models for somitogenesis have
emerged that specifically incorporate the action of the experimentally observed clock
and wavefront in the formation of the somite pre-pattern1 in the anterior-most PSM.
Prominent examples are the recent models by Baker, Schnell, and Maini [57, 59, 63],
which are a partial reformulation of earlier somitogenesis models based on the cell
cycle [50]. These models are still largely phenomenological, especially with respect
to the clock. However, they have successfully reproduced experiments in which the
wavefront is perturbed [57]
Baker and coworkers have also recently formulated a biologically informed RD
model of the wavefront, which is understood to involve an anterior-posterior (AP)
gradient of Fibroblast Growth Factor (FGF) in the PSM [64]. Decreasing levels of
FGF are at least partially responsible for triggering somite formation [65]. However,
1
The somite pre-pattern refers to the stable bands of high-low gene expression in the anterior-most
PSM, which form before visible morphological segmentation occurs.
16
they have not yet incorporated this model into the above model for pre-patterning.
Still other recent RD models describe the segmentation of the tissue that occurs after
pre-patterning, which involves cell rearrangement and changes in cell adhesion [49,
57, 58, 66, 67].
Cell-Based Models
In the clock and wavefront mechanism, modeling of the clock oscillations has been
dominated by cell-based models. This predominance can likely be traced back to
early ordinary differential equation (ODE) models such as those by Goodwin [16, 18]
and Griffith [19, 20], where the discovery of intracellular genetic regulatory mechanisms made compartmentalized, cell-based approaches for homeostasis and biological
rhythms a viable alternative to continuum, tissue-based approaches [17]. However,
phenomenological clock models continue to be proposed, most of which are premised
on cell-based phase oscillators where the clock does not have a specific biochemical
basis.
Phase Oscillators
In 1997, based on the discovery of oscillatory gene expression in chick by Palmeirim
and coworkers, Lewis [35] developed a clock and wavefront model that treated an
axial line of mesodermal cells as uncoupled phase oscillators. By prescribing an
anteriorly slowing frequency of phase oscillations in cells along the PSM tissue, the
model produced anteriorly traveling phase waves that emanated from a steady, clocklike oscillation in the tailbud. The wavefront was associated with the frequency
of the phase oscillators decreasing to zero, and phases of successive blocks of cells
were thereby arrested anteriorly into an alternating high-low pattern. However, the
17
model contained no direct biological mechanism for the clock, the wavefront, or their
interaction.
More recent phase oscillator models have also been proposed. As a discrete reformulation of the continuum-based Flow-Distributed Oscillator models of Kaern and
coworkers [68], Jaeger and Goodwin [36] developed a cell-based, uncoupled phase
oscillator model that was similar to Lewis’s earlier model. However, the authors did
not view their Cellular Oscillator model as an implementation of a clock and wavefront
mechanism. The general setting of the model made it capable of producing multiple
kinds of fixed stripe patterns by arresting a traveling spatiotemporal wave.
The most recent phase oscillator models have included intercellular coupling [69,
70]. Intercellular signaling pathways are known to be active in PSM cells during
somitogenesis [71]. Using coupled systems of ordinary [69] or delay [70] differential
equations, these models focus on phase coupling while abstracting the details of the
oscillator mechanism in the cells. This can simplify the mathematical analyses, allowing easier examination of how coupling makes pattern formation robust to the effects
of system noise.
Ordinary Differential Equation (ODE) Models
After the discovery of oscillatory expression of c-hairy1 in the PSM of chick in
1997, a plethora of other genes with oscillatory cellular expression were soon found in
zebrafish, chick, and mouse [3, 5]. In all three model organisms, two dominant genetic
motifs for the clock emerged from these investigations. The first was an intracellular
self-repression loop of so-called basic Helix-Loop-Helix (bHLH) genes, and the second
was the intercellular positive feedback mechanism of the Notch pathway. The various
mathematical models of the clock have typically focused on one or both of these
motifs.
18
One of the first mathematical models of a clock-gene was presented in 2002 by
Hirata and coworkers [26], which investigated oscillatory expression of the bHLH Hes1
protein in mouse. They established experimentally that the Hes1 protein acted as
a self-repressing transcription factor by inhibiting hes1 mRNA production, and that
sustained oscillations required fast decay of the Hes1 protein by ubiquitin-proteasomemediated degradation. They complemented their experimental investigations with the
following ODE model:
dx
= B y − C x − A x z,
dt
dy
E
=
− D y,
dt
1 + x2
dz
F
− G x − A x z,
=
dt
1 + x2
(1)
(2)
(3)
where x is the Hes1 protein concentration in the cell, y is the hes1 mRNA concentration, and z is a presumed “Hes1-interacting factor” that allows the system to
sustain oscillations [26]. A–G are positive parameters affecting production and decay
rates. Note that the x2 term in the denominator of the production terms for y and z
represents some form of interaction of the Her1 protein with itself, perhaps through
homodimerization or cooperative binding at multiple DNA binding sites.
Since this first model was published, additional ODE models have been put forward. An early model of Cinquin [72] was one of the first to question whether oscillations were truly cell-autonomous as opposed to requiring intercellular signaling to be
sustained. A more recent multicellular model by Tiedemann and coworkers [29] had
some success incorporating a wavefront mechanism into the clock to arrest oscillations
into a pattern. Like the above model by Hirata et al., this model introduced a third
equation for separate tracking of the Hes1 protein in the cytosolic and nuclear compartments, which enabled the system to exhibit sustained oscillations. This model
also incorporated a nonlinear, rate-limited protein decay mechanism in the nuclear
19
compartment only, while additional results with intercellular signaling were only preliminary. Another recent model took the same subcellular compartment approach
in tracking the Hes1-related clock-gene Hes7 [73], and it was also shown that ratelimiting decay mechanisms play a dual role with the number of repressor binding sites
in the generation of sustained oscillations.
Lastly, it should be noted that in the past five years considerable genetic complexity has been uncovered in the mouse oscillator [3, 7, 74]. Oscillations are currently
believed to be coupled between three interacting signaling pathways (Notch, Wnt,
and FGF), each with multiple biomolecular agents. Goldbeter and Pourquié [75]
have recently published a comprehensive ODE model of the oscillator, which for a
single cell requires sixteen ODEs with another sixteen auxiliary algebraic equations
and some 77 parameters!
Delay Differential Equation (DDE) Models
The requirement of the somewhat mysterious third state variable z for sustained
oscillations in the ODE model by Hirata et al. lead to the introduction of delay
differential equation (DDE) models of the clock. These models introduced biologically
realistic transcription, translation, and transport delays into the production terms
of protein and mRNA, which allowed sustained oscillations in a system with only
these two dependent variables and a reduced number of model parameters. Negative
feedback with time delay DDE models have arguably become the most prominent
models of clock oscillations in somitogenesis.
Sustained Hes1 oscillations in mouse cells produced by a DDE model of gene
regulation were initially reported in 2003 by Jensen and coworkers [76] and also by
Monk [77]. In the same journal issue as Monk, Lewis [27] published a similar model
of delayed autoinhibition induced oscillations of homologous bHLH clock-genes in
20
zebrafish. The simplest version of Lewis’s model was the following system of DDEs:
dp(t)
= a m(t − Tp ) − b p(t),
dt
dm(t)
k
=
2 − c m(t),
dt
m)
1 + p(t−T
p0
(4)
(5)
where p is the protein concentration in the cell produced with delay Tp > 0, and m
is the mRNA concentration produced with delay Tm > 0. The positive parameters
a, b, k and c affect production and decay rates. po is a critical concentration of the
protein at which mRNA production is half its maximum value k.
Equations (4)–(5) are essentially the same as equations (1)–(2) above with z = 0
and if Tp = Tm = 0. Dulac’s Criterion can be used to show that solutions to (4)–(5)
cannot be nontrivially periodic if Tp = Tm = 0 (see Appendix A). With sufficiently
long total delay Tp + Tm and sufficiently large decay constants b and c, this system
exhibits sustained oscillations for a large range of the remaining parameter values a,
k, and p0 , however, the period of oscillation is sensitive to the total delay [27].
Lewis extended this basic model with the addition of intercellular positive feedback on the clock-gene via Notch coupling, and showed that synchronization of two
coupled cells with different natural frequencies was possible. Lewis also considered the
role of a second self-repressing clock-gene that heterodimerizes with the first clockgene. Followup experimental and modeling work by Lewis and coworkers measured
certain key parameters of the model [4] and established that Notch signaling acts as
a coordinator of clock oscillations in zebrafish, but not as a fundamental driver of
oscillations in individual cells2 [2].
2
Actually, a very simple DDE model for Notch coupled oscillations by Jiang and Lewis slightly
predates this 2003 model [78].
21
A 2007 paper by Cinquin [30] developed a related two clock protein model with
intercellular activation that required thirteen differential equations for each cell. The
model development required that numerous parameters be estimated, but was significant in that it extended Lewis’s two coupled cell model to a one-dimensional,
anterior-to-posterior (AP) line of coupled cells. Furthermore, the model included an
AP graded control protein that interacted with the clock-proteins via heterodimerization. The heterodimers repressed clock-protein production alongside the other clock
protein dimers. The model is notable in that it generated spatiotemporal waves of
expression of the clock-genes across the PSM. However, these waves did not arrest
anteriorly. Previously to this, the only multicellular extension of Lewis’s model was
an examination of lateral synchronization of oscillators [28, 79], which did not consider
axial control of the oscillation rate.
Along with Hes1, other clock-genes such as Hes7, Lfng, Axin2, Notch, and Wnt
oscillate in the PSM of mouse [3]. In parallel with the above DDE modeling developments in zebrafish, a series of DDE models for Hes and other oscillators in mouse
has been published. These models have focused on various aspects of the oscillator
such as the instability of the protein [80], the number of repressor binding sites [81],
the role of co-repressors [82], the instability of cell-autonomous oscillations [83], the
interaction between multiple signaling pathways [84], and the interaction with the
determination wavefront [25].
A limited number of the above models have had stochastic components, specifically [27, 30, 83]. In [27], Lewis showed that a certain level of transcriptional noise
added to his deterministic model could help sustain oscillations in a parameter regime
that produced damped oscillations in the corresponding deterministic model, a phenomenon known as stochastic resonance. The proper accounting of internal and external noise during somitogenesis is an area of active interest [85].
22
Finally, the amount of mathematical analyses that have accompanied these models
has been relatively limited. Even simple linear analyses, such as the computation of
Hopf bifurcations, are complicated by the presence of delays [56]. Some very recent
progress has been made [86, 87, 88, 89, 90]. The work by Verdugo and Rand [88] is
notable in that they were able to continue the periodic solution of the system (4)–(5)
away from the Hopf bifurcation point via an asymptotic expansion using Lindstedt’s
method, as well as find closed form expressions for the period and amplitude of the
approximate solutions with respect to the parameters.
Modeling Scopes and Multiple Scales
Biological systems are complex. Careful handling of this complexity is necessary
when developing useful mathematical models of biological systems, such as the somitogenesis models discussed above. Two issues of particular importance are the choice
of modeling scope and the consideration of multiple scales [32, 33, 56]. These issues
are somewhat interrelated.
The complexity of mathematical models of a biological system typically grows as
more information about the system becomes available. Unfortunately, the information
about biological systems can be simultaneously expansive yet incomplete. Making
smart modeling decisions in the face of this dichotomy requires careful consideration
of the scientific questions being addressed.
The choice of modeling approach used to meet the scientific objectives may be
divided (somewhat artificially) into top-down vs. bottom-up. Put simply, a bottom-up
approach tries to identify all the pieces of a system and their interconnections, so that
when put together the emergent properties are those observed experimentally. On
the other hand, a top-down approach begins with large-scale experimentally observed
23
phenomenon, and tries to determine how the overall system function depends on the
cooperative interaction of the principle subsystems. The inside workings of these
subsystems can remain very poorly understood, and, as such, may be treated as
“black boxes”.
There are trade-offs to each approach, and each approach (or a hybrid of the two)
is appropriate depending upon the circumstances [56]. For example, a prominent class
of somitogenesis models involves oscillations of gene expression on the cellular level.
In some models these oscillations are prescribed as phase oscillators, without regard
to the biochemical mechanism driving them, because the scientific focus is on the role
of oscillator slowing and/or coupling in pattern formation [70]. In other models, the
focus itself is on the detailed genetic circuits that drive oscillatory expression within
each cell [87]. The former model is simpler with respect to the myriad genes that
are involved in the clock, while the latter model may identify specific experimental
targets for testing hypotheses about the oscillator mechanism.
The presence of multiple scales is a main reason for the coexistence of top-down
and bottom up approaches in mathematical models of biological systems. Biological
systems exist across a broad spatial spectrum: from populations to individuals to
organs to tissues to cells to organelles to molecules to ions. There are also a broad
range of timescales: from inter-generational evolution to seasonal growth cycles to
circadian rhythms to neural impulses. Appropriate consideration of multiple scales
typically offers significant opportunity for simplification of mathematical models in
the face of biological complexity (e.g., Michaelis-Menten kinetics), while integration
of models at different scales becomes an additional consideration [32, 33].
Finally, another complication in mathematically modeling biological systems is
the existence of multiple model organisms used for studying phenomena in the life
sciences. For example, the prominent model organisms in somitogenesis are zebrafish,
24
chick, and mouse, each with its own genetics, epigenetics, and evolutionary history.
Consideration of a given mathematical model requires some understanding of the
model organism(s) to which the mathematical model is applicable. This is particularly
critical when trying to draw conclusions about one organism (e.g., human) from
another organism (e.g., mouse).
With these modeling issues in mind, the first model of this work is presented in
the next chapter.
25
A MULTI-STABLE PHASE OSCILLATOR MODEL OF SOMITOGENESIS
As discussed in the previous chapter, the essential features of somitogenesis pattern formation may be produced with a relatively simple, cell-based phase oscillator
model. The main drawback of such a model is that it typically does not incorporate
a biochemical mechanism, and thus provides only a phenomenological description of
the process. Nonetheless, such phase oscillator models offer a useful proof of concept
of the clock and wavefront mechanism of somite formation [35, 36], as well as for
investigations of oscillator synchronization [69].
In this chapter, a simple phase oscillator model is presented that extends the
early modeling work done by Lewis [35] and Jaeger and Goodwin [36], and offers
certain improvements on these models. Unlike a more recent phase oscillator model by
Riedel-Kruse and coworkers [69], which focused on oscillation synchronization through
coupling, the present model does not include intercellular coupling of phase oscillators.
Instead, the model is used to inform the development, in the next chapter, of a
biologically grounded model of the posterior formation of waves of gene expression in
the presomitic mesoderm (PSM). These waves of gene expression are a key component
(the clock) of the clock and wavefront mechanism.
Model Description
Experimental observations of somitogenesis in several model organisms has lead
to the following understanding of the elongating embryo [3, 5, 11, 91, 92].
The posterior tailbud consists of a progenitor zone, where the majority of new
cells are added to the tailbud. Rapid cell division in a region dorsal to this progenitor
zone continually supplies new mesoderm-destined cells to the posterior-most tailbud.
26
Cell division and mixing continues as these cells move towards the PSM through the
initiation zone of the anterior tailbud, where coherent steady or periodic expression
of certain somitogenesis genes begins.
At any point in time, the presomitic mesoderm can be divided into posterior
PSM and anterior PSM. Cell division and rearrangement diminishes considerably as
cells exit the tailbud and initially traverse the posterior PSM. While in the posterior
PSM, which is about two-thirds of the presomitic tissue, cells remain in an uncommitted state, capable of forming any part of a future somite. Once cells pass into
the anterior PSM they commit to become a specific part of a nascent somite, which
has not yet physically segmented. At any point in time, the anterior PSM contains
approximately two nascent somites. Segmentation steadily converts the anterior-most
PSM tissue into somites (becoming somitic mesoderm) as new presomitic mesoderm
is simultaneously added to the posterior-most PSM.
The clock and wavefront mechanism incorporates several experimental observations of somitogenesis [35, 63]. Cells in the initiation zone of the anterior tailbud have
periodic clock-gene expression that oscillates in phase. After leaving the tailbud and
entering the posterior PSM, cells’ oscillatory expression rate substantially decreases as
they move toward the anterior PSM. The sequentially slowing oscillation rates across
many cells produce anteriorly traveling waves of gene expression across the PSM
tissue. As the determination wavefront passes posteriorly through the anterior PSM,
oscillations arrest in blocks of cells in an alternating high-low expression pattern,
where each block represents a nascent, polarized somite.
The present multi-stable phase oscillator (MPO) model represents a simple phenomenological implementation of the clock and wavefront mechanism. The clock is
generated by prescribing a constant phase oscillation frequency in the tailbud. The
wavefront mechanism moving posteriorly across the PSM slows oscillations into an
27
apparent traveling phase wave (termed the clock-wave), and eventually arrests oscillations into constant phases that are monotonically increasing across groups of cells in a
stepwise fashion (multi-stability). When these stepped phases are composed with an
appropriate periodic function, an alternating high-low pattern in the anterior-most
PSM is realized.
The MPO model is constructed for a line of K total cells along the medial anteriorto-posterior (AP) axis of the mesoderm and tailbud. Each cell is associated with a
phase oscillator, with φk (t) denoting the phase of the k th cell, 1 ≤ k ≤ K. The phase
may be interpreted as the state of expression of a given cell’s clock-gene(s).
A key experimental observation is that the frequency of synchronized oscillations
in the tailbud is equal to the somite formation rate in the anterior-most PSM, which
is approximately constant over a significant portion of developmental time [5, 93].
To be as general as possible, time is normalized to the period of tailbud oscillation.
That is, one unit of time is equal to the period of oscillation in the tailbud, which is
equal to the formation time of one somite in the anterior-most PSM. Likewise, space
is normalized to the AP length of one somite, so that one spatial unit equals the AP
length of a single somite.
Cell division and rearrangement decrease considerably after cells exit the initiation
zone in the anterior-most tailbud [5, 11, 85, 91]. Thus, cells are assumed to remain
stationary relative to each other as they pass through the PSM. Individual cells
are assumed to exit the anterior-most tailbud and enter the posterior-most PSM
sequentially at a constant rate, traverse the PSM, and ultimately be incorporated
into somites forming in the anterior-most PSM. The time when the k th cell enters the
PSM, Tk , is given by the linear relationship
Tk =
k−1
,
λµ
(6)
28
where λ is the number of cells per AP somite length, and µ is the somite formation
rate, assumed here to be the normalized constant µ = 1 somite per time unit. λ
varies with the organism under consideration, and is assumed constant. Although
experimental evidence suggests µ and λ may slowly change, especially towards the end
of somite formation, the constant value assumptions should be a valid approximation
for the majority of somites produced during somitogenesis [93].
To capture the somitogenesis patterning phenomenon, the dynamics of the k th
cell’s phase, φk (t), is given by the following differential equation:
φ̇k (t) = 1 − s t − Tk ; τ 1 sin2 (2π φk ),
(7)
2
where s is the following continuously differentiable function that reflects the time the
cell has been in the PSM:
s(t; τ 1 ) =
2
0,
„
«.
2
t−τ
t
1
2
1 e
,
2
0 < t ≤ τ1 ,
2
«.„
„
2
1
1
−
e
2
1,
t ≤ 0,
t−τ 1
2
«
t−2 τ 1
2
, τ1 ≤ t < 2 τ1 ,
2
2
2 τ 1 ≤ t,
2
where τ 1 > 0 is a half-life parameter. The function s is sigmoidal, non-decreasing,
2
concave up for 0 < t < τ 1 , and concave down for τ 1 < t < 2 τ 1 . The function s may
2
2
2
be regarded as giving the maturity of a cell in the PSM, or, equivalently, as indicating
a cell’s susceptibility to differentiate into part of a somite at a given time t. Zero represents fully immature/unsusceptible, while one represents fully mature/susceptible.
Figure 5a shows s with the half-life parameter τ 1 = 3.
2
The function s represents the wavefront in the clock and wavefront mechanism.
Biochemical candidates for the wavefront include morphogen gradients, which are
spatially and/or temporally graded concentrations of one or more chemicals [43, 64].
29
(a)
y=s(t−Tk; 3), k=1,...,60
(b)
y=s(t; 3)
0.8
0.8
0.6
0.6
t=6
t=8
y
1
y
1
0.4
0.4
0.2
0.2
0
0
0
2
4
6
t (unitless time)
0
20
40
k (cell #)
60
Figure 5: Maturity/susceptibility plots. (a) A plot of y = s(t; 3) showing the change
in a cell’s maturity/susceptibility over time, with half-life τ 1 = 3. The cell is fully
2
immature/unsusceptible (y = 0) for t ≤ 0 and fully mature/susceptible (y = 1) for
t ≥ 6 = 2 τ 1 . (b) Spatial profiles of maturities/susceptibilities of an anterior-to2
posterior line of sixty cells at times t = 6 and t = 8. k = 1 is anterior.
Because cells exit the tailbud in a temporal order, the composition s(t − Tk ; τ 1 )
2
represents the maturity/susceptibility of the k
th
cell at time t. Figure 5b shows the
resulting spatially graded profiles of the levels of s(t − Tk ; τ 1 ) across an AP line of
2
sixty cells at two different times.
The function s depends on time, which makes the differential equation (7) nonautonomous. However, for any k, s(t − Tk ) is constant outside the compact transition
interval
h
i
Ik := Tk , Tk + 2τ 1 ,
2
allowing autonomous analysis of the dynamics outside this interval. Consider the
k th cell, so that s(t − Tk ) is constant outside the transition interval Ik . For t ≤ Tk ,
s(t − Tk ) = 0, and so φ̇k (t) = 1 and the model reproduces the periodic oscillations
in the tailbud (the initial clock mechanism). For t ≥ Tk + 2 τ 1 , s(t − Tk ) = 1 and so
2
30
y=dφ1/dt, τ1/2=3, T1=0
1
y
0.8
0.6
0.4
t≤0
t=3
t≥6
0.2
0
0
0.5
1
φ1
1.5
2
Figure 6: Phase portrait snapshots for the multi-stable phase oscillator model of the
first cell given by the non-autonomous differential equation (7), with k = 1, τ 1 = 3,
2
and T1 = 0. The middle (grey) curve at t = 3 is transient, but represents the slowing
phase oscillations at the instant the cell is halfway to full maturity/susceptibility.
φ̇k (t) = 1 − sin2 (2π φk ) has semi-stable equilibria when sin2 (2π φ∗k ) = 1, i.e., when
1 3 5
φ∗k = ± , ± , ± , . . . .
4 4 4
(8)
See Figure 6.
For clarity in the computation of equilibria and discussion of the resulting multistability, a non-generic case has been considered. Note that this example can be made
generic, and hence more robust, by stretching the range of the function s by a small
amount. For example, making the range of s to be [0, 1 + ], for some > 0, would
suffice.
The positivity of φ̇k during the transition interval guarantees that all solutions
eventually approach one of these phase angles monotonically from below. Which of
these equilibria is approached depends on both the initial phase φk (0) and the slowing
31
of phase oscillations during the transition interval, which itself depends on the choice
of the half-life parameter τ 1 and shape of the function s.
2
Initial conditions for each cell may be chosen so that all cells oscillate in phase
before the first cell exits the tailbud at T1 = 0. A convenient choice that properly
arranges the cell phases in the first and subsequent somites is φk (0) = 14 , 1 ≤ k ≤ K.
Furthermore, τ 1 is chosen to correspond to the lifetime of a cell in the PSM for a
2
given model organism, which is approximately 2 τ 1 . Identical initial conditions and
2
monotonicity of solutions, when combined with the ordering of the Tk , lead to the
emanation of anteriorly traveling phase waves from synchronized tailbud oscillations.
In addition phase wave oscillations arrest blocks of cells in an alternating high-low
pattern in the anterior PSM.
Figure 7 shows numerical solutions for the first six cells of the phase oscillator
model described above, where six cells per AP somite length is assumed (λ = 6) and
the maturity/susceptibility half-life is three tailbud oscillation periods (τ 1 = 3). Two
2
groups of three cells reach distinct limiting phase values, with each cell taking slightly
longer than 2τ 1 = 6 oscillation periods to essentially reach full maturity/susceptibility.
2
Specifically,
lim φ1 (t) = lim φ2 (t) = lim φ3 (t) =
17
,
4
lim φ4 (t) = lim φ5 (t) = lim φ6 (t) =
19
.
4
t→∞
t→∞
t→∞
and
t→∞
t→∞
t→∞
The limiting phase difference between the two cohorts of cells represent the polarity
between the anterior and posterior half of the first somite. Figure 8 shows the longterm, grouped behavior of the phase solutions of sixty cells across the PSM. Solutions
were computed using Matlab’s ode45 solver [94]. The code that generated the
solutions and figures may be found in Appendix C.
32
y=φk(t), τ1/2=3, Tk=(k−1)/6, k=1,...,6
5
φ1(t)
4
φ2(t)
3
φ3(t)
y
φ4(t)
2
φ5(t)
φ6(t)
1
0
0
1
2
3
4
5
t (unitless time)
6
7
8
Figure 7: Computed solutions of the multi-stable phase oscillator model (7) for the
first somite (six total cells) with initial condition φk (0) = 14 , k = 1, 2, . . . , 6. Groupings
of three cells into constant limiting phases can be seen as cells sequentially reach full
maturity/susceptibility. Specifically, limt→∞ φ1 (t) = limt→∞ φ2 (t) = limt→∞ φ3 (t) =
17
and limt→∞ φ4 (t) = limt→∞ φ5 (t) = limt→∞ φ6 (t) = 19
.
4
4
y=φk(60.00), k=1,2,...,60
14
12
y
10
8
6
4
0
2
4
6
x (somite #)
8
10
Figure 8: Long-term computed solution behavior of the multi-stable phase oscillator
model (7) for ten somites (sixty total cells) with initial condition φk (0) = 41 , for
k = 1, 2, . . . , 60. All cells have reached full maturity/susceptibility, and grouping into
constant limiting phases is apparent.
33
The spatial formation of the somitogenesis pattern may be realized by composing
an appropriate 1-periodic function with the phase solutions to the differential equation (7). A convenient choice for converting phase to expression level in the the k th
cell, pk (t), is
pk (t) =
1
1 + sin (2π φk (t)) .
2
(9)
The expression levels pk (t) are normalized between zero and one. Figure 9 shows the
development of the resulting somitogenesis pattern over two oscillation cycles in the
tailbud. Figure 10 shows the computed approximation of the stable somitogenesis
pattern that forms asymptotically.
Comparison to Existing Phase Oscillator Models
Although the MPO model does not incorporate intercellular coupling, it still offers
certain advantages over two existing, uncoupled phase oscillator models of somitogenesis. These are discussed in turn below.
Lewis’s Phase Oscillator Model
The 1997 paper by Palmeirim and coworkers [14] presented the first experimental
evidence of a gene with oscillatory expression in the chick PSM that was independent
of the cell-cycle. In a supplement to this paper, Lewis presented a phase oscillator
model of somitogenesis [35]. The temporal rate of change in phase φ(x, t) of the cell
34
y=pk(6.00)
y
(a)
1
1
0.5
0.5
0
0
0
y
(b)
10
0
(f)
1
0.5
0.5
0
0
5
10
0
(g)
y=pk(6.50)
1
0.5
0.5
0
0
(d)
5
10
0
(h)
y=pk(6.75)
1
0.5
0.5
0
0
5
x (somite #)
10
5
10
5
10
y=pk(7.75)
1
0
10
y=pk(7.50)
1
0
5
y=pk(7.25)
1
(c)
y
5
y=pk(6.25)
0
y
y=pk(7.00)
(e)
0
5
x (somite #)
10
Figure 9: Reproduction of the spatiotemporal somitogenesis pattern across ten somite
lengths by the MPO model, given by equations (9), where the phases φk (t) were
computed numerically using equations (7) with initial conditions φk (0) = 14 , for
k = 1, 2, . . . , 60. (a)–(h) show two oscillation cycles in the tailbud. Red cells are
immature/insusceptible, green cells are mature/susceptible, and grey cells are in transition. Polarized somites, each with six cells, form anteriorly as more posterior cells
continue to oscillate.
35
y=pk(60.00)
1
0.8
y
0.6
0.4
0.2
0
0
2
4
6
x (somite #)
8
10
Figure 10: Reproduction of the long-term somitogenesis pattern across ten somite
lengths by the MPO model, given by equation (9), where the phases φk (60) were
computed numerically using equations (7) with initial conditions φk (0) = 41 , for k =
1, 2, . . . , 60. All cells are green, indicating full maturity/susceptibility. Polarized
somites, each with six cells, have formed stably across the entire PSM.
at position x ≤ 0 at time t ≥ 0 was given by the following initial value problem1 :
∂φ
1
=
x+t ,
∂t
1+e 2
φ(x, 0) = 0,
which may be solved by simple integration to give the solution
Z
φ(x, t) =
0
1
t
1
1+e
x+s
2
ds = t + ln
x
1 + e2
1+e
x+t
2
2
.
Lewis uses more negative values of the continuous position variable x to signify more posterior
positions in the PSM, so that somites form from right to left. Non-dimensional spatial units are in
somite lengths, and non-dimensional temporal units are in tailbud oscillation periods.
36
y=φ∞(x), Lewis Phase Oscillator
12
10
y
8
6
4
2
0
−10
−8
−6
−4
x (somite #)
−2
0
Figure 11: Asymptotic phase solutions for Lewis’s phase oscillator model [35]. The
posterior-most cell is at x = 0. Phases are monotonically distributed but not grouped
into polarized somites.
The asymptotic spatial phase pattern φ∞ (x) produced by this model is given by
φ∞ (x) := lim φ(x, t)
t→∞
= lim
t→∞
t + ln
x
1 + e2
x+t
2 !
1+e 2
!
x 2
2
1
+
e
= lim ln et + ln
x+t
t→∞
1+e 2
!
x 2
2
1
+
e
= lim ln et
x+t
t→∞
1+e 2
!
x 2
2
1
+
e
= ln lim et
x+t
t→∞
1+e 2
x
= ln (e−x + 2 e− 2 + 1).
See Figure 11 for a plot of φ∞ (x).
37
In Lewis’s model, the realization of the high-low expression of groups of cells is
given by the following function h(x, t):
h(x, t) = f (m(x, t)) w(z(φ(x, t))),
(10)
where
f (m) =
1
m ,
1+e2
m(x, t) = x + t,
w(z) =
1
,
1 + e10z
z(φ) = cos (2π φ).
In the product of the functions f ◦ m and w ◦ z in (10), the composition f ◦ m
eliminates expression in mature cells after a certain time2 , while w◦z acts to normalize
the expression pattern between zero and one. The asymptotic spatial pattern given
by w(z(φ∞ (x)), shown in Figure 12, does not capture the high-low limiting phase
behavior quite as well the MPO model (compare to Figure 10).
Although the MPO model is not as easily solved analytically, it improves upon
Lewis’s model in several ways. First, the construction of the MPO model greatly
simplifies the realization of normalized oscillatory expression (compare equation (9)
to equation (10)). Second, the asymptotically grouped phases of cell cohorts in the
MPO model is more robust (compare Figures 8 and 11). Finally, the MPO model’s
biologically motivated maturation/susceptibility function s, with the half-life parameter τ 1 , makes the model readily tunable to different species.
2
2
This transient expression in the paraxial mesoderm occurs for some genes, but not for others.
38
y=w(z(φ∞(x))), Lewis Phase Oscillator
1
0.8
y
0.6
0.4
0.2
0
−10
−8
−6
−4
x (somite #)
−2
0
Figure 12: Asymptotic somitogenesis pattern for Lewis’s phase oscillator model [35].
The posterior-most cell is at x = 0. Note that some cells are not well polarized.
Jaeger and Goodwin’s Cellular Oscillator Model
In 2001, Jaeger and Goodwin presented “A Cellular Oscillator Model for Periodic
Pattern Formation” [36]. In some sense, their Cellular Oscillator (CO) model generalized Lewis’s earlier phase oscillator model discussed above. However, the authors did
not recognize their model as an implementation of a clock and wavefront mechanism3 .
Instead, the authors cast their model as a cell-based version of the fluid-based Flow
Distributed Oscillator model of Kaern and coworkers [68].
The MPO model shares several key features with the CO model. Both models use
an AP line of cells with synchronized, periodic oscillations in the tailbud (called the
initiation zone in the CO model). Both models also have sequentially slowing phase
oscillations that create traveling phase waves. The MPO model has the advantage
3
Cooke and Zeeman’s earliest version of the clock and wavefront model did not anticipate the
anteriorly traveling waves of gene expression in the PSM [12, 51].
39
that the phase waves are completely arrested, whereas the CO model’s waves are
not [36, see p.175].
The construction and analysis of the MPO model makes its connection to the clock
and wavefront mechanism more transparent than the CO model. The CO model does
allow more complex striped patterns to be formed by non-autonomously altering the
frequency of oscillation in the initiation zone and/or changing the rate at which cells
exit this zone. Although not implemented above, such features could be added to the
MPO model without great difficulty.
Discussion
Admittedly, many of the choices in the construction of the MPO model are somewhat arbitrary. While phenomenologically sound, these choices are not necessarily
the only or best ones that reproduce the key qualitative features of pattern formation
in somitogenesis.
For example, the squared sine function in equation (7) is a convenient choice
for producing the desired phase monotonicity and slowing conditions, as well as the
distribution of phase equilibria. Other choices with similar qualitative shape are
certainly possible, which is also the case for the maturity/susceptibility function s.
The combined roles of these two functions during the transition interval affect the
specific formation of the transitional clock-wave in the PSM.
The MPO model is primarily used as a proof of concept for a more biologically
grounded, and hence more scientifically useful, model developed in the next chapter. Thus, further analysis concerning these function choices is not carried out here.
Likewise, the non-generic semi-stability of the equilibria given in (8) can easily be
40
remedied into more robust full stability, but little would be gained from the effort
given the purpose at hand.
Perhaps the best extension of the MPO model would be the addition of intercellular coupling of the phase oscillators. This would allow the extension of the work
by Riedel-Kruse and coworkers [69], in which the effect of coupling on phase synchronization in the initiation zone of the tailbud was investigated. Extending the present
model would allow a more complete analysis of the effect of coupling on clock-wave
formation and the arresting of oscillations in the presence of noise. In fact, very recent
work by Morelli and coworkers [70] has presented and analyzed such a model, which
incorporates biologically realistic delays in the intercellular coupling.
Finally, it has been tacitly assumed that coupling between cells is a coordination/synchronization mechanism operating solely to overcome system noise. There is
some evidence that, depending on the organism, the coupling may form a core component of the oscillator mechanism itself [2, 3, 28, 69, 71, 74, 78, 83, 84, 85, 95, 96].
Thus, eliminating the coupling in some systems may dampen or abolish oscillations
altogether. This situation again reveals the limitations of phenomenological phase
oscillator models that oversimplify the underlying biological mechanism of action.
The next chapter offers a partial remedy of this problem by presenting a biologically
informed model of posterior clock-wave formation during somitogenesis.
41
A DELAY DIFFERENTIAL EQUATION MODEL OF POSTERIOR
CLOCK-WAVE FORMATION
A good mathematical model is like a good map: it has just enough detail
to help you get where you need to go.
Although the multi-stable phase oscillator model presented in the previous chapter
reproduces the key spatiotemporal dynamics of somitogenesis patterning, it lacks a
specific biological mechanism of action. There are no underlying genetics or biochemistry. This can potentially limit the model’s usefulness in formulating and checking
scientific hypotheses concerning somitogenesis.
This chapter presents a biologically informed mathematical model for clock-wave
formation in the posterior presomitic mesoderm (PSM), for which the function of
many of the genetic and biochemical elements is reasonably well-established in several
model organisms. Recall that the clock-wave is an experimentally observed spatiotemporal wave of gene expression in the PSM that arises from coordinated oscillations
in gene expression at the cellular level. The clock-wave is understood to embody the
clock in the clock and wavefront mechanism of somite formation. The present model
reproduces the posterior portion of the waves of gene expression in the PSM that
emanate from steady oscillations in the tailbud. The anterior fixation of the waves
into bands of gene expression is not reproduced. Henceforth, the present model will
be called the posterior clock-wave (PCW) model.
In several model organisms, the genetic and biomolecular mechanisms of experimentally observed gradients in the PSM are now being established [7, 43, 64]. These
spatiotemporal gradients move in accordance with the wavefront of determination
that sweeps posteriorly across the anterior PSM, triggering the successive formation
42
of somites. Furthermore, several of these gradients have been directly implicated in
the susceptibility of cells in the PSM to form somites [7, 97, 65, 98, 42]. Thus, these
gradients are excellent candidates for the wavefront. As such, the wavefront can be
appropriately be termed a gradient-wavefront.
At this point in time, the biological elements that join the clock-wave and gradientwavefront to fully realize somite formation are not well-understood [7, 99]. However,
some mathematically informed hypotheses are beginning to emerge [25, 29, 63, 100,
101]. The PCW model does not incorporate any elements of the gradient-wavefront
in the clock and wavefront mechanism, and thus cannot be expected to reproduce the
entire process of somitogenesis. However, the PCW does employ an experimentally
observed posterior gradient of a so-called control protein, which controls the rate of
oscillations by interacting with the clock protein via heterodimerization.
Given the situation of current knowledge, the present work focuses on the initial
formation of the clock-wave in the posterior PSM. While keeping the model on a
firm biological foundation, an effort is made to incorporate a minimum of biological
components of the cell-based clocks whose aggregate behavior generates the posterior
clock-wave in the PSM. More specifically, a minimal genetic circuit is shown to control
the rate of oscillatory gene expression across coupled cells in such a way that allows
proper formation of the posterior clock-wave in the PSM.
Previous delay differential equation (DDE) models by Lewis and coworkers [2,
4, 27] and Cinquin [30] are the primary motivation for the PCW model presented
below. Having only four differential equations per cell, the PCW model maintains
the simplicity and clarity of Lewis’s coupled two cell model, yet the PCW model can,
like Cinquin’s model but with a much more transparent formulation, reproduce the
dynamical waves of gene expression that form in the posterior PSM of zebrafish.
43
Furthermore, the new model suggests a novel biological mechanism underlying
how competitive dimerization controls the oscillation rate. While other models have
focused on saturating protein decay kinetics as an additional source of nonlinearity [29, 73, 75], the PCW model suggests that differential decay between clock protein
monomer and dimer, including heterodimer with the control protein, is an important
mechanism for slowing oscillations and generating the clock-wave in the posterior
PSM.
The Biological Components of the Clock
The clock-wave arises from coordinated oscillatory gene-expression across cells
in the PSM. In various model organisms, experiments have revealed multiple genes
involved in oscillatory gene expression [3, 5, 74]. This has lead many investigators to
distinguish components of the clock from outputs of the clock [14, 102]. Components
of the clock include genes required to generate the clock’s oscillations as well as genes
necessary to coordinate these oscillations. The expression of a component gene may
or may not itself oscillate.
Outputs of the clock include genes downstream from the component genes. Outputs typically oscillate as they are driven passively by the components of the clock.
This distinction does not preclude outputs from being necessary for proper somite
formation. Furthermore, intracellular and intercellular feedback loops in the genetic
circuitry complicate the experimental identification of component vs. output [74].
With consideration of the above, the PCW model is composed of the interaction
between three key, experimentally identified components that generate the clockwaveform. These cell-level components are: 1) a clock, 2) a control protein, and
3) a coordinating signal. Each component is discussed in turn below. Because the
44
present work was largely motivated by experiments and previous modeling work in
the zebrafish model organism, a general discussion of each component is first given,
followed by specific considerations of the component in zebrafish.
The Clock
Multiple oscillatory genes have been identified in the PSM of several model organisms, such as zebrafish, chick, and mouse [3, 5, 14, 103]. These genes typically
exhibit some redundancy in function, complicating the characterization of a central
pacemaker that drives the oscillatory expression of the remaining genes. Depending
upon the model organism, the pacemaker may be a cell-autonomous genetic oscillator requiring single or multiple genes, or, it may require an intercellular network of
genes [6].
In zebrafish, for example, expression of the basic helix-loop-helix (bHLH) her
(hairy and enhancer-of-split related) genes her1, her7, her11, her12, and her15 oscillates in the PSM. Some of these genes are expressed across the entire PSM (her1 and
her7), while others are expressed only anteriorly (her 11) or anteriorly and posteriorly
with an interim space (her12 and her15) [5]. Gene knockout and protein morpholino
knockdown experiments have suggested that there is at least some redundancy in
the function of these genes, whose proteins are understood to act as transcriptional
repressors after homo- or heterodimerization with other bHLH proteins [5]. The remaining bHLH genes her4, her6, her13.2 and hey1 are expressed in a non-oscillatory
fashion in the zebrafish PSM [5]. With the exception of the posteriorly-to-anteriorly
graded her13.2, these genes are expressed anteriorly in bands that indicate nascent
somites [5].
It should be noted that several of the bHLH genes are active in other developing
tissues in the zebrafish embryo [103]. Many also have homologues in higher organisms
45
such as mouse [103], where they are referred to as Hes genes. Several Hes genes
have direct analogues in human. Furthermore, the bHLH structure is well-preserved
evolutionarily from more primitive organisms, and was originally discovered in the
developmental genes of the fruit fly Drosophila [104].
The three oscillatory genes her1, her7, and her12 have the most prominent effect
upon posterior clock-wave formation in zebrafish [5, 105], and previous models have
considered one or two clock-genes (her1 and/or her7) [2, 4, 27]. The PCW model
assumes a cell-autonomous clock with a single clock-gene. Use of a single clock-gene
is justified on the grounds that her genes show some redundancy in zebrafish, and
that Hes7 in mouse oscillates independently of, for example, Hes1 [106], which apparently cannot sustain independent oscillations [83]. Admittedly, this is a considerable
simplification of the genetic circuits of all model organisms. In the next chapter, this
minimal assumption of a single clock-gene is shown to be sufficient for generating a
biologically realistic clock-wave, which raises the interesting scientific question as to
the reason for the presence of multiple clock-genes across several model organisms.
Given the above considerations, the PCW model tracks both mRNA and protein
levels of a single clock-gene. The clock protein is assumed to form a homodimer
that represses its mRNA production after a delay. Three main sources of delay are
the transcription and post-transcriptional processing of mRNA, the translation delay in protein production, and the transport of molecules between the nuclear and
cytosolic compartments within a cell or between the cytosolic compartments of adjacent cells. This system is capable of autonomous, sustained oscillatory gene expression [27, 76, 77, 80]. The relative amounts of clock protein monomers, homodimers,
and heterodimers with the control protein are explicitly tracked, allowing different
decay rates for each [107].
46
Control of clock mRNA transcription is modeled using the approach of Shea and
Ackers [23, 108, 109], which includes parameters such as DNA binding energies and
cooperativity between various protein transcription factors bound to the promoter
at multiple binding sites. This modeling formalism is a significant simplification
of eukaryotic transcription process [34, 108], but represents an initial step towards
biological realism as compared to existing models such as [2, 27, 30]. The additional modeling details are particularly appropriate because of the presence of both
repressors and activators controlling the clock-gene, such that monotonicity of the
mRNA transcription rate with respect to the levels of transcription factors is no
longer ensured [110].
The Control Protein
Several spatiotemporally graded biomolecules have been identified in the PSM of
multiple model organisms. In zebrafish, the posteriorly-to-anteriorly graded Her13.2
protein interacts with at least one of the clock proteins (Her1) and affects the formation of the clock-wave [111, 112]. Even though a direct interaction of oscillatory and
graded bHLH proteins has not been verified in chick or mouse, the her13.2 gene is
homologous to Hes6 in mouse [111, 113], and several biomolecular gradients have been
identified in chick and mouse that could interact with one or more clock-genes [3, 7].
Motivated by experiments in zebrafish [30, 111, 112], we suppose that the graded
control protein interacts with the clock protein by heterodimerization.
Because
Her13.2 protein has a truncated amino acid sequence normally used for DNA binding [111], it is assumed that neither control protein homodimers nor heterodimers
with clock protein can repress clock-gene transcription. This is in contrast to the
model of Cinquin [30], in which heterodimers may also repress transcription. In fur-
47
ther contrast to the model of Cinquin [30], because Her13.2 protein has a functional
dimerization domain [111], homodimerization of the control protein is allowed.
The control protein level is prescribed as an external control on the system, with
a maximum value in the tailbud that deceases anteriorly in the PSM. The graded
level of control protein, combined with competitive dimerization between control and
clock proteins, results in slowing oscillation rates in successively anterior cells and
the formation of a clock-wave. Furthermore, simulations show that the amount of
slowing is a key factor in clock-wave formation, which agrees with experimental investigations [4]. Thus, the control protein influences the rate of oscillation generated
by clock protein self-repression through competitive dimerization, which is a control
mechanism seen in related bHLH networks [113, 114].
The Coordinating Signal
In multiple model organisms, experiments have revealed components of the clock’s
genetic circuit that are responsible for coordinating oscillations of clock-gene expression between cells through intercellular signaling. In all model organisms this
intercellular coupling is achieved through the notch signaling pathway [3, 5], with
more complex coupling pathways occurring in higher vertebrates such as chick and
mouse [3, 7, 74, 115]. This has lead to considerable scientific debate concerning
whether the coupling acts weakly to coordinate otherwise cell-autonomous oscillations, or, alternatively, whether the coupling plays a prominent role in generating
oscillations [2, 72, 83, 96]. In the first case, a clock-gene’s expression continues to
oscillate in the absence of intercellular signaling but loses coherence with other cells
because of system noise. In the second case, inhibition of intercellular signaling
causes oscillations to cease altogether, perhaps after transient, incoherent damping.
Experiments precise enough to allow differentiation of these alternatives have been
48
difficult to conduct [99]. Furthermore, different scenarios could happen depending
on the model organism, e.g., amniotes such as chick and mouse vs. lower vertebrates
such as zebrafish.
In zebrafish, the role of intercellular coupling is currently thought to be one of
coordination, not generation, of oscillations [2, 5, 27, 28, 69, 78, 85, 95]. Constitutively
expressed Notch proteins are released from a cell’s membrane into the cytoplasm (as
Notch Intercellular Domain (NICD)) after extracellular binding with a Delta ligand
(also a protein) presented through the membrane of an adjacent cell. In zebrafish,
NICD has been shown to activate expression of bHLH genes such as her1 and her7,
and the Notch ligand DeltaC oscillates in phase with these two clock-genes in the
PSM [5]. The Notch ligand DeltaD does not oscillate, however, but it is necessary
for coordinated oscillations and is constitutively expressed in the posterior PSM of
zebrafish along with other required genes in the Notch pathway such as Su(H) [91,
116].
Informed by the oscillatory expression of DeltaC synchronized with clock-gene
expression in zebrafish, the PCW model incorporates a single coordinating-signal
gene and assumes that production of coordinating signal mRNA is repressed by clock
protein homodimer and that the clock mRNA is activated by signaling protein from
adjacent cells [27]. Because Notch signaling is non-diffusive and contact-dependent,
the effect of signaling is confined to nearest neighbors. This arrangement is very
similar to the one proposed in [27]. Following [2, 27], the effect of the coupling signal
on the clock is assumed to be weaker than the clock-gene’s self-repression. Thus,
the model anticipates that intercellular coupling coordinates oscillations instead of
helping to drive them. While likely true for zebrafish [2, 28, 69, 78, 85, 95], this may
not be a valid assumption in chick or mouse [3, 71, 74, 83, 84].
49
Modeling Posterior Clock-Wave Formation: Uncoupled Cells
The PCW model is premised on a cell-autonomous oscillator, with a gradient
controlled oscillation rate and weak coupling for coordination of oscillations. Thus,
an uncoupled model is presented first, followed by an extension of the model that
adds intercellular coupling via a coordinating-signaling gene. The elements of the
uncoupled model and its construction are discussed next.
PSM Growth
As with the phase oscillator model in Chapter 3, a line of K total cells along the
medial anterior-to-posterior (AP) axis of the mesoderm is considered. Cells are assumed to enter the posterior-most PSM from the initiation zone in anterior-most tailbud at regularly spaced time intervals, without volume change or rearrangement [11].
Tk denotes the time of entry of the k th cell, and is given by the linear relationship in
equation (6) from Chapter 3, page 27, which is repeated here:
Tk :=
k−1
,
λµ
(11)
where λ is the number of cells per AP somite length (assumed constant), and µ is
the somite formation rate in somites per minute, which is equal to the oscillation
frequency in the tailbud [5] (also assumed constant). These steady growth and oscillation assumptions should be a reasonable approximation over a significant portion
of developmental time [93].
Model Variables
On the cellular level, the dynamics of the oscillator are realized by a clock-gene and
a control protein that interacts with the clock protein by competitive dimerization.
50
For each cell, the model’s state variables are the amount of total clock protein and
mRNA, whereas the amount of total control protein is prescribed non-autonomously
as an external control on the system.
For the k th cell, let ck (t) represent the cytosolic copy number of clock mRNA as a
function of time and define the nuclear copy number of total clock protein and total
signal protein, respectively, as follows:
bk (t) := Ck (t) + 2 C:C k (t) + C:Gk (t),
C
(12)
bk (t) := Gk (t) + 2 G:Gk (t) + C:Gk (t),
G
(13)
where Ck , C:C k , Gk , G:Gk , and C:Gk are the nuclear copy number of clock protein monomer, clock protein homodimer, control protein monomer, control protein
homodimer, and clock protein heterodimer with control protein, respectively.
The Control Protein
bk (t) is assumed to be controlled
The amount of nuclear total control protein G
by an external system that is not explicitly modeled here. Before cell k leaves the
bk (t) is assumed to be maintained at
tailbud and enters the posterior PSM (t < Tk ), G
bmax . After entering the PSM (t ≥ Tk ), the total control protein
a maximal constant G
bk (t) is
is assumed to decrease monotonically to zero with half-life τ 1 . Altogether, G
2
given by
bk (t) = G
bmax · g(t − Tk ; τ 1 ),
G
2
(14)
51
with the continuously differentiable function g given by
1,
t ≤ 0,
„
«.
2 t−τ 1
t
2
1 − 1 e
,
0 < t ≤ τ1 ,
2
2
„
«.„
«
g(t; τ 1 ) =
2
2 t−τ 1
t−2 τ 1
1
2
2 ,
e
τ1 ≤ t < 2 τ1 ,
2
2
2
0,
2 τ 1 ≤ t,
(15)
2
where τ 1 > 0 is a half-life parameter. The function g is sigmoidal, non-increasing,
2
concave down for 0 < t < τ 1 , and concave up for τ 1 < t < 2 τ 1 . When viewed axially
2
2
2
along an AP line of PSM cells, the resulting spatial profile agrees qualitatively with
those computed in [64]. See Figure 13.
Other gradient profiles are certainly possible, such as the following simple exponential profile [43]
1,
gexp (t; τ 1 ) =
ln 2
2
−
t
τ1
2
e
t ≤ 0,
(16)
0 < t.
Note that gexp is continuous but not differentiable at t = 0, and therefore the resulting
spatial profile is not smooth where cells exit the tailbud.
The Intracellular Clock
Clock protein is produced as monomer at the ribosomes in the cytosol, but is active
as a homodimerized repressor in the nucleus. Clock mRNA is produced at the DNA in
the nucleus, but it is translated into protein in the cytosol. Following Lewis [27], the
units of protein and mRNA concentrations are copy number per nuclear and cytosolic
compartment, respectively.
52
Temporal Gradient,
Cells 1 and 10
Cell 1
Cell 10
1
0.8
0.6
0.4
0.2
0
0
50
100
150
Time (min.)
Spatial Gradient,
Time= 84 min.
(b)
200
Normalized Total Control Protein
Normalized Total Control Protein
(a)
1
0.8
0.6
0.4
0.2
0
1
10
20
30
40
50
Cell # (1 is anterior)
Figure 13: Total control protein gradients. The spatiotemporal gradient of the nork (t)
malized total control protein, G
b max = g(t − Tk ; 30), where t is time, Tk is the time
G
that cell k exits the tailbud, and τ 1 = 30 minutes is the half-life. (a) Spatiotemporal
2
gradient as a function of time in cell one, g(t − T1 ; 30), and cell ten, g(t − T10 ; 30).
Cell ten, which exits from tailbud later than the cell one (T1 < T10 ), maintains the
high level of total control protein for a longer time. (b) Spatiotemporal gradient as a
function of cell position given by g(84 − Tk ; 30), k = 1 to 50. As tailbud growth adds
cells to the PSM, the tailbud-PSM boundary moves right, and the spatial gradient
profile follows this moving boundary.
b
53
Competitive dimerization of the clock and control proteins is represented by the
following three possible bidirectional dimerization reactions:
2C
+
kC:C
−
kC:C
C:C,
2G
+
kG:G
−
kG:G
G:G,
and C + G
+
kC:G
−
kC:G
C:G,
(17)
where the forward and reverse (strictly) positive reaction rates are given above and
below the arrows, respectively.
Using mass action kinetics for the protein dimerization reactions (17), and considering the production and decay of all forms of the clock protein and of clock mRNA,
leads to the following system of delay differential equations that describe the dynamics
of the clock-gene in the k th cell:
−
Ċk (t) = αC ck (t − τC ) − βC Ck (t) + 2 kC:C
C:C k (t) −
+
−
+
2 kC:C
(Ck (t))2 + kC:G
C:Gk (t) − kC:G
Ck (t) Gk (t),
˙ k (t) = −βC:C C:C k (t) − k − C:C k (t) + k + (Ck (t))2 ,
C:C
C:C
C:C
(18)
(19)
−
G:Gk (t) −
Ġk (t) = Pk (t) − βG G(t) + 2 kG:G
+
−
+
2 kG:G
(Gk (t))2 + kC:G
C:Gk (t) − kC:G
Ck (t) Gk (t),
(20)
˙ k (t) = −βG:G G:Gk (t) − k − G:Gk (t) + k + (Gk (t))2 ,
G:G
G:G
G:G
(21)
˙ k (t) = −βC:G C:Gk (t) − k − C:Gk (t) + k + Ck (t) Gk (t),
C:G
C:G
C:G
(22)
ċk (t) = γc Hn (C:C k (t − τc )) − δc ck (t),
(23)
where the function Hn represents the nonlinear effect of the repressive homodimer
C:C on mRNA production and is described in detail below. Greek letters represent
non-negative model parameters. The parameter αC is the production rate of protein
(as monomer) from mRNA occurring with delay τC . Parameters βC , βC:C , and βC:G
are the linear decay rates of clock protein in monomer, homodimer, and heterodimer
54
forms, respectively. The parameter γc affects the mRNA production rate, which
occurs with delay τc . The parameter δc is the linear mRNA decay rate. It is assumed
that all forms of control protein may decay linearly with rates βG , βG:G , βC:G . Furthermore, the production of control protein monomer, given by the function Pk (t),
is assumed to be chosen such that the total control protein in cell k is prescribed
by (14).
Taking a time derivative of (12) and using equations (18)–(19) gives:
b˙ k (t) = Ċk (t) + 2 C:C
˙ k (t) + C:G
˙ k (t)
C
= αC ck (t − τC ) − βC Ck (t) − 2 βC:C C:C k (t) − βC:G C:Gk (t).
Pre-appending this differential equation for total clock protein to the above system
gives the following:
b˙ k (t) = αC ck (t − τC ) − βC Ck (t) − 2 βC:C C:C k (t) − βC:G C:Gk (t),
C
(24)
−
Ċk (t) = αC ck (t − τC ) − βC Ck (t) + 2 kC:C
C:C k (t) −
−
+
+
(Ck (t))2 + kC:G
C:Gk (t) − kC:G
Ck (t) Gk (t),
2 kC:C
˙ k (t) = −βC:C C:C k (t) − k − C:C k (t) + k + (Ck (t))2 ,
C:C
C:C
C:C
(25)
(26)
−
Ġk (t) = Pk (t) − βG Gk (t) + 2 kG:G
G:Gk (t) −
+
−
+
2 kG:G
(Gk (t))2 + kC:G
C:Gk (t) − kC:G
Ck (t) Gk (t),
(27)
˙ k (t) = −βG:G G:Gk (t) − k − G:Gk (t) + k + (Gk (t))2 ,
G:G
G:G
G:G
(28)
˙ k (t) = −βC:G C:Gk (t) − k − C:Gk (t) + k + Ck (t) Gk (t),
C:G
C:G
C:G
(29)
ċk (t) = γc Hn (C:C k (t − τc )) − δc ck (t).
(30)
55
Note that the special case where βC = βC:C = βC:G =: βCb gives linear decay of
total clock protein, i.e., equation (24) becomes
b˙ k (t) = αC ck (t − τC ) − βC Ck (t) − 2 βC:C C:C k (t) − βC:G C:Gk (t)
C
= αC ck (t − τC ) − βCb (Ck (t) + 2 C:C k (t) + C:Gk (t))
bk (t).
= αC ck (t − τC ) − βCb C
(31)
Differential decay of protein monomers and dimers has been shown to have important
implications on system dynamics [107], thus nonlinear decay of total protein, in which
βC , βC:C , and βC:G are not all equal, is considered during model validation in the next
chapter.
System (24)–(30) can be simplified considerably by making a fast dimerization
approximation [107, 109]. This approximation relies upon the assumption that the
dimerization reactions (17) occur on a much faster timescale than all other production
and decay processes [117]. The details of this approach are discussed further below.
Clock-Gene Regulation by a
Single Repressive Transcription Factor
The transcription factor C:C is assumed to down-regulate clock mRNA production
through one or more cis binding sites at the gene’s promoter1 , and the number of
such binding sites can be a key factor in the generation of sustained oscillations [81].
The binding of this transcription factor to DNA is assumed to be in thermodynamic
equilibrium, and the approach of Shea and Ackers [23, 108, 109] is used to derive the
function Hn in equation (30), where n = 1 or 2 binding sites. This function models
the probability that the RNA polymerase II (RNAP-II) transcription complex is
1
A cis binding site is on the same side of the DNA as the gene’s promoter region.
56
assembled on a gene’s promoter. Specifically,
Hn (C:C) =
ZON
,
ZON + ZOFFn
where ZON and ZOFFn are sums of terms representing states where RNAP-II complex is bound or unbound, respectively, to the gene’s promoter. The ratio gives the
probability that the promoter is in the RNAP-II bound state in which clock mRNA
transcription occurs. In this case,
ZON = ρP P
and
ZOFFn = 1 + Ψn (C:C),
so that
Hn (C:C) =
ρP P
,
ρP P + (1 + Ψn (C:C))
(32)
The term in the numerator in (32) represents the single RNAP-II bound state in which
mRNA transcription occurs, while the terms in denominator represents all possible
states of the promoter. The function Ψn depends upon the number n of cis binding
sites for the repressor C:C and is described more fully next.
The terms in the numerator and denominator of (32) are derived from the DNA
binding energies and concentrations of various biomolecules. For example, the binding
probability of a single RNAP-II complex to the gene’s promoter is represented by the
product of the binding affinity ρP for RNAP-II complex and the copy number P
of the complex. The binding affinity has units of inverse copy number, and may
be computed from the binding energy ∆GP < 0 of the RNAP-II complex using the
thermodynamically derived formula ρP := e
∆GP
RT
, where R is the ideal gas constant
and T is the temperature [23]. In the denominator of (32), the zero energy state with
∆G0
nothing bound to DNA (∆G0 = 0) is represented by the constant 1 = e0 = e R T .
57
While binding energies have been measured in certain model prokaryotic systems [23, 108, 109, 118], to this author’s best knowledge they have not been measured for any of the eukaryotic cells in which somitogenesis is studied. Thus, binding
affinity parameters must be estimated from a biologically realistic range during model
validation, as discussed in the next chapter. Furthermore, the unitless parameter
ρ := ρP P
is estimated instead of estimating both the binding affinity ρP for RNAP-II complex
and the copy number P of the complex, which is assumed constant. The lumped
parameter ρ incorporates the assembly of the RNAP-II complex, a process which,
in eukaryotes, can involve several intermediate steps [119, see Fig. 6-16], and which
are assumed fast relative to other processes. This modeling formalism is a significant simplification of the eukaryotic transcription process [34], but it represents an
appropriate step towards biological realism as compared to existing models [108].
The repression of the C:C dimer is assumed to be ideal, i.e., the binding of repressor
and RNAP-II complex is assumed to be mutually exclusive. The function Ψn (C:C)
represents the states of self-repression of the clock mRNA by clock protein homodimer,
C:C, given n binding sites. See Figure 14 a–b. For one binding site,
Ψ1 (C:C) := ρC:C C:C,
(33)
where ρC:C is the binding affinity for the C:C dimer to the single binding site, with
units of inverse copy number. For two binding sites,
Ψ2 (C:C) = ρC:C- C:C + ρ-C:C C:C + ωC:C (ρC:C- C:C) (ρ-C:C C:C) ,
(34)
where ρC:C- and ρ-C:C are the binding affinities for the C:C homodimer to the first
and second binding site, respectively. ωC:C ≥ 0 is the unitless cooperativity between
58
Figure 14: Binding site configurations. Possible cis binding site configurations at
the clock-gene promoter. P is RNAP-II transcription complex which binds to the
blue region. C:C is the clock protein homodimer which prevents P from binding
by binding to the red region(s). N is an activator which binds to the green region
adjacent to the P and C:C binding sites. (a) One repressor binding site with no
activation. (b) Two repressor binding sites with no activation. (c) One repressor
binding site with activation. (d) Two repressor binding sites with activation.
two simultaneously bound C:C homodimers, which accounts for any additional energy
released (or required) when two homodimers bind simultaneously. For simplicity in
the face of lack of knowledge, each site is assumed equally likely to be bound by C:C
dimer, so that
ρC:C- = ρ-C:C =: ρC:C ,
and equation (34) reduces to
Ψ2 (C:C) := 2 (ρC:C C:C) + ωC:C (ρC:C C:C)2 .
(35)
Altogether, gene regulation with one or two binding sites for a single, ideal repressor is given by the function
Hn (C:C) :=
ρ
,
ρ + (1 + Ψn (C:C))
(36)
with n = 1 or 2, and the functions Ψ1 and Ψ2 given by equations (33) and (35),
respectively. The function Hn can be viewed as a generalization of the monotonic Hill
59
functions typically used to model mRNA transcription [110], such as in differential
equation (5) of the Lewis model on page 20.
Modeling Posterior Clock-Wave Formation: Coupled Cells
The model developed above does not include intercellular signaling. Coupling
between cells is now added to produce the full PCW model.
Intercellular Signaling
The dynamics of the coordinating signal protein, Sk , and mRNA, sk , in the k th cell
are given by the following equations, which are respectively similar to (24) and (30):
Ṡk (t) = αS sk (t − τS ) − βS Sk (t),
ṡk (t) = γs H1 (C:C k (t − τs )) − δs sk (t),
with Greek letters representing the analogous parameters and H1 as given in equation (36). To the best of this author’s knowledge, there are no experimental data
concerning the precise action of repressive clock protein dimers at the coordinatingsignal gene’s promoter. Following [2, 27], one binding site is assumed. Note that the
coordinating signal protein S acts in the nuclei of adjacent cells. Thus, τS includes
the cell membrane exchange time for the coordinating signal protein, and is typically
an order of magnitude larger than τC .
Clock-Gene Regulation by Both
Repressive and Activating Transcription Factors
The coordinating signal protein influences adjacent cells by up-regulation of clockgene expression in adjacent cells [120]. The combined effect of self-inhibition and
60
activation from adjacent cells requires replacing the dynamics of the clock mRNA
(equation (30)) with the following delay differential equation:
ċk (t) = γc Fn (C:C k (t − τc ), Nk (t − τc )) − δc ck (t),
where
X
Nk (t) :=
Si (t)
(37)
{i : cell i adjacent to cell k}
is the total signal from neighboring cells, and is called the signaling protein in cell k.
Following [2, 27], this summation is a simplification of the membrane-mediated signal
transduction of the Notch intercellular communication pathway [121]. In particular,
the possible nonlinear effects of membrane receptor saturation are not considered.
The function Fn represents the combined effect of the repressor C:C and activator
N on clock mRNA production. Again using the approach of Shea and Ackers [23,
108, 109], Fn is given by the following probability ratio:
Fn (C:C, N ) =
ZON
,
ZON + ZOFFn
where ZON and ZOFFn are sums of terms representing states where RNAP-II complex
is bound or unbound, respectively, to the gene’s promoter. The positive integer n
is the number of cis binding sites for the repressor, while the activator is assumed
to have only one binding site. See Figure 14 c–d. As before, the binding of the
repressor and RNAP-II complex is mutually exclusive, however binding of repressor
and activator is not assumed to be mutually exclusive. Transcription proceeds only
in those states in which RNAP-II complex is bound, which may or may not occur
cooperatively with activator.
61
Consideration of the two possible RNAP-II bound states gives
ZON = ρP P 1 + ωN (ρN N )
= ρP P (1 + ωN -P (ρN N ))
= ρ (1 + ωN (ρN N )) ,
where ρ := ρP P , as before. ρN is the binding affinity of the activator N induced by
adjacent cells’ coordinating signal protein S (recall equation (37)). The assumption
that intercellular coupling is weak relative to intracellular negative feedback means
that ρN ρC:C . ωN > 1 is the cooperativity between the activator N and the
RNAP-II complex.
For one repressor binding site, consideration of all the RNAP-II unbound states
gives
ZOFF1 = 1 + ρC:C C:C + ρN N + ωN -C:C (ρN N ) (ρC:C C:C) ,
where ωN -C:C represents the cooperativity between simultaneously bound repressor
and activator molecules. Consideration of two repressor binding sites gives
ZOFF2 = 1 + ρC:C- C:C + ρ-C:C C:C + ωC:C (ρC:C- C:C) (ρ-C:C C:C) +
ρN N + ωN -C:C- (ρN N ) (ρC:C- C:C) + ωN --C:C (ρN N ) (ρ-C:C C:C) +
ωN -C:C-C:C (ρN N ) (ρC:C- C:C) (ρ-C:C C:C) ,
where ωN -C:C- , ωN --C:C , and ωN -C:C-C:C represent the cooperativity between simultaneously bound repressor and activator molecules, with the first, second, and both
repressor site(s) occupied, respectively. As before, ρC:C- and ρ-C:C are the binding
affinities for the C:C homodimer to the first and second binding site, respectively,
with simultaneous binding cooperativity ωC:C .
To the best of the this author’s knowledge, no experimental data are available on
the binding affinities and cooperativities for the repressor and activator in PSM cells.
62
In response to this lack of knowledge, two straightforward, simplifying assumptions
are as follows:
1. The activator N and the repressor C:C bind independently of each other, so
that for one binding site
ωN -C:C = 1,
while for two binding sites
ωN -C:C- = ωN --C:C = 1 and ωN -C:C-C:C = ωC:C .
2. For two binding sites, the binding affinity of the repressor at each binding site
is the same, i.e.,
ρC:C- = ρ-C:C =: ρC:C .
Applying these assumptions on the parameters for one binding site gives
ZOFF1 = 1 + ρC:C C:C + ρN N + (ρN N ) (ρC:C C:C)
= 1 + ρC:C C:C + ρN N (1 + ρC:C C:C)
= (1 + ρN N ) (1 + ρC:C C:C)
= (1 + ρN N ) (1 + Ψ1 (C:C)) ,
where Ψ1 is given as before by equation (33). For two binding sites
ZOFF2 = 1 + 2 (ρC:C C:C) + ωC:C (ρC:C C:C)2 +
ρN N + 2 (ρN N ) (ρC:C C:C) + ωC:C (ρN N ) (ρC:C C:C)2
= 1 + 2 (ρC:C C:C) + ωC:C (ρC:C C:C)2 +
ρN N 1 + 2 (ρC:C C:C) + ωC:C (ρC:C C:C)2
= (1 + ρN N ) 1 + 2 (ρC:C C:C) + ωC:C-C:C (ρC:C C:C)2
= (1 + ρN N ) (1 + Ψ2 (C:C)) ,
63
where Ψ2 is given as before by equation (35). Altogether, for n = 1 or 2, the function
Fn is given by
Fn (C:C, N ) :=
ρ (1 + ωN ρN N )
.
ρ (1 + ωN ρN N ) + (1 + ρN N ) (1 + Ψn (C:C))
(38)
The same repressor binding affinity of clock dimers, ρC:C , is used in both Fn and
Hn (equation (36) on page 58), even though in the full PCW model these functions
represent activity at different genes. While this and the other simplifying assumptions
may restrict the PCW model’s behavior, it is a reasonable compromise given the lack
of measured parameter data, and it significantly reduces the number of parameters
that must be estimated during model validation.
64
Interim Model Summary
Altogether, the full model is given by the following system of delay differential
equations in each cell:
b˙ k (t) = αC ck (t − τC ) − βC Ck (t) − 2 βC:C C:C k (t) − βC:G C:Gk (t),
C
(39)
−
Ċk (t) = αC ck (t − τC ) − βC Ck (t) + 2 kC:C
C:C k (t) −
+
+
−
(Ck (t))2 + kC:G
C:Gk (t) − kC:G
Ck (t) Gk (t),
2 kC:C
˙ k (t) = −βC:C C:C k (t) − k − C:C k (t) + k + (Ck (t))2 ,
C:C
C:C
C:C
(40)
(41)
−
Ġk (t) = Pk (t) − βG Gk (t) + 2 kG:G
G:Gk (t) −
+
−
+
2 kG:G
(Gk (t))2 + kC:G
C:Gk (t) − kC:G
Ck (t) Gk (t),
(42)
˙ k (t) = −βG:G G:Gk (t) − k − G:Gk (t) + k + (Gk (t))2 ,
G:G
G:G
G:G
(43)
˙ k (t) = −βC:G C:Gk (t) − k − C:Gk (t) + k + Ck (t) Gk (t),
C:G
C:G
C:G
(44)
ċk (t) = γc Fn (C:C k (t − τc ), Nk (t − τc )) − δc ck (t),
(45)
Ṡk (t) = αS sk (t − τS ) − βS Sk (t),
(46)
ṡk (t) = γs H1 (C:C k (t − τs )) − δs sk (t),
(47)
where
Nk (t) =
X
Si (t),
(48)
{i : cell i adjacent to cell k}
and subject to the following constraints:
bk (t) = Ck (t) + 2 C:C k (t) + C:Gk (t),
C
(49)
bk (t) = Gk (t) + 2 G:Gk (t) + C:Gk (t).
G
(50)
65
where the total control protein is prescribed by
bk (t) = G
bmax · g(t − Tk ; τ 1 ).
G
2
(51)
The next section explains how a fast dimerization approximation is used to significantly simplify this model, enabling efficient numerical solution and analysis of the
model, as well as validation of the model in the next chapter.
The Fast Dimerization Approximation
The above system will now be non-dimensionalized in order to make a fast dimerization approximation. In this approximation, the protein dimerization reactions (17)
are assumed to be much faster than the other production and decay processes of clock
and control protein. Intuitively, this approximation makes the equilibration of the
dimerization reactions (17) instantaneous as the levels of total control protein and
total clock protein fluctuate in time. To this end, time is rescaled to dimensionless
time τ with respect to the maximum production or decay rate of clock and control
protein. Specifically,
τ := ν t,
where
ν := max {αC , βC , βC:C , 1, βG , βG:G , βC:G }
(52)
has units of inverse time. Without loss of generality, the cell number subscripts are
b
omitted, and the dependent variables are rescaled to the dimensionless variables X,
66
X, X:X, Yb , Y , Y :Y , X:Y , x, W ,
b τ
C
ν
b ) :=
X(τ
,
κC:C
b τ
G
ν
Yb (τ ) :=
,
κG:G
C:G ντ
X:Y (τ ) :=
,
κC:G
S ντ
Z(τ ) := γs ,
Z, and z as follows:
C ντ
X(τ ) :=
,
κC:C
G ντ
Y (τ ) :=
,
κG:G
c ντ
x(τ ) :=
,
κC:C
s ντ
z(τ ) := γs ,
αS
C:C ντ
X:X(τ ) :=
,
κC:C
G:G ντ
Y :Y (τ ) :=
,
κG:G
N ντ
W (τ ) := γs ,
αS
αS
where the positive dimerization dissociation constants are defined as follows:
κC:C :=
−
kC:C
+ ,
kC:C
each with units of copy number.
κG:G :=
γs
,
αS
−
kG:G
+ ,
kG:G
and κC:G :=
−
kC:G
+ ,
kC:G
also with units of copy number, is a convenient
choice for rescaling the coordinating signal protein, mRNA, and signaling protein.
This choice does not affect the derivation of the fast dimerization approximation.
The equations in system (39)–(47) are rewritten in the new dimensionless variables
as follows:
b˙ τ
C
ν
b 0 (τ ) =
X
ν κC:C
C
αC c τ −τ
− βC C ντ − 2 βC:C C:C ντ − βC:G C:G ντ
ν
=
ν κC:C
τC
βC
βC:C
βC:G κC:G
αC
=
x τ−
−
X(τ ) − 2
X:X(τ ) −
X:Y (τ ),
ν
ν
ν
ν
ν κC:C
Ċ ντ
0
X (τ ) =
νκ
C:C τ τ τ 2
τ − τC
−
+
C
= αC c
− βC C
+ 2 kC:C
C:C
− 2 kC:C
+
ν
ν
ν
ν
,
τ τ τ −
+
kC:G
C:G
− kC:G
C
G
(ν κC:C )
ν
ν
ν
−
αC τC βC
kC:C
=
x τ−
−
X(τ ) + 2
X:X(τ ) − (X(τ ))2 +
ν
ν
ν
ν
−
kC:G
κC:G
κG:G
X:Y (τ ) −
X(τ ) Y (τ ) ,
ν
κC:C
κC:G
67
˙ τ
C:C
ν
X:X (τ ) =
ν κC:C
−βC:C C:C
=
0
2
−
+
− kC:C
C:C ντ + kC:C
C ντ
ν κC:C
−
k
X2 (τ ) − C:C X2 (τ ) − (X1 (τ ))2 ,
ν
τ
ν
βC:C
ν
Ġ ντ
0
Y (τ ) =
νκ
G:G
τ τ τ τ 2
−
+
= P
− βG G
+ 2 kG:G
G:G
− 2 kG:G
G
+
ν
ν
ν
ν
τ τ τ .
+
−
− kC:G C
G
kC:G C:G
(ν κG:G )
ν
ν
ν
−
P ντ
βG
kG:G
=
−
Y (τ ) + 2
G:G(τ ) − (Y (τ ))2 +
ν κG:G
ν
ν
−
kC:G κC:G
κC:C
X:Y (τ ) −
X(τ ) Y (τ ) ,
ν
κG:G
κC:G
=−
˙ τ
G:G
ν
Y :Y (τ ) =
ν κG:G
−βG:G G:G
=
0
2
−
+
− kG:G
G:G ντ + kG:G
G ντ
ν κG:G
−
βG:G
k
=−
Y :Y (τ ) − G:G Y :Y (τ ) − (Y (τ ))2 ,
ν
ν
˙ τ
C:G
0
ν
X:Y (τ ) =
ν κC:G
−
+
−βC:G C:G ντ − kC:G
C:G ντ + kC:G
C ντ G ντ
=
ν κC:G
−
βC:G
kC:G
κC:C κG:G
=−
X:Y (τ ) −
X:Y (τ ) −
X(τ ) Y (τ ) ,
ν
ν
κ2C:G
τ
ν
τ
ċ
ν
x0 (τ ) =
ν κC:C
c
c
γc Fn C:C τ −τ
, N τ −τ
− δc c ντ
ν
ν
=
ν κC:C
γc
τc αS
τc δc
=
Fn κC:C X:X τ −
W τ−
− x(τ ),
,
ν κC:C
ν
γs
ν
ν
68
Ṡ
Z(τ ) =
ν
=
τ
ν
γs
αS
αS s
τ −τS
ν
ν
− βS S
τ
ν
γs
αS
αS τS βS
z τ−
Z(τ ),
−
ν
ν
ν
ṡ ντ
z(τ ) = γs
ν αS
s
γs H1 C:C τ −τ
− δs s ντ
ν
=
ν αγSs
τs δs
αS
H1 κC:C X:X τ −
− z(τ ).
=
ν
ν
ν
=
69
Altogether, this gives the following equivalent non-dimensionalized system, which
has small parameters if the dimerization reactions (17) are fast:
b 0 (τ ) = αC x τ − τC − βC X(τ ) −
X
ν
ν
ν
βC:C
βC:G κC:G
X:X(τ ) −
X:Y (τ ),
2
ν
ν κC:C
ν
αC τC βC
0
X
(τ
)
=
x
τ
−
− − X(τ ) + 2 X:X(τ ) − (X(τ ))2 +
−
−
ν
kC:C
kC:C
kC:C
−
kC:G κC:G
κG:G
X:Y (τ ) −
X(τ ) Y (τ ) ,
−
κC:C
κC:G
kC:C
ν
βC:C
X:X(τ ) − X:X(τ ) − (X(τ ))2 ,
−
kC:C
1 P ντ
ν
βG
0
Y (τ ) = −
− − Y (τ ) + 2 Y :Y (τ ) − (Y (τ ))2 +
−
kG:G
kG:G κG:G
kG:G
−
kC:G
κC:C
κC:G
X:Y (τ ) −
X(τ ) Y (τ ) ,
−
κG:G
κC:G
kG:G
−
kC:C
ν
X:X 0 (τ ) = −
βG:G
Y :Y (τ ) − Y :Y (τ ) − (Y (τ ))2 ,
−
kG:G
ν
βC:G
κC:C κG:G
0
X(τ ) Y (τ ) ,
X:Y (τ ) = − − X:Y (τ ) − X:Y (τ ) −
−
κ2C:G
kC:G
kC:G
τc γc
τc αS
δc
0
x (τ ) =
− x(τ ),
Fn κC:C X:X τ −
,
W τ−
ν κC:C
ν
γs
ν
ν
αS τS βS
Z(τ ) =
z τ−
Z(τ ),
−
ν
ν
ν
αS
τs δs
z(τ ) =
H1 κC:C X:X τ −
− z(τ ).
ν
ν
ν
−
kG:G
Y :Y 0 (τ ) = −
Given the following unitless parameter definitions:
ν
εX :=
− ,
kC:C
εY :=
− ,
kG:G
εX:Y :=
− ,
kC:G
ν
ν
εαC :=
ε1 :=
εβC:G :=
αC
− ,
kC:C
1
− ,
kG:G
βC:G
− ,
kC:G
εβC :=
βC
− ,
kC:C
εβC:C :=
βC:C
− ,
kC:C
εβG :=
βG
− ,
kC:G
εβG:G :=
βG:G
− ,
kC:G
70
by the definition of ν (see (52)), it follows that
ε∗ ≤ εX ,
for ∗ ∈ {αC , βC , βC:C },
ε∗ ≤ εY ,
for ∗ ∈ {1, βG , βG:G },
εβC:G ≤ εX:Y .
(Note that the output of the function P is defined to have units of copy number, so
that the constant 1 in the above definition of ε1 has units of inverse time, corresponding to the use of this constant in the definition of ν in (52).)
Rewriting yet again, the system becomes:
b 0 (τ ) = αC x τ − τC − βC X(τ ) −
X
ν
ν
ν
βC:G κC:G
βC:C
X:X(τ ) −
X:Y (τ ),
2
ν
ν κC:C
τC − εβC X(τ ) + 2 X:X(τ ) − (X(τ ))2 +
εX X 0 (τ ) = εαC x τ −
ν
−
kC:G
κG:G
κC:G
X:Y (τ ) −
X(τ ) Y (τ ) ,
−
κC:C
κC:G
kC:C
εX X:X 0 (τ ) = −εβC:C X:X(τ ) − X:X(τ ) − (X(τ ))2 ,
P ντ
0
− εβG Y (τ ) + 2 Y :Y (τ ) − (Y (τ ))2 +
εY Y (τ ) = εPG
κG:G
−
kC:G
κC:G
κC:C
X:Y (τ ) −
X(τ ) Y (τ ) ,
−
κG:G
κC:G
kG:G
εY Y :Y 0 (τ ) = −εβG:G Y :Y (τ ) − Y :Y (τ ) − (Y (τ ))2 ,
κC:C κG:G
0
X(τ ) Y (τ ) ,
εX:Y X:Y (τ ) = −εβC:G X:Y (τ ) − X:Y (τ ) −
κ2C:G
γc
τc αS
τc δc
0
x (τ ) =
Fn κC:C X:X τ −
,
W τ−
− x(τ ),
ν κC:C
ν
γs
ν
ν
αS τS βS
Z(τ ) =
z τ−
Z(τ ),
−
ν
ν
ν
αS
τs δs
z(τ ) =
H1 κC:C X:X τ −
− z(τ ).
ν
ν
ν
(53)
(54)
(55)
(56)
(57)
(58)
(59)
(60)
(61)
71
−
The fast equilibrium approximation of this system is given in the limit as kC:C
→
−
−
∞, kG:G
→ ∞, and kC:G
→ ∞ such that
−
kC:G
−
kC:C
and
−
kC:G
−
kG:G
both remain bounded. In
these limits,
ε∗ → 0 for all ∗ ∈ {X, Y, X:Y , αC , βC , βC:C , 1, βG , βG:G , βC:G }.
−
−
−
Thus, as kC:C
→ ∞, kG:G
→ ∞, and kC:G
→ ∞, all ε’s go to zero, and the system
simplifies to
b 0 (τ ) = αC x τ − τC − βC X(τ ) − 2 βC:C X:X(τ ) − βC:G κC:G X:Y (τ ), (62)
X
ν
ν
ν
ν
ν κC:C
X:X(τ ) = (X(τ ))2 ,
(63)
Y :Y (τ ) = (Y (τ ))2 ,
(64)
κC:C κG:G
X(τ ) Y (τ ),
κ2C:G
τc αS
τc γc
δc
0
Fn κC:C X:X τ −
,
W τ−
− x(τ ),
x (τ ) =
ν κC:C
ν
γs
ν
ν
τS βS
αS z τ−
−
Z(τ ),
Z(τ ) =
ν
ν
ν
αS
τs δs
z(τ ) =
H1 κC:C X:X τ −
− z(τ ).
ν
ν
ν
X:Y (τ ) =
(65)
(66)
(67)
(68)
Note that the right hand sides of (54) and (56) are both zero in the limit under the
assumption that both the ratios
−
kC:G
−
kC:C
and
−
kC:G
−
kG:G
remain bounded and by considering
the limiting relationships given by (55), (57), and (58).
Both dimerization reaction directions are indeed fast in this approximation because assuming
−
kC:C
→ ∞,
−
kG:G
→ ∞,
−
kC:G
→∞
and that κC:C , κG:G , and κC:G remain bounded, implies
+
kC:C
=
−
kC:C
→ ∞,
κC:C
+
kG:G
=
−
kG:G
→ ∞,
κG:G
+
kC:G
=
−
kC:G
→ ∞.
κG:G
72
Furthermore, in practice,
ν = max {αC , βC , βC:C , 1, βG , βG:G , βC:G } = 4.5 minute−1 ,
so that
εX =
εY =
εX:Y =
ν
−
kC:C
ν
−
kG:G
ν
−
kC:G
−
1 when kC:C
4.5 minute−1 ,
−
1 when kG:G
4.5 minute−1 ,
−
4.5 minute−1 .
1 when kC:G
Not only are the dimerization reaction rates typically unknown, the dissociation constants are also typically unknown. Thus, the parameters κC:C , κG:G , and κC:G , which
remain after the fast dimerization approximation, often must be estimated from a
biologically realistic range of values.
By considering the non-dimensionalized versions of (49)–(50), system (62)–(68) is
subject to the following constraints:
b ) = X(τ ) + 2 X:X(τ ) + κC:G X:Y (τ ),
X(τ
κC:C
κC:G
Yb (τ ) = Y (τ ) + 2 Y :Y (τ ) +
X:Y (τ ).
κG:G
Appending these constraints and renaming parameters as follows:
τX :=
τC
,
ν
αX :=
τx :=
τc
,
ν
γx :=
τZ :=
τS
,
ν
αZ :=
αC
,
ν
βX :=
βC
,
ν
βX:X :=
βC:C
,
ν
βX:Y :=
βC:G
,
ν
γc
,
ν κC:C
δx :=
δc
,
ν
κX :=
κC:C
,
κC:G
κY :=
κG:G
,
κC:G
αS
,
ν
βZ :=
βS
,
ν
τz :=
τs
,
ν
δz :=
δs
,
ν
73
gives the following equivalent non-dimensionalized system after some algebraic simplification and reintroduction of cell indexing:
bk0 (τ ) = αX xk (τ − τX ) − βX Xk (τ ) −
X
2
2 βX:X Xk (τ ) − βX:Y κY Xk (τ ) Yk (τ ),
2 α
S
0
xk (τ ) = γx Fn κC:C Xk (τ − τx ) ,
Wk (τ − τx ) − δx xk (τ ),
γs
(69)
(70)
Zk (τ ) = αZ zk (τ − τZ ) − βZ Zk (τ ),
2 − δz zk (τ ),
zk (τ ) = αZ H1 κC:C Xk (τ − τz )
(72)
bk (τ ) = Xk (τ ) + 2 Xk (τ ) 2 + κY Xk (τ ) Yk (τ ),
X
(73)
2
Ybk (τ ) = Yk (τ ) + 2 Yk (τ ) + κX Xk (τ ) Yk (τ ),
(74)
(71)
where
Wk (τ ) =
X
Zi (τ ).
(75)
{i : cell i adjacent to cell k}
If the last two nonlinear algebraic equations in this system ((73)–(74)) can be solved
bk (τ ) and Ybk (τ ), then this solution can be
to give Xk (τ ) and Yk (τ ) as functions of X
used to remove dependence on Xk (τ ) and Yk (τ ) in the remaining equations.
Algebraic Solution of the Fast Dimerization
Equations (73)–(74) can be solved algebraically for Xk (t) and Yk (t) in terms of
bk (t) and Ybk (t). Omitting the cell index and dimensionless time from the notation,
X
these two equations can be rewritten as
b
0 = 2 X 2 + (1 + κY Y ) X − X,
(76)
0 = 2 Y 2 + (1 + κX X) Y − Yb .
(77)
74
The quadratic formula can be used to solve equation (77) for the non-negative
root Y in terms of X giving
Y =
− (1 + κX X) +
1
=
4
q
(1 + κX X)2 − 4 · 2 (−Yb )
2·2
q
b
(1 + κX X) + 8 Y − (1 + κX X) .
2
Substituting this expression for Y into equation (76) gives
q
κY
2
2
b
(1 + κX X) + 8 Yb − (1 + κX X)
X − X.
0 = 2X + 1 +
4
(78)
Squaring both sides of the following equivalent equation:
q
κY
2
b
b − 2 X 2 − 1 − κY (1 + κX X) X
(1 + κX X) + 8 Y X = X
4
4
gives the following quartic polynomial equation in X:
κ κ
Y X
+ κY − 4 x3 +
2
!
b κY
κ2Y Yb
κY κX X
b − 1 X2 +
−
+
+ 4X
2
2
2
0 = (κY κX − 4) X 4 +
b
b − κY X
2X
2
!
b 2 . (79)
X −X
A non-negative solution to equation (78) is also a solution to the quartic equation (79), however existence and uniqueness of such a solution is not immediately
transparent and extraneous roots may be introduced. Furthermore, the formula for
the four roots of equation (79) is somewhat complicated and automating root selection
in software requires a rather elaborate algorithm.
An alternative approach employs a simple iterative numerical scheme to solve the
nonlinear system (76)–(77). As shown in the next section, this scheme has certain
theoretical and computational advantages: under a broad range of parameters it
gives efficient convergence to a unique non-negative root of the system (76)–(77). A
noteworthy advantage of this iterative numerical technique is that it extends nicely
to competitive dimerization of three or more proteins, whereas a purely algebraic
approach becomes intractable (see Appendix B).
75
An Iterative Numerical Scheme
for Computing the Fast Dimerization
The derivation of an iterative scheme for solving the system (76)–(77) begins by
using the quadratic formula to solve each equation for the non-negative roots X and
Y , respectively. This gives
q
1
2
b
X=
(1 + κY Y ) + 8 X − (1 + κY Y ) ,
4
q
1
2
b
(1 + κX X) + 8 Y − (1 + κX X) ,
Y =
4
2
which defines a map f from the non-negative quadrant R+ into itself, namely,
Xn+1
Xn
(80)
=f
Yn+1
Yn
whose component functions f1 and f2 are given by
q
1
2
b
(1 + κY Yn ) + 8 X − (1 + κY Yn ) ,
Xn+1 = f1 (Yn ) :=
4
q
1
2
b
Yn+1 = f2 (Xn ) :=
(1 + κX Xn ) + 8 Y − (1 + κX Xn ) ,
4
(81)
(82)
A fixed point of this map solves (76)–(77), and the special structure of the component
functions f1 and f2 of the map f gives the following relationships:
xn+2 = (f1 ◦ f2 )(xn ),
yn+2 = (f2 ◦ f1 )(yn ).
A solution to (76)–(77) is found if each of these composition maps can be shown to
converge to a unique fixed point from any non-negative initial condition. Recall the
well-known Banach Contraction Principle:
Theorem 1 (Banach Contraction Principle). Let W be a closed subset of a Banach
space (V, k · k). If f is a contraction map from W into itself, then f has a unique
76
fixed point p in W . Furthermore, if w is any element of W , then
lim f n (w) = p,
kf n (w) − pk ≤ cn kw − pk,
and
n→∞
where c < 1 is the Lipschitz constant for the contraction map f .
Under certain conditions on the parameters, the composition maps f1 ◦ f2 and
f2 ◦ f1 are contraction maps from R+ into itself, where R+ := [0, ∞) is a closed subset
of the Banach space (R, | · |). To this end, consider the following first derivatives of
f1 and f2 :
1 + κY Y
κY
q
− 1 < 0,
4
2
b
(1 + κY Y ) + 8 X
1 + κX X
κX
q
− 1 < 0.
f20 (X) =
4
(1 + κX X)2 + 8 Yb
f10 (Y ) =
Furthermore, the second derivatives of f1 and f2 are given by
− 3
b (1 + κY Y )2 + 8 X
b 2 ≥ 0,
f100 (Y ) = 2 κ2Y X
− 23
00
2 b
2
b
f2 (X) = 2 κX Y (1 + κX X) + 8 Y
≥ 0,
so that f10 and f20 are negative, non-decreasing, and bounded above by zero. Thus,
the maximum absolute values of these derivatives occur at zero, and |(f1 ◦ f2 )0 | may
be bounded as follows:
|(f1 ◦ f2 )0 (X)| = |f10 (f2 (X)) f20 (X)|
= |f10 (f2 (X))| · |f20 (X)|
≤ |f10 (0)| · |f20 (0)|
=
=
κY
4
κY κX
16
!!
1
1− p
b
1 + 8X
1− p
1
b
1 + 8X
κX
4
1− p
!
1− p
1
!!
1
1 + 8 Yb
!
1 + 8 Yb
,
77
with the same bound holding for |(f2 ◦ f1 )0 |. Thus, if this bound is strictly less than
one, then iterations of (80) converge from any non-negative initial condition to a
unique fixed point in the non-negative quadrant, which is the unique solution to (76)–
(77) in the non-negative quadrant. Furthermore, the convergence of the iterations is
exponential.
With respect to the original parameters, the condition for a contractive map becomes
κC:C κG:G
1
1 − q 1
< 1.
1− q
2
b
b
16 κC:G
C
G
1 + 8 κC:C
1 + 8 κG:G
In particular, the iterative scheme is guaranteed to converge to a unique solution
whenever κC:C = κG:G = κC:G . However, if κC:G is much smaller than the product of
κC:C and κG:G , then this condition is not satisfied.
In practice, convergence is observed under less stringent conditions on the dissociation constants and is verified by ensuring a sufficiently small residual in (76)–(77).
Once the monomer concentrations X and Y are approximated with sufficient accuracy
using the iterative scheme, the dimer concentrations X:X, Y :Y , and X:Y can be computed directly using equations (63)–(65). Physical intuition suggests the existence of
a unique fixed point of the map (80) under more general conditions, representing a
unique non-negative globally attracting equilibrium of the fast dimerization. This
leads to the following conjecture:
Conjecture 1. The iterative scheme given by the map (80), with component functions (81)–(82), converges to a unique fixed point in the closed positive quadrant for
any non-negative initial condition and for any choice of strictly positive dissociation
constants and non-negative total concentrations.
An extension of the iterative numerical scheme to the case of competitive dimerization of three proteins is given in Appendix B.
78
Model Summary
Converting the non-dimensionalized model (69)–(74) back into the original variables and parameters, the full PCW model is given by the following system of delay
differential-algebraic equations in each cell:
2
Ck (t)
Ck (t) Gk (t)
˙
b
− βC:G
,
C k (t) = αC ck (t − τC ) − βC Ck (t) − 2 βC:C
κC:C
κC:G
!
2
Ck (t − τc )
ċk (t) = γc Fn
, Nk (t − τc ) − δc ck (t),
κC:C
Ṡk (t) = αS sk (t − τS ) − βS Sk (t),
2 !
Ck (t − τs )
ṡk (t) = γs H1
− δs sk (t),
κC:C
2
C
Ck (t) Gk (t)
k (t)
bk (t) = Ck (t) + 2
C
+
,
κC:C
κC:G
2
G
Ck (t) Gk (t)
k (t)
bk (t) = Gk (t) + 2
G
+
,
κG:G
κC:G
(83)
(84)
(85)
(86)
(87)
(88)
where
Nk (t) =
X
Si (t),
(89)
{i : cell i adjacent to cell k}
and the total control protein is prescribed by
bk (t) = G
bmax · g(t − Tk ; τ 1 ).
G
2
(90)
Note that, after computing monomer levels, Ck (t) and Gk (t), from a given amount
bk (t) and C
bk (t), the corresponding
of total clock protein and total control protein, C
79
dimer levels are given by
2
Ck (t)
C:C k (t) =
,
κC:C
2
Gk (t)
G:Gk (t) =
,
κG:G
Ck (t) Gk (t)
C:Gk (t) =
.
κC:G
(91)
(92)
(93)
This completes the construction of the PCW model. In the next chapter, the
PCW model is validated with respect to posterior clock-wave formation in the zebrafish model organism. The validation process involves parameter estimation and
model selection, and it shows that the model is capable of reproducing experimentally
observed results. Significantly, validation also suggests specific biological mechanisms
that are responsible for posterior clock-wave formation.
80
MODEL VALIDATION: AN APPLICATION TO ZEBRAFISH SOMITOGENESIS
The posterior clock-wave (PCW) model described in the previous chapter is developed with substantial consideration of the zebrafish model organism. In this chapter, the PCW model is validated against experimental data in zebrafish. Validation
demonstrates that the PCW model is capable of reproducing the experimentally observed posterior clock-wave in zebrafish, accomplishing a main goal of the modeling
endeavor.
The PCW model verifies an existing scientific hypothesis as well as suggesting a
new one. In validating the PCW model, the experimentally observed aggregate behavior of the clock-wave is shown to be sufficient in constraining the genetic control
mechanisms underlying the somitogenesis clock in zebrafish. Model validation confirms the necessity of multiple repressor binding sites for the clock-gene autoinhibition
in zebrafish, as identified in this and other model organisms [81, 114, 122]. Results
also reveal that, in addition to delayed, nonlinear mRNA production processes, differential decay of clock protein monomers and dimers is an important component of the
gradient controlled competitive dimerization mechanism of clock-wave generation.
Recall that linear decay (i.e., first order kinetics) is assumed in the PCW model
for each of the monomer, homodimer, and heterodimer clock protein species. Viewed
with regard to the total clock protein, different decay constants for each of these forms
(i.e., differential decay) leads to nonlinear decay of total clock protein. (Recall the
discussion concerning equation (31) on page 55.) While this additional nonlinearity
complicates analysis of the model, the computational analyses developed in this work
nonetheless establish its critical importance in model behavior.
81
Validation of the PCW model indicates that dimerization-related nonlinear total
protein decay mechanisms should be addressed experimentally as an alternative or
complement to rate-limited decay mechanisms that have been recently considered in
other somitogenesis models [29, 73]. In these models, monomer and dimer forms of
the clock protein are not separately considered, and the decay of clock protein C is
given by the following expression:
η
C
,
C∗ + C
(Michaelis-Menten Kinetics)
where η is the maximum (or saturating) decay rate, which occurs asymptotically for
high levels of C, and C∗ is the critical concentration of C where the decay rate is half
maximum [21]. This expression can represent, for example, the limited availability of
protein decay machinery within a cell (e.g., proteosomes).
Computational Considerations for Validation
Model validation proceeds through the following steps:
1. Identification of model parameters that have been measured experimentally.
2. Estimation of the model parameters that have not been measured.
3. Model selection based on the results of parameter estimation.
4. Sensitivity analysis for the estimated parameters.
5. Robustness verification by perturbation of all parameters, measured and estimated.
The medial axis of the anterior-to-posterior (AP) presomitic mesoderm (PSM) in
zebrafish contains about fifty cells. Thus, validation of a one-dimensional model of
82
posterior clock-wave formation requires on the order of fifty cells. The PCW model
uses four delay differential equations to represent the dynamics of each cell, giving a
system of 200 differential equations to be analyzed and solved during model validation. In addition, computation of fifty solutions of the adjoining nonlinear system of
algebraic equations is necessary for the fast dimerization approximation. During numerical computations, the algebraic system must be solved for each evaluation of the
right hand side of the system of differential equations. A more complete accounting
of the effect of intercellular coupling requires modeling a two- or three-dimensional
array of cells. For instance, a two-dimensional model on a rectangular lattice with
fifty cells along the AP axis and five cells along the medial-to-lateral axis gives a
system of 1000 differential equations and 250 systems of algebraic equations.
Ultimately, model validation rests upon the ability of the PCW model to reproduce
the experimentally observed clock-waveform in the posterior PSM. The solution of the
resulting large system of equations may only be numerically approximated, and wellestablished computer codes exist for this task [123]. The minimal construction of the
PCW model, combined with modern computational power, makes model simulation
efficient enough for validation tasks. Furthermore, model robustness can be examined
using object-oriented software tools that allow accurate and simultaneous perturbation of all model parameters (some 1100 parameters for a fifty cell simulation).
A task in model validation that is especially computationally intensive is the estimation of model parameters that have not been experimentally determined. Monte
Carlo methods are useful for estimating parameters from high-dimensional, biologically informed parameter distributions, so long as simulation is efficient enough to
allow sufficient sampling of the parameter space [124]. For this purpose, a two cell
PCW model can be used to initially evaluate parameter fitness, as this is the smallest
83
system that includes the effects of intercellular coupling1 . Fortunately, the numerous simulations (numbering in the tens of thousands) may be carried out in parallel
across multiple computer processors. A useful by-product of this estimation procedure is information on model sensitivity to the various estimated parameters. This
initial parameter estimation can then be fully validated in simulations with many
cells, which can further test model robustness by perturbing all model parameters,
including the experimentally determined ones.
Model validation involves considerable software development. In addition to routines for simulating the model itself, code must be written to handle parameter sampling and estimation, model selection, and parameter perturbation. The Matlab
codes employed for these purposes are available in Appendix C.
Clock-Wave Formation in Zebrafish
The general PCW model developed in the previous chapter is now applied specifically to posterior clock-wave formation in the zebrafish model organism. Initially,
this requires designating specific biological components to general model components,
identifying validation criteria for zebrafish, and assigning known parameters. Inevitably, certain questions remain open after this preliminary work, which typically
take the form of unknown model parameters. Not surprisingly, parameter estimation
comprises a significant portion of the remaining validation work. In association with
parameter estimation, several modeling scenarios are considered during validation of
the PCW model, each of which reflects a different underlying biological mechanism
that could be at work, such as multiple repressor binding sites at the clock-gene or
1
As a further reduction, assuming intercellular coupling is so weak as to be negligible allows
analysis and initial validation of a single cell model involving only two differential equations.
84
differential decay for clock protein monomers and dimers. Altogether, this process is
termed model selection.
Assignment of Model Components
As discussed in the previous chapter, three basic helix-loop-helix (bHLH) genes
play the most prominent role in the posterior PSM of the zebrafish, namely her1,
her7, and her12. The respective, and at least partially redundant, roles of these
genes in forming the clock-wave are not entirely clear [5, 102, 122, 125]. Furthermore, experimental evidence suggests her1 and her7 may be differentially regulated
in the posterior and anterior PSM [5, 102, 122, 125]. Given a somewhat bewildering
array of possibilities, her7 is chosen to represent the single clock-gene in the PCW
model because: 1) unlike her12, her7 expression extends across the entire PSM, and
2) the posterior clock-wave still forms during Her1 protein knockdown, even though
expression fails to propagate anteriorly [102, 122].
In zebrafish, expression of the bHLH protein Her13.2 is highest in the tailbud and
decreases anteriorly in the PSM [111, 112]. Her13.2 protein is chosen to represent
the externally prescribed control protein in the PCW model because it likely heterodimerizes with other bHLH proteins such as Her7 [111]. Because Her13.2 proteins
have a truncated amino acid sequence normally used for DNA binding [111], it is
assumed that neither Her13.2 homodimers nor heterodimers with Her7 can repress
her7 mRNA transcription. Thus, Her13.2 influences Her7 self-repression only through
competitive dimerization, as seen in related bHLH networks [113, 114].
DeltaC is chosen to represent the coordinating-signal gene in the PCW model
because its expression in the PSM is oscillatory and synchronized with her7 expression [91]. DeltaC ligands presented at a cell’s membrane activate Notch protein
receptors in adjacent cells’ membranes, releasing Notch Intracellular Domain (NICD)
85
proteins that up-regulate clock-gene expression, including her7 [91, 126]. Following
the models in [2, 27], equation (89) is a simplified representation of this intercellular
signaling process. It is assumed that the effects of other constitutively expressed genes
in the Notch signaling pathway, such as DeltaD and Su(H) [5], may be subsumed into
the appropriate model parameters.
Model Validation Criteria
Experimental data for zebrafish provide the following constraints on the clockwave behavior:
(i) The putative oscillation period in the tailbud is 30 minutes at 28◦ C, which is the
same time it takes for each somite to form in the anterior PSM [3, 5, 69, 85, 93].
(ii) The oscillation rate in more anterior PSM cells slows sufficiently in order to
generate a clock-wave with two to three traveling bands of gene expression,
which emanate from the tailbud and narrow as they reach the anterior-most
PSM [4, 5].
(iii) Oscillatory expression of the clock and coordinating-signal genes is synchronized [127].
With regard to these three constraints on the proper formation of the clock-wave,
the effect of two biological control mechanisms, which have been implicated in other
systems, is examined:
1. Differential decay rates of the clock protein monomers and dimers [107].
2. The number of binding sites for the repressive clock protein homodimer, including the cooperativity between simultaneous binding to multiple sites [118, 128].
86
Experimental evidence exists both for clock protein dimerization [111] and for multiple
cis regulatory sites for clock-genes in zebrafish (as well as in chick and mouse) [30,
81, 114, 122].
Given the biological potential for differential decay and multiple binding sites for
clock protein, four different model scenarios (I–IV) are considered:
I A single binding site for the self-repressing clock protein homodimer (n = 1
in (38) on page 63) and monomer-only clock protein decay (βC:C = βC:G = 0
in (83) on page 78).
II A single binding site for the self-repressing clock protein homodimer (n = 1
in (38)) and equal decay rates for clock protein monomer, homodimer, and heterodimer with the control protein (βC = βC:C = βC:G in (83)).
III Two binding sites for the self-repressing clock protein homodimer (n = 2 in (38))
and monomer-only clock protein decay (βC:C = βC:G = 0 in (83)).
IV Two binding sites for the self-repressing clock protein homodimer (n = 2 in (38))
and equal decay rates for clock protein monomer, homodimer, and heterodimer
with the control protein (βC = βC:C = βC:G in (83)).
Numerical simulation with Matlab’s delay differential equation solvers dde23
and ddesd2 [123] are used to assess the ability of model scenarios I–IV to:
2
dde23 was developed before ddesd for systems of delay differential equations in normal form with
fixed delays in the state variables (but not in their derivatives). ddesd handles the more general case
of state dependent delays, of which fixed delays represent a special case. Because of implementation
differences concerning discontinuity tracking in solution derivatives, dde23 is a somewhat faster
algorithm when there are fewer overall system delays, while ddesd is a considerably faster algorithm
when there are many delays. The latter case occurs when examining model robustness, in which the
nominal delays in each cell are perturbed, giving a different set of four delays in each cell.
87
(a) Produce synchronized periodic expression of the clock and coordinating-signal
genes in the tailbud within 10% of the experimentally observed value (30 minutes).
(b) Produce sufficient decrease in the oscillation rate between the tailbud (high level
of total control protein Her13.2) and the more anterior PSM (low level of total
control protein Her13.2).
(c) Produce a realistic posterior clock-wave in a simulated AP line of cells with two
properly spaced, posterior-most expression bands of the clock and coordinatingsignal mRNA.
(d) Exhibit robustness of clock-wave formation with respect to heterogeneity in the
parameters across the cell population.
Where possible, model parameters are assigned experimentally determined nominal values. For nine parameters, ranges of biologically realistic values are initially
determined for purposes of parameter estimation, which is described further below.
Given any of the model scenarios I–IV, a parameter selection from the set of feasible
parameter values is considered to produce a valid fit to experimental data if the
corresponding model satisfies criteria (a)–(d).
Parameter Value and Range Selection
Below, all parameters whose nominal values in zebrafish are experimentally determined are first given, including citations for each choice3 . After these known parameters, range selections for estimated parameters are given and explained. Estimated
3
Copy numbers per compartment can be converted to molar concentration using Avogadro’s
constant with a nuclear volume Vnuc = 43 π (2.5 µm)3 = 65.4 µm3 and a cytosolic volume Vcyt =
4
3
3
= 458 µm3 , where the nuclear diameter is 5 µm [27] and the cell diameter
3 π (5 µm) − (2.5 µm)
is assumed to be 10 µm.
88
parameters are particularly important with respect to the model scenarios I–IV being
investigated, and most have not been determined experimentally, to the best of this
author’s knowledge. However, two parameters (τc and βC ) are estimated because
of sensitivity considerations. A joint uniform distribution is constructed from these
parameter ranges and sampled for combinations of values conducive to clock-wave
formation, with respect to model scenarios I–IV, as described above.
Experimentally Determined Parameter Values: The following parameters have
been experimentally determined in the literature:
λ The number of cells per AP somite length. While this number is not strictly
constant across the PSM in zebrafish, the constant value λ = 5 cells per somite
is a reasonable approximation over a good portion of the PSM tissue [5].
µ The somite formation rate in somites per minute in the anterior PSM. While
the formation rate is not strictly constant throughout somite development in
zebrafish, µ is roughly constant over a good portion of developmental time [11,
93]. The somite formation rate is observed to be equal to the frequency of
oscillation in the tailbud [5]. In all clock-wave simulations, an initial settling
time occurs in which oscillations stabilize before any cells enter the PSM, which
correlates with the initiation of synchronized oscillatory expression in the early
embryo before somitogenesis begins. After the settling time, the oscillation
period, T , is computed and this computed period is used to determine µ =
1
.
T
µ determines the exit times Tk of cells from the tailbud using (11):
Tk =
k−1
.
λµ
In zebrafish, the oscillatory gene expression in the tailbud has a period of 30 minutes at 28◦ C [5], so at this temperature µ =
1
30
somites per minute. If for any
89
reason the oscillation period cannot be computed after the settling time, the
default value µ =
1
30
somites per minute is used.
τC The delay, in minutes, arising from clock protein production at the ribosomes
and subsequent transport into the nucleus. The value τC = 1.7 minutes from [27]
is used in all simulations.
αC , αS The production constants, in protein copies per mRNA copy per minute, of
clock and coordinating signal protein, respectively. The common value αC =
αS = 4.5 protein copies per mRNA copy per minute from [27] is used in all
simulations.
γc , γs Production constants, in mRNA copies per minute, of clock and coordinating
signal mRNA, respectively. γc = γs in all simulations, and the common value
is computed from the chosen value of ρ (see below) so that the baseline mRNA
production rate in the absence of repressors or activators is the following value
cited in [27]:
γc
ρ
ρ+1
= γs
ρ
ρ+1
= 33 mRNA copies/minute.
ρN The promoter binding affinity of the activator N , with units of inverse copy
number. To the best of this author’s knowledge, this constant has not been measured in zebrafish. In the generation of oscillatory gene expression in zebrafish,
the intercellular signaling does not dominate the intracellular self-repression of
the clock-gene [4]. Following the critical concentration parameters used in [2],
the activator N is given one tenth the binding affinity of the repressor C:C in
all simulations, i.e., ρN =
ρC:C
.
10
ωN The unitless cooperativity between simultaneously bound activator N and
RNAP-II complex at the promoter. This number is strictly greater than one.
90
To the best of this author’s knowledge, this parameter has not been measured
in zebrafish, so a typical value ωN = 25 is used in all simulations, which is taken
from [108].
δc The decay constant, in inverse minutes, of clock mRNA. In all simulations the
value used is δc = 0.206 minute−1 from [2], which is estimated from experiments
on zebrafish [4].
τS The delay, in minutes, arising from coordinating signal protein production at
the ribosomes and subsequent transport through the cell membrane into the
cytosol and then nuclei of adjacent cells. The value τS = 20 minutes from [27]
is used in all simulations.
βS The decay constant, in inverse minutes, of coordinating signal protein. The
value βS = 0.23 minute−1 from [27] is used in all simulations.
τs The delay, in minutes, arising from coordinating signal mRNA transcription at
the DNA, post-transcriptional processing, and subsequent transport into the
cytosol. The value τs = 12.2 minutes is used in all simulations. Note that
this choice reflects a minor (accidental) variation from the value 12.4 minutes
used in [2], which itself was taken from an experimentally determined range
for zebrafish [4]. This range is obtained as a sum of a non-measured initiation
delay, which is assumed to be in the range of 3 to 8 minutes in [4], and an
experimentally determined range of 8.4 ± 1.2 minutes. In [2] the initiation delay
is taken to be 4 minutes resulting in value 8.4 + 4 = 12.4 minutes. Using the
full range of initiation delays and full range of measured delays gives the range
[7.2 + 3, 9.6 + 8] = [10.2, 17.6] minutes, which contains 12.2 minutes.
91
δs The decay constant, in inverse minutes, of coordinating signal mRNA. In all
simulations the value used is δs = 0.273 minute−1 from [2], which is estimated
from experiments on zebrafish [4].
Parameters Estimated from a Range of Values: The following parameters must
be estimated from a range of values:
bmax The maximum level of total control protein, which occurs in the tailbud and
G
posterior-most PSM and has units of copy number per nucleus. In zebrafish,
this parameter represents the maximum level of total nuclear Her13.2 protein.
To the best of this author’s knowledge, quantitative data for this parameter in
bmax is estimated from the range 0 to 2000 copies
zebrafish are unavailable, so G
per nucleus. This allows the maximum control protein level to be on the same
order of magnitude as the typical mean level of the oscillatory total clock protein.
τ 1 The half-life, in minutes, of the total control protein in a given cell, which is
2
spatiotemporally graded in the PSM [5, 111, 112]. The half-life is the time after
a cell leaves the tailbud when the level of total nuclear control protein is reduced
to
1
2
bmax . To the best of this author’s knowledge, quantitative data for this
G
parameter for the control protein Her13.2 in zebrafish are unavailable. Multicellular simulations have shown that a range of values is typically acceptable,
e.g., 25–60 minutes. The choice of τ 1 can affect both the initial shape and axial
2
positioning of the clock-wave. Unless otherwise noted, simulations in this work
use τ 1 = 30 minutes, so that the half-life is of a similar order of magnitude as
2
the oscillation period in the zebrafish tailbud.
ρ The unitless product of the binding affinity ρP for RNAP-II complex to a gene’s
promoter and the copy number P of the complex, i.e., ρ := ρP P . The assembly
92
of the RNAP-II complex is assumed to be a fast process at equilibrium so that P ,
and therefore ρ, may be treated as constant. To the best of this author’s knowledge, data for zebrafish are unavailable, so ρ is estimated from the range
1
3
to 3,
which can be interpreted as a 25–75% probability of the complex binding in the
absence of any other repressors or activators. Note that the lumped parameter
ρ may include effects of several constitutively expressed transcription factors in
the PSM, such as DeltaD and Su(H), at the clock-genes in zebrafish [91, 116].
However, for simplicity in the face of a lack of measured data, the present work
uses the same value for ρ for both the clock-gene and the coordinating-signal
gene.
τc The delay, in minutes, arising from clock mRNA transcription at the DNA, posttranscriptional processing, and subsequent transport into the cytosol. Previous
work [27] has shown that the existence and period of sustained oscillations is
sensitive to the total system delay. Because of this sensitivity, τc is estimated
from the range 2.3 to 8.1 minutes. This range is derived from the experiments
in [4], which estimated τc = 3.7 ± 1.4 + Tinit minutes, where Tinit is a (nonmeasured) initiation delay assumed to be in the range of 3 to 8 minutes. Because
of differences in the PCW model from the model in [2], Tinit is taken in this work
to be in the range 0 to 3 minutes, giving the minimum value 3.7 − 1.4 + 0 =
2.3 minutes, and the maximum value 3.7 + 1.4 + 3 = 8.1 minutes. Subsequent
analysis of parameter sensitivities verified that this is an adequate range for
modeling oscillations in zebrafish with the PCW model (see Figure 16).
ρC:C The binding affinity of the C:C repressor to a gene’s promoter, with units of
inverse copy number. To the best of this author’s knowledge, this constant
has not been measured in zebrafish, so ρC:C is estimated from the biologically
93
realistic range 0.01 to 1 copy−1 , which is taken from the range given in [108],
and which includes the (roughly equivalent) critical concentration used in [27].
ωC:C The unitless cooperativity between two C:C repressors simultaneously bound to
a gene’s promoter. To the best of this author’s knowledge, this constant has not
been measured in zebrafish, so ωC:C is estimated from the biologically realistic
range 1 to 100 taken from [108].
βC The decay constant, in inverse minutes, of clock protein monomer. Previous
work [27, 80] has shown that the existence and period of sustained oscillations
is sensitive to the protein decay rate. Because of this sensitivity and the fact
that the PCW model explicitly tracks the distribution of total clock protein
as monomer, homodimer, and heterodimer, βC is estimated from the range
0.2 to 0.5 minute−1 , which includes the value 0.23 minute−1 from [27] corresponding to a protein monomer half-life of
ln 2
0.23
= 3 minutes.
κG:G , κC:C ,
The protein dimer dissociation constants with units of copy number. To the best
κC:G
of this author’s knowledge, values for these constants have not been measured
in zebrafish, so each dissociation constant is independently estimated from the
range 10 to 1000 copies, which is based on the range cited in [107].
Parameters for Model Scenarios I–IV: The following parameters are set differently in model scenarios I–IV:
βC:C , βC:G The respective decay constants, in inverse minutes, of clock protein homodimer
and heterodimer with control protein. In all simulations, these parameters share
a common value, i.e., βC:C = βC:G . Attention to the findings in [107], concerning differential decay of monomers and dimers, suggests consideration of two
94
limiting cases. In models I and III, βC:C = βC:G = 0 corresponds to monomer
only clock protein decay and thus nonlinear decay of total clock protein. In
models II and IV, βC:C = βC:G = βC corresponds to decay of all clock protein
forms and gives linear decay of total clock protein.
n The number of binding sites for C:C repressor at the clock-gene’s promoter.
Previous work [81] has shown that the number of binding sites can be a key
parameter affecting oscillations produced through delayed negative feedback.
In models I and II, n = 1 corresponds to a single binding site. In models III
and IV, n = 2 corresponds to two binding sites.
Parameter Estimation and Model Selection
The following important observation allows validation in two stages: formation
of a realistic clock-wave depends on the key criterion ∆T , defined as the maximum
change in the period of oscillatory gene expression observed over the total control
protein range. Investigations for the present work show that parameter combinations
giving
∆T & ∆T∗ := 15 minutes
generate a biologically realistic clock-wave for the posterior PSM in zebrafish. In the
first validation stage, parameters are sought for which model scenarios I–IV satisfy
the above criteria (a) and (b) with ∆T ≥ 15 minutes using two identical, coupled
cells. A two cell model includes intercellular signaling effects, yet is much faster to
simulate than an entire line of cells representing the PSM. In the second stage, a
line of coupled cells with spatiotemporally graded total control protein is simulated
95
in order to verify that the selected model(s) produce a realistic clock-wave in the
absence (c) and presence (d) of a heterogeneous cell population.
Stage One Validation
In the first stage, a random sample of size 40,000 is taken from a uniform joint
distribution of the eight parameters ρ, τc , ρC:C , ωC:C , βC , κG:G , κC:C , and κC:G . As
described below, the range of the last free parameter, the total control protein in
bmax , determines ∆T . Zero-valued history functions are initially used for
the tailbud G
bk (t), ck (t), Sk (t), and sk (t) (equations (83)–(86)), k = 1 or 2,
the state variables C
and solutions are allowed to settle to steady-state behavior for at least 500 minutes.
At each time step of the differential equation solver the nonlinear algebraic system
(87)–(88) is solved for the monomer copy numbers Gk (t) and Ck (t) in terms of the
bk (t) = G
bmax and the state variable C
bk (t).
constant total control protein level G
For each sampled parameter combination the model is initially simulated in the
bmax = 0). If this initial solution settles into susabsence of total control protein (G
tained oscillations, then the effect of increasing the maximum total control protein
bmax is increased in steps
level on the period is determined, i.e., ∆T is computed. G
of ten until either sustained oscillations cease or a maximum value of 2000 copies
per nucleus is reached. For efficiency, previous solutions are used as initial history
bmax . To minimize possible stochastic effects not
functions while stepping through G
considered by this deterministic model, only periodic solutions with sufficiently large
amplitudes in each state variable are accepted (≥ 5 copies, peak-to-peak).
For each model scenario I–IV, the following table lists the number of parameter
selections out of a random sample of 40,000 total selections that produced periodic
bmax = 0, and 2) any nontrivial period for
solutions with 1) any nontrivial period for G
96
(a)
Model Comparison
(b)
(c)
∆T Distributions
Solutions for ∆Tmax
& ∆Tmax
III
IV
25
50
40
30
20
Model III
Model IV
20
15
∆T = 15
*
10
Model III
Model IV
70
Period (min.)
Periodic
30±3 min.
60
% of Simulations
% of Simulations
70
60
50
max
∆TIII
40
5
∆Tmax
= 8.7
IV
10
0
I
II
III
IV
0
0
5
10
15
20
∆T (min.)
Model Number
25
30
30
= 29.3
0
500
1000
1500
2000
Total Control Protein (copy #)
Figure 15: Model selection. (a) Dark bars: the percentage of periodic solutions
bmax = 0 copies/nucleus. Light bars: the percentage of
in 40,000 simulations when G
periodic solutions in 40,000 simulations also exhibiting the tailbud period of 30±3 min
bmax in the range 0–2000 copies/nucleus. (b) Stacked distributions
for some level of G
of ∆T for models III and IV for those solutions exhibiting the tailbud period. ∆T &
∆T∗ = 15 is a key requirement for proper clock-wave formation in zebrafish. (c) Period
as a function of total control protein for the best parameter selections for models III
max
max
and IV, which produce ∆TIII
= 29.3 min and ∆TIV
= 8.7 min, respectively. The
max
b
curve for model III stops at G
= 700 copies/nucleus, after which the amplitude of
the periodic solution drops below 5 copies/nucleus, peak-to-peak. Periodic solutions
cease altogether after about 960 copies/nucleus.
bmax = 0 and a period of 30 ± 3 minutes for some level of G
bmax in the range 0 to
G
2000 copies per nucleus:
Solution Model
I
II
III
IV
Periodic
22
0
19314
27812
30 ± 3 min.
0
0
4882
14888
Figure 15a presents the same data using percentages rather than raw counts.
Only models III and IV, with two binding sites, exhibit sustained oscillations with
a period of 30 ± 3 minutes, satisfying criterion (a) on page 87 for model validation.
For these two models, Figure 15b shows the distribution of ∆T for those simulations
that produced a period of 30 ± 3 minutes. The raw data are provided in the following
table:
97
Model
∆T
Interval
[0, 2.5)
[2.5, 5)
[5, 7.5)
[7.5, 10)
III
966
910
1049
893
516
292
IV
8987
5026
853
22
0
0
Model
∆T
Interval
[15, 17.5) [17.5, 20)
[10, 12.5) [12.5, 15)
[20, 22.5) [22.5, 25)
[25, 27.5) [27.5, 30)
III
140
67
28
16
3
2
IV
0
0
0
0
0
0
The important observation is that even though model IV produces the required period
of oscillation 30±3 minutes for almost 40% of parameter selections (as opposed to 12%
in model III, see Figure 15a), the maximum period change ∆T for model IV is only
max
= 8.7 minutes. This is considerably smaller than the minimum ∆T necessary
∆TIV
for realistic clock-wave formation, which is ∆T∗ = 15 minutes. In contrast, for model
III, 22% (1064 out of 4882) of the parameter values that produce period 30±3 minutes
also produce ∆T ≥ 10, and 5% (256 out of 4882) produce ∆T ≥ 15. The maximum
max
= 29.3 minutes, satisfying critierion (b) on
period change for model III is ∆TIII
page 87 for model validation.
Figure 15c displays the period of the oscillation as a function of the total control
max
protein level in models III and IV using the parameter selections producing ∆TIII
max
and ∆TIV
, respectively. These selections are listed in the following table:
∆T Param.
ρC:C
ωC:C
βC
κG:G
κC:C
κC:G
bmax
G
max
∆TIII
= 29.3 0.639
5.66 0.944
30.9
0.497
22.5
155
584
700
max
∆TIV
= 8.7
6.39 0.718
30.6
0.250
98.5
738
851
1000–2000
ρ
0.447
τc
Note that a more biologically realistic clock-wave is typically generated by choosing,
bmax that still produces sufwhen possible, a smaller value of total control protein G
max
ficiently large ∆T . This is done with the clock-wave simulation with ∆TIV
where
bmax = 1000 copies per nucleus is used instead of 2000 copies per nucleus. As seen
G
98
bmax curve for model IV is very flat in this interval, and
in Figure 15c, the ∆T vs. G
the reduction in ∆T from using 1000 instead of 2000 copies per nucleus is less than
one minute.
For the given sample size of 40,000, no parameter selections for models I and II
produce oscillations with the required period 30 ± 3 minutes. Thus, according to the
PCW model, one may conclude that a single repressor binding site at the clock-gene
is incapable of producing the experimentally observed oscillations in the zebrafish
tailbud, regardless of the clock protein decay mechanism. Furthermore, model IV,
with equal monomer and dimer clock protein decay rates (linear total protein decay)
cannot produce sufficiently large ∆T (Figure 15b–c). Only model III, combining two
binding sites and monomer-only clock protein decay, admits a significant number
of parameter combinations that produce sufficiently large ∆T with the potential to
generate a biologically realistic clock-wave.
Parameter Sensitivities: The above parameter estimation procedure also provides parameter sensitivity information for the eight estimated parameters (ρ, τc ,
ρC:C , ωC:C , βC , κG:G , κC:C , and κC:G ) with respect to the criteria (a)–(b) on page 87.
Figures 16 and 17 provide sensitivity histograms for these parameters for the selected
model III. Each of these parameters is initially drawn independently from a uniform
distribution so that deviation from a uniform distribution indicates sensitivity. Each
figure column represents one parameter and the three histograms from top to bottom
represent number of instances in 40,000 random parameter selections in which the
corresponding solution satisfies increasingly stringent requirements:
1. (top figure row) The number of instances that the solution is periodic in the
absence of total control protein.
99
2. (middle figure row) The number of instances that the solution is periodic in the
absence of total control protein, and has a period of 30 ± 3 minutes for some
level of total control protein in the range 0 to 2000 copies per nucleus.
3. (bottom figure row) The number of instances that the solution is periodic in
the absence of total control protein, has a period of 30 ± 3 minutes for some
level of total control protein in the range 0 to 2000 copies per nucleus, and has
a period change ∆T ≥ 15 minutes over a range of total control protein.
Note that the vertical axes of the histograms change as the requirements become more
stringent. In general, the largest selective pressure is applied by the last requirement
∆T ≥ ∆T∗ = 15 minutes, which is typically sufficient for generating a biologically
realistic clock-wave for zebrafish. Also, these histograms are 1-dimensional projections
of an 8-dimensional parameter space, and thus do not indicate correlations between
different parameters.
The following is a summary of the sensitivity analysis from Figures 16 and 17:
1. The delay τc is the most sensitive parameter. The requirement that solutions
have period 30 ± 3 minutes shifts the range of admissible values lower, while the
additional requirement ∆T ≥ 15 minutes shifts the set of admissible values of
τc toward larger values in the sub-range (Figure 16). The mean preferred value
is somewhat smaller than τc = 7.7 minutes used in [2].
2. Model selection not very sensitive to parameters ρC:C and ωC:C , however there
are substantial differences between histograms in middle row and the bottom
row of Figure 16 for these two parameters. While there are many low parameter
values that produce a periodic orbit of preferred period, there are substantially
fewer low parameter values that support a sufficient change in period ∆T .
100
Periodic
Periodic
Periodic
Periodic
4000
4000
10000
10000
2000
2000
5000
5000
0
0.331
2
ρ
30±3 min
3
0
2.3 4 5 6 8.1
τc
0
0.01 0.5
ρC:C
30±3 min
1
0
30±3 min
2000
2000
2000
500
1000
1000
1000
2
3
ρ
∆T≥15 min
0
2.3 4 5 6 8.1
τ
c
0
0.01 0.5
ρ
1
0
∆T≥15 min
∆T≥15 min
100
40
40
50
50
20
20
ρ
2
3
0
2.3 4 5 6 8.1
τ
c
0
0.01 0.5
ρ
C:C
50 100
ω
C:C
100
0
0.331
1
C:C
∆T≥15 min
50 100
ωC:C
30±3 min
1000
0
0.331
1
1
0
1
50 100
ω
C:C
Figure 16: Parameter sensitivities for model III, part 1: ρ, τc , ρC:C , and ωC:C . Graphs
give the number of simulated solutions out of 40,000 total simulations satisfying the
specified criteria. All parameters are drawn independently from uniform distributions
whose ranges are given by the x-axis limits.
101
Periodic
5000
0
0.2 0.35 0.5
βC
Periodic
Periodic
Periodic
10000
4000
10000
5000
2000
5000
0
10 500 1000
κG:G
30±3 min
0
10 500 1000
κC:C
30±3 min
0
10 500 1000
κC:G
30±3 min
30±3 min
2000
2000
400
2000
1000
1000
200
1000
0
0.2 0.35 0.5
β
0
10 500 1000
κ
0
10 500 1000
κ
0
10 500 1000
κ
∆T≥15 min
∆T≥15 min
∆T≥15 min
∆T≥15 min
C
G:G
200
200
100
100
0
0.2 0.35 0.5
β
C
C:C
50
0
10 500 1000
κ
G:G
C:G
50
0
10 500 1000
κ
C:C
0
10 500 1000
κ
C:G
Figure 17: Parameter sensitivities for model III, part 2: βC , κG:G , κC:C , and κC:G .
Graphs give the number of simulated solutions out of 40,000 total simulations satisfying the specified criteria. All parameters are drawn independently from uniform
distributions whose ranges are given by the x-axis limits.
102
3. Smaller values of the binding affinity parameter ρ are preferred under the largest
selective pressure applied by the requirement ∆T ≥ 15 minutes (Figure 16).
4. Faster decay rates for protein monomer, i.e., larger βC , are favored under all
three requirements (Figure 17). These values are somewhat larger than βC =
0.23 minute−1 used in [2].
5. κC:C is the most sensitive dissociation constant in the competitive dimerization,
with larger ∆T favoring a tighter range of values for κC:C than for κG:G or κC:G
(Figure 17). For the remaining parameters κG:G and κC:G , the preferred values
are on the lower end of their range (Figure 17).
Stage Two Validation
The second validation stage addresses criteria (c) and (d) on page 87 for proper
clock-wave generation across a growing AP line of fifty cells in the axial PSM. Recall
that criterion (c) concerns the formation of a clock-wave with two properly spaced,
posterior-most bands of high gene expression in the PSM, and criterion (d) concerns
formation of this clock-wave in a heterogeneous cell population. In the simulations
b is spatiotemporally graded across
for this validation stage, the total control protein G
the cells, as given by equation (90) on page 78.
b c, S, and s, Figures 18 and 19 compare the simulated
For the state variables C,
max
clock-waves for model III and model IV with the parameters that produce ∆TIII
max
and ∆TIV
, respectively. In both figures the cell populations are homogeneous, i.e.,
each cell has the exact same parameters as every other cell. In Figure 18, the spacing
of the posterior-most bands of clock mRNA expression is about twelve cells, close
to the mean value of approximately 10.5 cells measured experimentally [129, see
Fig. 3]. In contrast, the spacing in Figure 19 is about 25 cells, which is much larger
0
10
20
30
40
50
0
600
1000
400
500
200
0
0
10
20
30
40
50
0
Cell # (1 is anterior)
600
150
400
100
200
50
0
0
10
20
30
40
50
80
0
600
60
400
40
200
20
0
0
10
20
30
40
50
0
b protein (copy #)
G
0
200
b Protein (copy #)
G
200
c mRNA (copy #)
400
2000
time= 265.0 min
s mRNA (copy #)
600
4000
b protein (copy #)
G
time= 265.0 min
b protein (copy #)
G
S protein (copy #)
b protein (copy #)
C
103
Cell # (1 is anterior)
Figure 18: Simulated clock-wave in fifty homogeneous cells for the model III parammax
max
eter selection giving ∆TIII
= 29.3 min. Only model III, with large ∆TIII
, produces
properly spaced expression peaks in the posterior PSM. All plots occur 265 min after
the first cell exits the tailbud, and red bars represent cells still in the tailbud. In all
plots, the total control protein gradient (blue curve) travels from left to right as time
passes, while the bands of expression travel from right to left as cellular expression
oscillates.
than the experimentally observed spacing. Thus, as indicated by the first validation
stage, model III is capable of producing a biologically realistic clock-wave, satisfying
criterion (c) while model IV does not.
Model Robustness: The robustness of the clock-wave in model III with respect to heterogeneity in the cell population is examined by randomly perturbing
parameter values in all cells. Figure 20 represents model III from Figure 18, but
with normally distributed noise added independently to parameters in all cells with
a positivity constraint and so that 99.7% of the values are within ±1% of the nominal values. Clock-wave formation still proceeds in a reasonable fashion, satisfying
criterion (d).
0
10
20
30
40
50
0
1000
1000
500
500
0
0
10
20
30
40
Cell # (1 is anterior)
50
0
500
50
0
0
10
20
30
40
50
0
1000
60
40
500
20
0
0
10
20
30
40
50
0
b protein (copy #)
G
0
1000
100
b Protein (copy #)
G
500
500
c mRNA (copy #)
1000
time= 265.0 min
s mRNA (copy #)
1000
b protein (copy #)
G
time= 265.0 min
1500
b protein (copy #)
G
S protein (copy #)
b protein (copy #)
C
104
Cell # (1 is anterior)
Figure 19: Simulated clock-wave in fifty homogeneous cells for the model IV paramemax
max
ter selection giving ∆TIV
, does not produce
= 8.7 min. Model IV, with small ∆TIV
properly spaced expression peaks in the posterior PSM. All plots occur 265 min after
the first cell exits the tailbud, and red bars represent cells still in the tailbud. In all
plots, the total control protein gradient (blue curve) travels from left to right as time
passes, while the bands of expression travel from right to left as cellular expression
oscillates.
0
0
10
20
30
40
50
0
1500
600
1000
400
500
0
200
0
10
20
30
40
Cell # (1 is anterior)
50
0
600
150
400
100
200
50
0
0
10
20
30
40
50
0
60
600
40
400
20
200
0
0
10
20
30
40
50
0
b protein (copy #)
G
200
200
b Protein (copy #)
G
400
2000
c mRNA (copy #)
600
time= 265.0 min
s mRNA (copy #)
4000
b protein (copy #)
G
time= 265.0 min
b protein (copy #)
G
S protein (copy #)
b protein (copy #)
C
105
Cell # (1 is anterior)
Figure 20: Simulated clock-wave in fifty heterogeneous cells for the model III pamax
rameter selection giving ∆TIII
= 29.3 min. Normally distributed noise is added
independently to parameters in all cells with a positivity constraint and so that 99.7%
of the values are within ±1% of the nominal values. This noise does not disrupt clockwave formation. All plots occur 265 min after the first cell exits the tailbud, and red
bars represent cells still in the tailbud. In all plots, the total control protein gradient
(blue curve) travels from left to right as time passes, while the bands of expression
travel from right to left as cellular expression oscillates.
106
Quantitative data on the Her13.2 control protein gradient in the zebrafish PSM
are scarce. Therefore, the robustness of clock-wave formation to changes in the spatiotemporal gradient of the total control protein is now examined. Simulations of
max
model III for the parameters giving ∆TIII
= 29.3 minutes show that a range of
half-lives are allowable for the sigmoidal function g that prescribes the total control
protein (recall equation (15) on page 51). Smooth clock-waves form for τ 1 ranging
2
from about 25 to greater than 60 minutes, see Figure 21. Interestingly, the broadening
the gradient profile (i.e., larger τ 1 ) shifts the axial position of clock-wave formation
2
anteriorly, but does not greatly affect the spacing between expression bands. Shorter
half-lives are less conducive to proper clock-wave formation, where it appears that
the level of total control protein changes too quickly for the generation of smooth
expression bands, see Figure 22. Lastly, a simple exponential decay of total clock
protein is also conducive to clock-wave formation (recall equation (16) on page 51),
see Figure 23.
Reproduction of Experiments
The PCW model, with only a single clock-gene, has been shown to reproduce
a one-dimensional projection of the posterior clock-wave observed zebrafish somitogenesis. In morpholino knockdown of Her1 clock protein zebrafish, redundancy in
the remaining clock-genes (including her7, which is the PCW model’s putative clockgene) apparently maintains formation of the posterior clock-wave [122, see Fig. 4k].
However, it must be noted that the PCW model cannot reproduce the failure of the
clock-wave to propagate anteriorly in the absence of the protein product of the second
clock-gene her1. Thus, in the most strict interpretation, the PCW model captures
only posterior clock-wave formation.
0
10
20
30
40
50
0
600
1000
400
500
0
200
0
10
20
30
40
Cell # (1 is anterior)
50
0
600
400
100
200
0
0
10
20
30
40
50
0
100
600
400
50
200
0
0
10
20
30
40
50
0
b protein (copy #)
G
0
200
b Protein (copy #)
G
200
c mRNA (copy #)
400
2000
s mRNA (copy #)
600
4000
time= 265.0 min
b protein (copy #)
G
time= 265.0 min
b protein (copy #)
G
S protein (copy #)
b protein (copy #)
C
107
Cell # (1 is anterior)
Figure 21: Simulated clock-wave in fifty homogeneous cells for the model III paramemax
ter selection giving ∆TIII
= 29.3 min, where the half-life of the total control protein,
τ 1 , is doubled from 30 to 60 min. Formation of the clock-wave relative to the tailbud
2
is shifted anteriorly along the PSM. All plots occur 265 min after the first cell exits
the tailbud, and red bars represent cells still in the tailbud. In all plots, the total
control protein gradient (blue curve) travels from left to right as time passes, while
the bands of expression travel from right to left as cellular expression oscillates.
10
20
30
40
50
0
600
1000
400
500
200
0
0
10
20
30
40
Cell # (1 is anterior)
50
0
400
100
200
50
0
0
10
20
30
40
50
80
0
600
60
400
40
200
20
0
0
10
20
30
40
50
0
b protein (copy #)
G
0
600
150
b Protein (copy #)
G
0
200
c mRNA (copy #)
400
2000
time= 265.0 min
200
s mRNA (copy #)
600
4000
b protein (copy #)
G
time= 265.0 min
b protein (copy #)
G
S protein (copy #)
b protein (copy #)
C
108
Cell # (1 is anterior)
Figure 22: Simulated clock-wave in fifty homogeneous cells for the model III paramemax
ter selection giving ∆TIII
= 29.3 min, where the half-life of the total control protein,
τ 1 , is halved from 30 to 15 min. Formation of the clock-wave is disorganized and the
2
bands of expression are not smooth. All plots occur 265 min after the first cell exits
the tailbud, and red bars represent cells still in the tailbud. In all plots, the total
control protein gradient (blue curve) travels from left to right as time passes, while
the bands of expression travel from right to left as cellular expression oscillates.
0
10
20
30
40
50
0
600
1000
400
500
0
200
0
10
20
30
40
Cell # (1 is anterior)
50
0
600
400
100
200
0
0
10
20
30
40
50
80
0
600
60
400
40
200
20
0
0
10
20
30
40
50
0
b protein (copy #)
G
0
200
b Protein (copy #)
G
200
c mRNA (copy #)
400
2000
time= 265.0 min
s mRNA (copy #)
600
4000
b protein (copy #)
G
time= 265.0 min
b protein (copy #)
G
S protein (copy #)
b protein (copy #)
C
109
Cell # (1 is anterior)
Figure 23: Simulated clock-wave in fifty homogeneous cells for the model III pamax
rameter selection giving ∆TIII
= 29.3 min, where the half-life of the exponentially
decaying total control protein, τ 1 , is 30 min. Formation of the clock-wave relative to
2
the tailbud is shifted anteriorly along the PSM as compared to the simulation with
sigmoidally decaying total control protein with the same half-life. All plots occur
265 min after the first cell exits the tailbud, and red bars represent cells still in the
tailbud. In all plots, the total control protein gradient (blue curve) travels from left to
right as time passes, while the bands of expression travel from right to left as cellular
expression oscillates.
1
200
0
0
10
20
30
40
50
2000
0
600
400
1000
200
0
0
10
20
30
40
50
0
600
300
200
400
100
200
0
0
10
20
30
40
50
100
0
600
400
50
Cell # (1 is anterior)
200
0
0
10
20
30
40
50
0
b protein (copy #)
G
400
400
b Protein (copy #)
G
2
c mRNA (copy #)
600
s mRNA (copy #)
3
time= 265.0 min
b protein (copy #)
G
time= 265.0 min
4
b protein (copy #)
G
S protein (copy #)
b protein (copy #)
C
110
Cell # (1 is anterior)
Figure 24: Simulated clock-wave in fifty heterogeneous cells for the model III paramemax
ter selection giving ∆TIII
= 29.3 min, but with a 99.9% reduction in the production
b All plots occur 265 min after the first cell exits the
rate of total clock protein C.
tailbud, and red bars represent cells still in the tailbud. In all plots, the total control
protein gradient (blue curve) travels from left to right as time passes, but oscillatory
expression does not occur and bands of expression do not form.
Figure 24 shows a simulated Her1 and Her7 combined knockdown experiment
reported by Oates and Ho [102]. Reducing Her7 clock protein production in model
III to 0.1% of its original value (αC = 0.0045) abolishes oscillations and the clockwave completely, resulting in a graded distribution of her7 clock mRNA that agrees
qualitatively with experiement [102, see Fig. 9O].
The PCW model can be extended to more realistic two- and three-dimensional
lattices of cells. Figure 25 shows the simulated expression pattern in a fifty by five
simple rectangular lattice of heterogeneous cells for the model III nominal parameters
max
giving ∆TIII
= 29.3 minutes.
111
b Protein, time= 265.0 min
C
c mRNA, time= 265.0 min
1
Lateral Cell #
Lateral Cell #
1
2
3
4
5
3
4
5
0
10
20
30
40
50
0
10
20
30
40
50
Axial Cell # (1 is anterior)
Axial Cell # (1 is anterior)
S Protein, time= 265.0 min
s mRNA, time= 265.0 min
1
Lateral Cell #
1
Lateral Cell #
2
2
3
4
5
2
3
4
5
0
10
20
30
40
Axial Cell # (1 is anterior)
50
0
10
20
30
40
50
Axial Cell # (1 is anterior)
Figure 25: Simulated clock-wave in a rectangular lattice of fifty axial by five lateral
max
heterogeneous cells, for the model III parameter selection giving ∆TIII
= 29.3 min.
Darker grey indicates a higher expression level. Interior, edge, and corner cells are coupled to their eight, five, and three adjacent nearest neighbors, respectively. Normally
distributed noise is added independently to parameters in all cells with a positivity
constraint and so that 99.7% of the values are within ±1% of the nominal values.
The noise does not disrupt clock-wave formation. All plots occur 265 min after the
first cell exits the tailbud. The sigmoidal total control protein gradient (not depicted)
travels from left to right as time passes, while the bands of expression travel from
right to left as cellular expression oscillates.
112
The Mechanism of Gradient Controlled Oscillation Rate
The use of different combinations from a set of genes to control a developmental
process is referred to as combinatorial control [119]. The standard view of a combinatorial control mechanism is that multiple protein transcription factors control
mRNA production from one or more targeted genes, so that different transcription
factor combinations produce different expression patterns, and hence, different developmental outcomes. The PCW model has a distinct combinatorial control mechanism
that operates via differential decay of protein monomers and dimers that form by
competitive dimerization of the clock and control proteins. Oscillations in clock-gene
expression are generated through delayed, autoinhibitory feedback, while the competitive dimerization of clock and control protein controls the rate of clock oscillations.
A spatiotemporal gradient in the control protein slows oscillations in such a way as
to produce the experimentally observed clock-wave.
Validation of the PCW model for zebrafish reveals this new aspect of combinatorial control, namely, graded competitive dimerization with differential decay. Furthermore, validation shows that differential decay of monomers and dimers works
synergistically with multiple repressor binding sites for clock protein dimer in order
to produce both the proper oscillation rate in the tailbud (from multiple binding
sites for clock homodimer) and large enough rate slowing (from dimerization and
differential decay with a graded control protein) to form the clock-wave. The careful
model construction and selection process, in association with the sensitivity analysis
and robustness results for the model, suggests that this novel mechanism of oscillation
control is worthy of experimental investigation.
113
Comparison to Existing Zebrafish Models
The multicellular PCW model extends the two cell, single clock-gene model of
Lewis and coworkers [2, 4, 27], while retaining much of that model’s simplicity. Protein
dimerization and homodimer repression of mRNA production through multiple DNA
binding sites are handled more precisely in the PCW model. The PCW model also
incorporates an axially graded protein that controls the oscillation rate and initiates
a clock-wave in the posterior PSM.
Construction of the PCW model is much more transparent than the two clockgene model of Cinquin [30], and the resulting system of equations is considerably
simpler (four vs. thirteen differential equations per cell). While not precluding existence of multiple clock-genes, the present model requires only one. Furthermore,
heterodimers of the clock and control proteins do not bind DNA, which appears to be
more biologically grounded, at least in the case of zebrafish [111, 113] and [119, see
Fig. 7-26]. Unlike the Cinquin model, the PCW model is simplified through use of a
fast dimerization approximation and also includes control protein homodimerization,
which may affect the competitive dimerization mechanism shown here to be important
for oscillation rate control. Finally, the PCW model’s representation of juxtacrine intercellular signaling4 uses strictly nearest-neighbor coupling that has been tested on
a two-dimensional lattice of heterogeneous cells.
Applicability of the PCW Model
Recall that modeling results are, at best, only as valid as the underlying assumptions. Important assumptions of the PCW model are as follows:
1. Cellular processes may be treated deterministically.
4
Juxtacrine signaling is non-diffusive and requires direct cell-to-cell contact.
114
2. The PSM grows at a steady rate and minimal cell division and rearrangement
occur in the PSM.
3. Clock and control protein dimerization is very fast.
4. Control of gene expression may be modeled using the the approach of Shea and
Ackers, with a constant level of RNAP-II transcription complex.
5. Clock-gene redundancy allows the use of a single, representative clock-gene.
6. The clock-gene has one or two repressor binding sites and a single activator site.
7. Independent binding of repressors and activators occurs at the clock-gene.
8. The coordinating-signal gene has a single repressor binding site for clock protein
homodimers.
9. The clock-gene has equal binding affinities for different repressor binding sites.
10. Protein monomers, protein dimers, and mRNA undergo first order (i.e., linear)
decay kinetics.
11. Intercellular signaling is weak as compared to intracellular clock-gene autoinhibition.
12. Response to coordinating signal from adjacent cells is linear (i.e., nonsaturating).
While some of these assumptions have been verified experimentally, a considerable
number of technical difficulties (and monetary costs) would be involved in experimentally verifying all of these assumptions in zebrafish, chick, and mouse. More detailed
experimental information on dimer dissociation constants, binding site arrangements,
115
and binding site affinities/cooperativities would be particularly useful. With enough
time and effort, many of the above assumptions could be checked in silico, such as
the role of multiple clock-genes, rate-limited decay kinetics, or saturating intercellular
signaling, the effects which could be substantial. Lastly, it is hoped that the present
mathematical model can inform mathematical models of model organisms other than
zebrafish, such as chick and mouse.
Further Analyses and Future Directions
With complex feedback mechanisms at work during somitogenesis, intuition about
the operation of genetic circuits can be of limited use in the formulation of scientific
hypotheses and design of experiments [24, 130]. A complementary mathematical
model can help guide these endeavors. While many papers in the biological literature
report the necessity of a certain gene for somitogenesis via gene knockout/knockdown
experiments, unfortunately there is often little useful information included for revising
quantitative models [131], which could then inform the next round of experiments.
As noted by others [27, 30], investigating the Notch-mediated coupling with the
above combined methodology would be especially useful in further understanding
somitogenesis. This coupling system apparently has the interesting property whereby
it synchronizes oscillations in the tailbud, coordinates traveling wave oscillations in
the PSM, and accommodates fixed waves during segmentation. The PCW model
presents a minimal biologically plausible account of such phenomena, and may be
advantageous over recent phase oscillator models such as [70].
Because Notch coupling is understood to coordinate oscillations in zebrafish rather
than drive them, the present work did not estimate intercellular coupling parameters.
A multicellular, Monte-Carlo style simulation that investigates the effects of coupling
parameters on the robustness of clock-wave formation would be useful. Such inves-
116
tigation would have significant additional computational expense and would require
the design of new computational tools for automating the assessment of clock-wave
fitness. Furthermore, a more complete understanding and accounting of noise sources
in the system would be necessary.
Lastly, considerable effort has been made in the minimal construction of the PCW
model. The PCW model is somewhat more complicated than Lewis’s original model
in [27], because of finer attention to nonlinear production of mRNA, protein dimerization, and differential clock protein decay. More traditional dynamical systems
analyses of Lewis’s original, uncoupled model have been accomplished (recall system
(4)–(5) on page 20). These include location of Hopf bifurcations [88, 89, 90], and
parametrized continuations of periodic asymptotic solution approximations away from
the Hopf point [88]. Dimerization and differential decay in the PCW model complicate
such analysis, however, future work may reveal that the model is simple enough to
allow such analyses. This would nicely complement the present numerical analyses
of the mechanism of oscillation rate control via graded competitive dimerization with
differential decay.
117
CONCLUSION
Somitogenesis embodies many hallmarks of pattern formation in developmental biology. Modern scientific investigation has revealed a fascinating interplay of
biomolecular clocks, spatiotemporal gradients, and intercellular communication underlying somitogenesis. Mathematical models, such as the two presented in this work,
have become essential tools for understanding the interplay between the biological
components of somitogenesis. Even simple phenomenological models, such as the
multi-stable phase oscillator (MPO) model presented in Chapter 3, help reveal the
key components of somitogenesis and provide a proof-of-concept for mechanisms such
as the clock and wavefront.
The posterior clock-wave (PCW) model described in Chapter 4 focuses on clockwave formation in the clock and wavefront mechanism. The PCW model incorporates
a more detailed biological mechanism than the MPO model, yet maintains a relatively simple description. In striking a good balance between biological realism and
reduced complexity, the model incorporates three prominent, experimentally identified components: an intracellular clock, a graded control protein, and an intercellular
coordinating signal. The model accurately integrates three key sources of nonlinearity
in the genetic regulatory network: mRNA production, competitive dimerization, and
differential protein decay. A fast dimerization approximation provides considerable
reduction in the system of delay differential equations representing mRNA and protein
dynamics in each cell, but requires the addition of a system of two nonlinear algebraic
equations to represent the fast dimerization. This nonlinear algebraic system is solved
with a novel iterative scheme that is extendable to competitive dimerization of three
or more proteins.
118
During model validation in Chapter 5, multiple binding sites for clock protein
autoinhibition, dimerization of clock and control proteins, and differential decay of
clock protein are shown to act synergistically in the PCW model’s reproduction of
the posterior clock-wave in zebrafish. Careful parameter estimation and model selection indicate that 1) multiple transcription binding sites are necessary for matching
the period of oscillations in the tailbud, and 2) differential decay of clock protein
monomers and dimers is necessary to sufficiently slow oscillations in the posterior
PSM and generate the clock-wave. This differential decay mechanism represents a
novel type of combinatorial control, and is a source of nonlinearity different from the
rate-limited (i.e., Michaelis-Menten) decay mechanisms included in other mathematical models. The multicellular PCW model is reasonably robust to perturbation of all
parameters, and this warrants experimental investigation of the apparent differential
decay mechanism.
Future Directions
There are many exciting directions for future research, including the following:
1. Examining the role of multiple clock-genes, including competitive dimerization
among three or more proteins.
2. Comparing the effects of differential vs. rate-limited protein decay.
3. A closer examination of the intercellular coupling mechanism, including improved estimation of coupling parameters, the effects of coordinating signal saturation, and robustness of clock-wave formation to biologically realistic internal
and external noise.
119
4. Analyses to further understand the mechanism of oscillation rate control and
how parameter choices affect clock-wave formation.
5. Modeling the control protein gradient as a state variable instead of a prescribed,
external control.
6. Incorporating the determination wavefront that arrests oscillatory gene-expression
into a somite pre-pattern.
7. Examining and (possibly) including additional stochastic effects in the model.
The ultimate goal for the PCW model is its incorporation into a full model of the the
clock and wavefront mechanism, joining the clock-wave with the gradient-wavefront
to reproduce the complete pattern of somitogenesis.
120
REFERENCES CITED
121
[1] Frederic H Martini. Fundamentals of Anatomy and Physiology, Chapter 29.
Prentice Hall, edition fifth, 2001.
[2] Ertugrul M Özbudak, Julian Lewis. Notch signalling synchronizes the zebrafish
segmentation clock but is not needed to create somite boundaries. PLoS Genet,
4(2):e15, 2008.
[3] Mary-Lee Dequéant, Olivier Pourquié. Segmental patterning of the vertebrate
embryonic axis. Nat Rev Genet, 9(5):370–82, 2008.
[4] François Giudicelli, Ertugrul M Özbudak, Gavin J Wright, Julian Lewis. Setting the tempo in development: an investigation of the zebrafish somite clock
mechanism. PLoS Biol, 5(6):e150, 2007.
[5] Scott A Holley. The genetics and embryology of zebrafish metamerism. Dev
Dyn, 236(6):1422–49, 2007.
[6] François Giudicelli, Julian Lewis. The vertebrate segmentation clock. Curr
Opin Genet Dev, 14(4):407–14, 2004.
[7] Alexander Aulehla, Bernhard G Herrmann. Segmentation in vertebrates: clock
and gradient finally joined. Genes Dev, 18(17):2060–7, 2004.
[8] Olivier Pourquié. The segmentation clock: converting embryonic time into
spatial pattern. Science, 301(5631):328–30, 2003.
[9] Yumiko Saga, Hiroyuki Takeda. The making of the somite: molecular events in
vertebrate segmentation. Nat Rev Genet, 2(11):835–45, 2001.
[10] H L Stickney, M J Barresi, S H Devoto. Somite development in zebrafish. Dev
Dyn, 219(3):287–303, 2000.
[11] John P Kanki, Robert K Ho. The development of the posterior body in zebrafish. Development, 124(4):881–93, 1997.
[12] J Cooke, E C Zeeman. A clock and wavefront model for control of the number of
repeated structures during animal morphogenesis. J Theor Biol, 58(2):455–76,
1976.
[13] René Thom. Mathematical Models of Morphogenesis. Mathematics and its
applications. Ellis Horwood Limited, 1983.
[14] Isabel Palmeirim, Domingos Henrique, David Ish-Horowicz, Olivier Pourquié.
Avian hairy gene expression identifies a molecular clock linked to vertebrate
segmentation and somitogenesis. Cell, 91(5):639–48, 1997.
122
[15] Hans Meinhardt. Models of segmentation. Ruth Bellairs, Donald A Ede,
James W Lash, editors, Somites in Developing Embryos, pages 179–89. Plenum
Press, 1986.
[16] Brian C Goodwin. Temporal Organization in Cells: A dynamic theory of cellular
control processes. Academic Press, 1963.
[17] Company of Biologists. Homeostasis and Feedback Mechanisms, number 18
series Symposia of the Society for Experimental Biology. Academic Press, 1964.
[18] Brian C Goodwin. Oscillatory behavior in enzymatic control processes. Adv
Enzyme Regul, 3:425–38, 1965.
[19] J S Griffith. Mathematics of cellular control processes. I. Negative feedback to
one gene. J Theor Biol, 20(2):202–8, 1968.
[20] J S Griffith. Mathematics of cellular control processes. II. Positive feedback to
one gene. J Theor Biol, 20(2):209–16, 1968.
[21] Christopher P Fall, Eric S Marland, John M Wagner, John J Tyson, editors.
Computational Cell Biology, Volume 20 series Interdisciplinary Applied Mathematics. Springer, 2002.
[22] A V Hill. The possible effects of the aggregation of the molecules of haemoglobin
on its dissociation curves. J. Physiol., 40:iv–vii, 1910.
[23] Madeline A Shea, Gary K Ackers. The OR control system of bacteriophage
lambda. A physical-chemical model for gene regulation. J Mol Biol, 181(2):211–
30, 1985.
[24] Andrew C Oates, Nicole Gorfinkiel, Marcos Gonzalez-Gaitan, Carl-Philipp
Heisenberg. Quantitative approaches in developmental biology. Nat Rev Genet,
10(8):517–30, 2009.
[25] Moises Santillán, Michael C Mackey. A proposed mechanism for the interaction
of the segmentation clock and the determination front in somitogenesis. PLoS
ONE, 3(2):e1561, 2008.
[26] Hiromi Hirata, Shigeki Yoshiura, Toshiyuki Ohtsuka, Yasumasa Bessho,
Takahiro Harada, Kenichi Yoshikawa, Ryoichiro Kageyama. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science,
298(5594):840–3, 2002.
[27] Julian Lewis. Autoinhibition with transcriptional delay: a simple mechanism
for the zebrafish somitogenesis oscillator. Curr Biol, 13(16):1398–408, 2003.
123
[28] Kazuki Horikawa, Kana Ishimatsu, Eiichi Yoshimoto, Shigeru Kondo, Hiroyuki
Takeda. Noise-resistant and synchronized oscillation of the segmentation clock.
Nature, 441(7094):719–23, 2006.
[29] Hendrik B Tiedemann, Elida Schneltzer, Stefan Zeiser, Isabel Rubio-Aliaga,
Wolfgang Wurst, Johannes Beckers, Gerhard K H Przemeck, Martin Hrabe
de Angelis. Cell-based simulation of dynamic expression patterns in the presomitic mesoderm. J Theor Biol, 248(1):120–9, 2007.
[30] Olivier Cinquin. Repressor dimerization in the zebrafish somitogenesis clock.
PLoS Comput Biol, 3(2):e32, 2007.
[31] Joel E Cohen. Mathematics is biology’s next microscope, only better; biology
is mathematics’ next physics, only better. PLoS Biol, 2(12):e439, 2004.
[32] Santiago Schnell, Philip K Maini, Stuart A Newman, Timothy J Newman,
editors. Multiscale Modeling of Developmental Systems, Volume 81 series Curr
Top Dev Biol. Academic Press, 2008.
[33] Santiago Schnell, Ramon Grima, Philip K Maini. Multiscale modeling in biology. Am Sci, 95:134–42, 2008.
[34] Ido Golding, Edward C Cox. Eukaryotic transcription: what does it mean for
a gene to be ‘on’ ? Curr Biol, 16(10):R371–3, 2006.
[35] Julian Lewis. A clock-and-wavefront model simulates the observed pattern of
c-hairy1 expression during somitogenesis. Cell, 91(5):639–48 (appendix), 1997.
[36] Johannes Jaeger, Brian C Goodwin. A cellular oscillator model for periodic
pattern formation. J Theor Biol, 213(2):171–81, 2001.
[37] A M Turing. The chemical basis of morphogenesis. Phil Trans Roy Soc Lond,
B327:37–72, 1952.
[38] A Gierer, H Meinhardt. A theory of biological pattern formation. Kybernetik,
12(1):30–9, 1972.
[39] Philip K Maini. Mathematical models in morphogenesis. V Capasso, O Diekmann, editors, Mathematics Inspired by Biology, pages 151–89. Springer, 1997.
[40] L Wolpert. Positional information and the spatial pattern of cellular differentiation. J Theor Biol, 25(1):1–47, 1969.
[41] Avigdor Eldar, Ruslan Dorfman, Daniel Weiss, Hilary Ashe, Ben-Zion Shilo,
Naama Barkai. Robustness of the BMP morphogen gradient in Drosophila
embryonic patterning. Nature, 419(6904):304–8, 2002.
124
[42] Julien Dubrulle, Olivier Pourquié. fgf8 mRNA decay establishes a gradient
that couples axial elongation to patterning in the vertebrate embryo. Nature,
427(6973):419–22, 2004.
[43] Marta Ibanes, Juan Carlos Izpisúa Belmonte. Theoretical and experimental
approaches to understand morphogen gradients. Mol Syst Biol, 4:176, 2008.
[44] L Wolpert. One hundred years of positional information.
12(9):359–64, 1996.
Trends Genet,
[45] D R Primmett, W E Norris, G J Carlson, R J Keynes, C D Stern. Periodic
segmental anomalies induced by heat shock in the chick embryo are associated
with the cell cycle. Development, 105(1):119–30, 1989.
[46] Ruth E Baker, Santiago Schnell, Philip K Maini. Formation of vertebral precursors: past models and future predictions. J Theor Med, 5:23–35, 2003.
[47] A A Polezhaev. A mathematical model of the mechanism of vertebrate somitic
segmentation. J Theor Biol, 156(2):169–81, 1992.
[48] J R Collier, D Mcinerney, S Schnell, P K Maini, D J Gavaghan, P Houston,
C D Stern. A cell cycle model for somitogenesis: mathematical formulation and
numerical simulation. J Theor Biol, 207(3):305–16, 2000.
[49] Santiago Schnell, Philip K Maini, Daragh McInerney, David J Gavaghan, Paul
Houston. Models for pattern formation in somitogenesis: a marriage of cellular
and molecular biology. C R Biol, 325(3):179–89, 2002.
[50] D McInerney, S Schnell, R E Baker, P K Maini. A mathematical formulation
for the cell-cycle model in somitogenesis: analysis, parameter constraints and
numerical solutions. Math Med Biol, 21(2):85–113, 2004.
[51] J Cooke. A gene that resuscitates a theory–somitogenesis and a molecular
oscillator. Trends Genet, 14(3):85–8, 1998.
[52] Arthur T Winfree. The Geometry of Biological Time, Volume 8 series Biomathemtics. Springer-Verlag, 1980.
[53] Albert Goldbeter. Biochemical Oscillations and Cellular Rhythms. Cambridge
University Press, 1996.
[54] M Kerszberg, L Wolpert. A clock and trail model for somite formation, specialization and polarization. J Theor Biol, 205(3):505–10, 2000.
[55] Santiago Schnell, Philip K Maini. Clock and induction model for somitogenesis.
Dev Dyn, 217(4):415–20, 2000.
125
[56] Ruth E Baker, Santiago Schnell. How can mathematics help us explore vertebrate segmentation? HFSP J, 3(1):1–5, 2009.
[57] Ruth E Baker, Santiago Schnell, Philip K Maini. Mathematical models for
somite formation. Curr Top Dev Biol, 81:183–203, 2008.
[58] Paul M Kulesa, Santiago Schnell, Stefan Rudloff, Ruth E Baker, Philip K
Maini. From segment to somite: segmentation to epithelialization analyzed
within quantitative frameworks. Dev Dyn, 236(6):1392–402, 2007.
[59] R E Baker, S Schnell, P K Maini. A clock and wavefront mechanism for somite
formation. Dev Biol, 293(1):116–26, 2006.
[60] K J Dale, O Pourquie. A clock-work somite. Bioessays, 22(1):72–83, 2000.
[61] Hans Meinhardt. Models of Biological Pattern Formation. Academic Press,
1982.
[62] Hans Meinhardt. Models of biological pattern formation: from elementary steps
to the organization of embryonic axes. Curr Top Dev Biol, 81:1–63, 2008.
[63] R E Baker, S Schnell, P K Maini. A mathematical investigation of a Clock and
Wavefront model for somitogenesis. J Math Biol, 52(4):458–82, 2006.
[64] R E Baker, P K Maini. Travelling gradients in interacting morphogen systems.
Math Biosci, 209(1):30–50, 2007.
[65] Albert Goldbeter, Didier Gonze, Olivier Pourquié. Sharp developmental thresholds defined through bistability by antagonistic gradients of retinoic acid and
FGF signaling. Dev Dyn, 236(6):1495–508, 2007.
[66] James A Glazier, Ying Zhang, Maciej Swat, Benjamin Zaitlen, Santiago Schnell.
Coordinated action of N-CAM, N-cadherin, EphA4, and ephrinB2 translates
genetic prepatterns into structure during somitogenesis in chick. Curr Top Dev
Biol, 81:205–47, 2008.
[67] Nicola J Armstrong, Kevin J Painter, Jonathan A Sherratt. Adding adhesion
to a chemical signaling model for somite formation. Bull Math Biol, 71(1):1–24,
2009.
[68] M Kaern, M Menzinger, A Hunding. Segmentation and somitogenesis derived
from phase dynamics in growing oscillatory media. J Theor Biol, 207(4):473–93,
2000.
[69] Ingmar H Riedel-Kruse, Claudia Muller, Andrew C Oates. Synchrony dynamics during initiation, failure, and rescue of the segmentation clock. Science,
317(5846):1911–5, 2007.
126
[70] Luis G Morelli, Saul Ares, Leah Herrgen, Christian Schroter, Frank Julicher,
Andrew C Oates. Delayed coupling theory of vertebrate segmentation. HFSP J,
3(1):55–66, 2009.
[71] Padmashree C G Rida, Nguyet Le Minh, Yun Jin Jiang. A Notch feeling of
somite segmentation and beyond. Dev Biol, 265(1):2–22, 2004.
[72] Olivier Cinquin. Is the somitogenesis clock really cell-autonomous? A coupledoscillator model of segmentation. J Theor Biol, 224(4):459–68, 2003.
[73] S Zeiser, O Rivera, C Kuttler, B Hense, R Lasser, G Winkler. Oscillations
of Hes7 caused by negative autoregulation and ubiquitination. Comput Biol
Chem, 32(1):47–51, 2008.
[74] Mary-Lee Dequéant, Earl Glynn, Karin Gaudenz, Matthias Wahl, Jie Chen,
Arcady Mushegian, Olivier Pourquié. A complex oscillating network of signaling
genes underlies the mouse segmentation clock. Science, 314(5805):1595–8, 2006.
[75] Albert Goldbeter, Olivier Pourquié. Modeling the segmentation clock as a
network of coupled oscillations in the Notch, Wnt and FGF signaling pathways.
J Theor Biol, 252(3):574–85, 2008.
[76] M H Jensen, K Sneppen, G Tiana. Sustained oscillations and time delays in
gene expression of protein Hes1. FEBS Lett, 541(1-3):176–7, 2003.
[77] Nicholas A M Monk. Oscillatory expression of Hes1, p53, and NF-kappaB
driven by transcriptional time delays. Curr Biol, 13(16):1409–13, 2003.
[78] Yun-Jin Jiang, Julian Lewis. Notch as a synchroniser in somite segmentation.
Esmond J Sanders, James W Lash, Charles P Ordahl, editors, The Origin and
Fate of Somites, pages 71–9. IOS Press, 2001.
[79] Kana Ishimatsu, Kazuki Horikawa, Hiroyuki Takeda. Coupling cellular oscillators: a mechanism that maintains synchrony against developmental noise in
the segmentation clock. Dev Dyn, 236(6):1416–21, 2007.
[80] Hiromi Hirata, Yasumasa Bessho, Hiroshi Kokubu, Yoshito Masamizu, Shuichi
Yamada, Julian Lewis, Ryoichiro Kageyama. Instability of Hes7 protein is
crucial for the somite segmentation clock. Nat Genet, 36(7):750–4, 2004.
[81] Stefan Zeiser, H Volkmar Liebscher, Hendrik Tiedemann, Isabel Rubio-Aliaga,
Gerhard K H Przemeck, Martin Hrabe de Angelis, Gerhard Winkler. Number
of active transcription factor binding sites is essential for the Hes7 oscillator.
Theor Biol Med Model, 3:11, 2006.
127
[82] Samuel Bernard, Branka Cajavec, Laurent Pujo-Menjouet, Michael C Mackey,
Hanspeter Herzel. Modelling transcriptional feedback loops: the role of
Gro/TLE1 in Hes1 oscillations. Philos Transact A Math Phys Eng Sci,
364(1842):1155–70, 2006.
[83] Yoshito Masamizu, Toshiyuki Ohtsuka, Yoshiki Takashima, Hiroki Nagahara,
Yoshiko Takenaka, Kenichi Yoshikawa, Hitoshi Okamura, Ryoichiro Kageyama.
Real-time imaging of the somite segmentation clock: revelation of unstable
oscillators in the individual presomitic mesoderm cells. Proc Natl Acad Sci U
S A, 103(5):1313–8, 2006.
[84] J G Rodrı́guez-González, M Santillán, A C Fowler, Michael C Mackey. The
segmentation clock in mice: interaction between the Wnt and Notch signalling
pathways. J Theor Biol, 248(1):37–47, 2007.
[85] Andrew Mara, Scott A Holley. Oscillators and the emergence of tissue organization during zebrafish somitogenesis. Trends Cell Biol, 17(12):593–9, 2007.
[86] Hiroshi Momiji, Nicholas A M Monk. Dissecting the dynamics of the Hes1
genetic oscillator. J Theor Biol, 254(4):784–98, 2008.
[87] Hiroshi Momiji, Nicholas A M Monk. Oscillatory expresssion of Hes family
transcription factors: insights from mathematical modeling. Miguel Maroto,
Micholas A M Monk, editors, Cellular Oscillatory Mechanisms, Volume 641
series Advances in Experimental Medicine and Biology, Chapter 6, pages 72–87.
Springer, 2008.
[88] Anael Verdugo, Richard Rand. Hopf bifurcation in a DDE model of gene expression. Commun Nonlinear Sci Numer Simulat, 13:235–42, 2008.
[89] Nicole Radde. The impact of time delays on the robustness of biological oscillators and the effect of bifurcations on the inverse problem. EURASIP J
Bioinform Syst Biol, page 327503, 2009.
[90] Peng Feng, Menaka Navaratna. Modelling periodic oscillations during somitogenesis. Math Biosci Eng, 4(4):661–73, 2007.
[91] Andrew Mara, Joshua Schroeder, Cecile Chalouni, Scott A Holley. Priming,
initiation and synchronization of the segmentation clock by deltaD and deltaC.
Nat Cell Biol, 9(5):523–30, 2007.
[92] O Pourquié, P P Tam. A nomenclature for prospective somites and phases of
cyclic gene expression in the presomitic mesoderm. Dev Cell, 1(5):619–20, 2001.
[93] Christian Schroter, Leah Herrgen, Albert Cardona, Gary J Brouhard, Benjamin
Feldman, Andrew C Oates. Dynamics of zebrafish somitogenesis. Dev Dyn,
237(3):545–53, 2008.
128
[94] Mark W Reichelt, Lawrence F Shampine. ode45. MATLAB & Simulink,
R2008b. The MathWorks, Inc., Natick, MA USA, 2005. www.mathworks.com.
[95] Yun-Jin Jiang, Birgit L Aerne, Lucy Smithers, Catherine Haddon, David IshHorowicz, Julian Lewis. Notch signalling and the synchronization of the somite
segmentation clock. Nature, 408(6811):475–9, 2000.
[96] Miguel Maroto, J Kim Dale, Mary-Lee Dequéant, Anne-Cecile Petit, Olivier
Pourquié. Synchronised cycling gene oscillations in presomitic mesoderm cells
require cell-cell contact. Int J Dev Biol, 49(2-3):309–15, 2005.
[97] J Dubrulle, M J McGrew, O Pourquié. FGF signaling controls somite boundary
position and regulates segmentation clock control of spatiotemporal Hox gene
activation. Cell, 106(2):219–32, 2001.
[98] A Sawada, M Shinya, Y J Jiang, A Kawakami, A Kuroiwa, H Takeda.
Fgf/MAPK signalling is a crucial positional cue in somite boundary formation.
Development, 128(23):4873–80, 2001.
[99] Olivier Cinquin. Understanding the somitogenesis clock: what’s missing? Mech
Dev, 124(7-8):501–17, 2007.
[100] Paul Francois, Vincent Hakim, Eric D Siggia. Deriving structure from evolution:
metazoan segmentation. Mol Syst Biol, 3:154, 2007.
[101] K I Mazzitello, C M Arizmendi, H G E Hentschel. Converting genetic network
oscillations into somite spatial patterns. Phys Rev E Stat Nonlin Soft Matter
Phys, 78(2 Pt 1):021906, 2008.
[102] Andrew C Oates, Robert K Ho. Hairy/E(spl)-related (Her) genes are central
components of the segmentation oscillator and display redundancy with the
Delta/Notch signaling pathway in the formation of anterior segmental boundaries in the zebrafish. Development, 129(12):2929–46, 2002.
[103] Ryoichiro Kageyama, Toshiyuki Ohtsuka, Taeko Kobayashi. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development,
134(7):1243–51, 2007.
[104] B H Jennings, D M Tyler, S J Bray. Target specificities of Drosophila enhancer
of split basic helix-loop-helix proteins. Mol Cell Biol, 19(7):4600–10, 1999.
[105] Dirk Sieger, Diethard Tautz, Martin Gajewski. her11 is involved in the somitogenesis clock in zebrafish. Dev Genes Evol, 214(8):393–406, 2004.
[106] Y Bessho, G Miyoshi, R Sakata, R Kageyama. Hes7: a bHLH-type repressor
gene regulated by Notch and expressed in the presomitic mesoderm. Genes
Cells, 6(2):175–85, 2001.
129
[107] Nicolas E Buchler, Ulrich Gerland, Terence Hwa. Nonlinear protein degradation
and the function of genetic circuits. Proc Natl Acad Sci U S A, 102(27):9559–64,
2005.
[108] Nicolas E Buchler, Ulrich Gerland, Terence Hwa. On schemes of combinatorial
transcription logic. Proc Natl Acad Sci U S A, 100(9):5136–41, 2003.
[109] Moises Santillán, Michael C Mackey. Why the lysogenic state of phage lambda
is so stable: a mathematical modeling approach. Biophys J, 86(1 Pt 1):75–84,
2004.
[110] Tomas Gedeon, Konstantin Mischaikow, Kate Patterson, Eliane Traldi. When
activators repress and repressors activate: a qualitative analysis of the SheaAckers model. Bull Math Biol, 70(6):1660–83, 2008.
[111] Akinori Kawamura, Sumito Koshida, Hiroko Hijikata, Takuya Sakaguchi,
Hisato Kondoh, Shinji Takada. Zebrafish hairy/enhancer of split protein links
FGF signaling to cyclic gene expression in the periodic segmentation of somites.
Genes Dev, 19(10):1156–61, 2005.
[112] Dirk Sieger, Bastian Ackermann, Christoph Winkler, Diethard Tautz, Martin
Gajewski. her1 and her13.2 are jointly required for somitic border specification
along the entire axis of the fish embryo. Dev Biol, 293(1):242–51, 2006.
[113] S Bae, Y Bessho, M Hojo, R Kageyama. The bHLH gene Hes6, an inhibitor of
Hes1, promotes neuronal differentiation. Development, 127(13):2933–43, 2000.
[114] C Leimeister, K Dale, A Fischer, B Klamt, M Hrabe de Angelis, F Radtke, M J
McGrew, O Pourquié, M Gessler. Oscillating expression of c-Hey2 in the presomitic mesoderm suggests that the segmentation clock may use combinatorial
signaling through multiple interacting bHLH factors. Dev Biol, 227(1):91–103,
2000.
[115] J K Dale, M Maroto, M-L Dequ/’eant, P Malapert, M McGrew, O Pourqui/’e.
Periodic notch inhibition by lunatic fringe underlies the chick segmentation
clock. Nature, 421(6920):275–8, 2003.
[116] Dirk Sieger, Diethard Tautz, Martin Gajewski. The role of Suppressor of Hairless in Notch mediated signalling during zebrafish somitogenesis. Mech Dev,
120(9):1083–94, 2003.
[117] James P Keener, Joel E Keizer. Computational Cell Biology, Volume 20 series
Interdisciplinary Applied Mathematics, Chapter 4. Springer, 2002.
[118] Benno Muller-Hill. The lac operon. de Gruyter, 1996.
130
[119] Bruce Alberts, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts,
Peter Walter. Molecular Biology of the Cell, Chapter 7. Garland Science, edition
fourth, 2002.
[120] O Nakagawa, D G McFadden, M Nakagawa, H Yanagisawa, T Hu, D Srivastava,
E N Olson. Members of the HRT family of basic helix-loop-helix proteins act
as transcriptional repressors downstream of Notch signaling. Proc Natl Acad
Sci U S A, 97(25):13655–60, 2000.
[121] Sarah J Bray. Notch signalling: a simple pathway becomes complex. Nat Rev
Mol Cell Biol, 7(9):678–89, 2006.
[122] Martin Gajewski, Dirk Sieger, Burkhard Alt, Christian Leve, Stefan Hans,
Christian Wolff, Klaus B Rohr, Diethard Tautz. Anterior and posterior waves of
cyclic her1 gene expression are differentially regulated in the presomitic mesoderm of zebrafish. Development, 130(18):4269–78, 2003.
[123] Jacek Kierzenka, Lawrence F Shampine, Skip Thompson. dde23 and ddesd.
MATLAB & Simulink, R2008b. The MathWorks, Inc., Natick, MA USA, 2008.
www.mathworks.com.
[124] The Markov chain Monte Carlo revolution. Persi diaconis. Bull Amer Math
Soc, 46:179–205, 2009.
[125] Sunita S Shankaran, Dirk Sieger, Christian Schroter, Carmen Czepe, MarieChristin Pauly, Mary A Laplante, Thomas S Becker, Andrew C Oates, Martin
Gajewski. Completing the set of h/E(spl) cyclic genes in zebrafish: her12 and
her15 reveal novel modes of expression and contribute to the segmentation clock.
Dev Biol, 304(2):615–32, 2007.
[126] Andrew C Oates, Claudia Mueller, Robert K Ho. Cooperative function of deltac
and her7 in anterior segment formation. Dev Biol, 280(1):133–49, 2005.
[127] Dorthe Julich, Chiaw Hwee Lim, Jennifer Round, Claudia Nicolaije, Joshua
Schroeder, Alexander Davies, Robert Geisler, Julian Lewis, Yun-Jin Jiang,
Scott A Holley. beamter/deltaC and the role of Notch ligands in the zebrafish
somite segmentation, hindbrain neurogenesis and hypochord differentiation.
Dev Biol, 286(2):391–404, 2005.
[128] Mark Ptashne. A genetic Switch. Cold Spring Harbor Press, 2004.
[129] Scott A Holley, Robert Geisler, Christiane Nusslein-Volhard. Control of her1
expression during zebrafish somitogenesis by a delta-dependent oscillator and
an independent wave-front activity. Genes Dev, 14(13):1678–90, 2000.
131
[130] Julian Lewis. From signals to patterns: space, time, and mathematics in developmental biology. Science, 322(5900):399–403, 2008.
[131] Julian Lewis, Ertugrul M Özbudak. Deciphering the somite segmentation clock:
beyond mutants and morphants. Dev Dyn, 236(6):1410–5, 2007.
132
APPENDICES
133
APPENDIX A
IMPOSSIBILITY OF NONTRIVIAL PERIODIC SOLUTIONS IN LEWIS’S
UNCOUPLED DDE MODEL WITHOUT DELAYS
134
Theorem 2. Consider the following system of differential equations:
dp(t)
= a m(t − Tp ) − b p(t),
dt
dm(t)
k
=
2 − c m(t),
dt
m)
1 + p(t−T
p0
where a, b, k and c, and po are (strictly) positive constants. If Tp = Tm = 0, then the
Dulac criterion implies that nontrivial periodic solutions are impossible.
p
am − bp
Proof. Let x = and f (x) =
, so that ẋ = f (x). Suppose
k
m
− cm
1+(p/p0 )2
a nontrivial closed orbit ∂A, with outward normal n, bounds the nontrivial planar
region A. Green’s Theorem implies:
I
ZZ
f (x) · n ds =
∂A
Z
0
T
∇ · f (x) dA,
A
ZZ
∂
∂
(a m − b p) +
ẋ(t) · n dt =
∂m
A ∂p
ZZ
0 = −(b + c)
dA < 0,
A
which is a contradiction.
k
− cm
1 + (p/p0 )2
dA,
135
APPENDIX B
COMPETITIVE DIMERIZATION OF THREE PROTEINS
136
The prevalence of multiple clock-genes in several model organisms suggests the
future examination of competitive dimerization of multiple clock proteins. As an
example of the generalizability of the iterative scheme presented in Chapter 4, the
case for competitive dimerization of three proteins is briefly outlined.
Suppose there is competitive dimerization between three proteins C, D, and G.
Consideration of all forms of each of these proteins gives the following definitions of
total proteins:
b := C + 2 C:C + C:D + C:G,
C
b := D + 2 D:D + C:D + D:G,
D
b := G + 2 G:G + C:G + D:G.
G
Consideration of all the relevant chemical interactions gives the following six possible
bidirectional dimerization reactions:
2C
C +D
+
kC:C
−
kC:C
+
kC:D
−
kC:D
C:C,
2D
C:D,
C +G
+
kD:D
−
kD:D
+
kC:G
−
kC:G
D:D,
2G
C:G,
D+G
+
kG:G
−
kG:G
+
kD:G
−
kD:G
G:G,
D:G,
where the forward and reverse (strictly) positive reaction rates are given above and
below the arrows, respectively.
A fast dimerization approximation results in finding a non-negative equilibrium
for the following system of nine ordinary differential equations derived from mass
137
action kinetics:
−
+
−
Ċ = 2 kC:C
C:C − 2 kC:C
C 2 + kC:D
C:D −
+
−
+
kC:D
C D + kC:G
C:G − kC:G
C G,
˙ = −k − C:C + k + C 2 ,
C:C
C:C
C:C
−
+
−
Ḋ = 2 kD:D
D:D − 2 kD:D
D2 + kC:D
C:D −
+
−
+
kC:D
C D + kD:G
D:G − kD:G
D G,
˙ = −k − D:D + k + D2 ,
D:D
C:C
C:C
−
+
−
Ġ = 2 kG:G
G:G − 2 kG:G
G2 + kC:G
C:G −
+
−
+
kC:G
C G + kD:G
D:G − kD:G
D G,
˙ = −k − G:G + k + G2 ,
G:G
G:G
G:G
˙ = −k − C:D + k + C D,
C:D
C:D
C:D
˙ = −k − C:G + k + C G,
C:G
C:G
C:G
˙ = −k − D:G + k + D G.
D:G
D:G
D:G
Setting the left hand side of the above system equal to zero at equilibrium gives,
after some simplification, the following nonlinear algebraic system of six equations
with nine unknowns:
C:C =
C2
,
κC:C
C:D =
CD
,
κC:D
D:D =
D2
,
κD:D
C:G =
CG
,
κC:G
(94)
G2
DG
,
D:G =
,
κG:G
κD:G
where κC:C , κD:D , κG:G , κC:D , κC:G , and κD:G are the (strictly) positive dissociation
G:G =
constants for the various dimers. To these six equations the following constraints are
138
appended:
b = C + 2 C:C + C:D + C:G,
C
b = D + 2 D:D + C:D + D:G,
D
(95)
b = G + 2 G:G + C:G + D:G.
G
Using the relationships in (94) in the right hand sides of (95) gives
2
b = C +2 C + CD + CG,
C
κC:C κC:D κC:G
2
b = D +2 D + CD + DG,
D
κD:D κC:D κD:G
2
b = G+2 G + CG + DG.
G
κG:G κC:G κD:G
Rewriting each of these equations as quadratics in the respective monomer concentrations gives
D
G
b
0=
C + 1+
+
C − C,
κC:C
κC:D κC:G
C
2
G
2
b
0=
D + 1+
+
D − D,
κD:D
κC:D κD:G
C
D
2
2
b
G − G.
G + 1+
+
0=
κG:G
κC:G κD:G
2
2
Using the quadratic equation to solve for the non-negative roots of these equations
3
gives a mapping G from R+ into itself defined component-wise by
s
2
b
C
κC:C
Dn
Gn
Dn
Gn
+
+8
− 1+
+
Cn+1 =
1+
4
κC:D κC:G
κC:C
κC:D κC:G
s
2
b
κD:D
Cn
Gn
D
Cn
Gn
1+
+
+8
− 1+
+
Dn+1 =
4
κC:D κD:G
κD:D
κC:D κD:G
s
2
b
κG:G
Cn
Dn
G
Cn
Dn
Gn+1 =
+8
1+
+
− 1+
+
4
κC:G κD:G
κG:G
κC:G κD:G
139
This mapping defines an iterative numerical scheme for the computation of the
monomer concentrations C, D, and G, as a function of the total protein concentrations
b D,
b and G.
b Convergence has been observed in practice for typical values of the
C,
dissociation constants and a range of total protein levels.
140
APPENDIX C
MATLAB CODES
141
Multi-Stable Phase Oscillator (MPO) Model Code
somitogenesisMPO.m: This simulates the MPO model using Matlab’s
ode45 solver, and plots some useful results.
function somitogenesisMPO
% Usage :
%
somitogenesisMPO
%
% This s i m u l a t e s t h e m u l t i −s t a b l e p ha s e o s c i l l a t o r model o f ...
somitogenesis .
% Initialize
K= 6 0 ; % Number o f c e l l s
c e l l I n d i c e s= ( 1 :K) ’ ;
c e l l s P e r S o m i t e= 6 ;
tau= 3 ; % S u s c e p t i b i l i t y / m a t u r i t y h a l f − l i f e
T= ( c e l l I n d i c e s − 1 ) / c e l l s P e r S o m i t e ; % PSM c e l l e n t e r t i m e s
% S o l v e system o f ODEs from z e r o t o s i x t y time u n i t s
s o l= ode45 ( @rhs , [ 0 6 0 ] , 0 . 2 5 ∗ o n e s ( s i z e ( c e l l I n d i c e s ) ) , o d e s e t ( ...
’RelTol ’ , 1 0 e −9) ) ;
% Plots
f i g u r e H= f i g u r e ( 1 ) ; c l f ( f i g u r e H ) ;
set ( f i g u r e H , ’PaperSize ’ , [ 6 3 ] , ’PaperType ’ , ’<custom >’ , ’...
Units ’ , ’inches ’ ) ;
142
p o s i t i o n= get ( f i g u r e H , ’Position ’ ) ;
p o s i t i o n ( 3 : 4 )= [ 6 3 ] ;
set ( f i g u r e H , ’Position ’ , p o s i t i o n ) ;
axisH= subplot ( 1 , 2 , 1 ) ;
t G r i d= − 1 : 0 . 1 : 7 ;
s e r i e s H= plot ( axisH , tGrid , s ( tGrid , 3 ) , ’LineWidth ’ , 2 ) ;
ylim ( axisH , [ −0.1 1 . 1 ] ) ;
ylabel ( axisH , ’y’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
xlim ( axisH , [−1 7 ] ) ;
xlabel ( axisH , ’t ( unitless time)’ , ’FontSize ’ , 1 2 , ’...
FontWeight ’ , ’demi ’ ) ;
t i t l e ( axisH , ’y=s(t; 3) ’ , ’FontSize ’ , 1 4 , ’FontWeight ’ , ’...
demi ’ ) ;
set ( axisH , ’XMinorTIck ’ , ’on ’ , ’YMinorTIck ’ , ’on ’ , ’...
FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
set ( axisH , ’Position ’ , [ 0 . 1 2 0 . 1 7 5 0 . 3 5 0 . 6 7 5 ] ) ;
text ( −3 , 1 . 2 5 , ’(a)’ , ’FontSize ’ , 1 6 , ’FontWeight ’ , ’demi ’ ) ;
axisH= subplot ( 1 , 2 , 2 ) ;
s e r i e s H= plot ( axisH , c e l l I n d i c e s , s ( 6 − T, 3 ) , ’.-’ , ...
c e l l I n d i c e s , s ( 8 − T, 3 ) , ’.-’ , ’LineWidth ’ , 0 . 5 ) ;
ylim ( axisH , [ −0.1 1 . 1 ] ) ;
ylabel ( axisH , ’y’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
xlim ( axisH , [ 0 K+1]) ;
xlabel ( axisH , ’k (cell #) ’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’...
demi ’ ) ;
143
t i t l e ( axisH , [ ’y=s(t-T_k; 3) , k=1 ,... , ’ num2str (K) ] , ’...
FontSize ’ , 1 4 , ’FontWeight ’ , ’demi ’ ) ;
legend ( axisH , ’t=6 ’ , ’t=8 ’ ) ;
set ( axisH , ’XMinorTIck ’ , ’on ’ , ’YMinorTIck ’ , ’on ’ , ’...
FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
set ( axisH , ’Position ’ , [ 0 . 6 2 0 . 1 7 5 0 . 3 5 0 . 6 7 5 ] ) ;
text ( −16 , 1 . 2 5 , ’(b)’ , ’FontSize ’ , 1 6 , ’FontWeight ’ , ’demi ’ ) ...
;
f i g u r e H= f i g u r e ( 2 ) ; c l f ( f i g u r e H ) ;
set ( f i g u r e H , ’PaperSize ’ , [ 6 3 ] , ’PaperType ’ , ’<custom >’ , ’...
Units ’ , ’inches ’ ) ;
p o s i t i o n= get ( f i g u r e H , ’Position ’ ) ;
p o s i t i o n ( 3 : 4 )= [ 6 3 ] ;
set ( f i g u r e H , ’Position ’ , p o s i t i o n ) ;
axisH= subplot ( 1 , 1 , 1 ) ;
p hi G r i d= 0 : 0 . 0 0 5 : 2 ;
s e r i e s H= plot ( axisH , phiGrid , o n e s ( s i z e ( p hi G r i d ) ) , phiGrid ...
, 1 − 0 . 5 ∗ ( sin ( 2 ∗ pi ∗ p hi G r i d ) ) . ˆ 2 , phiGrid , 1 − ( sin ( 2 ∗ pi ∗ p hi G r id ...
) ) . ˆ 2 , ’LineWidth ’ , 2 ) ;
set ( s e r i e s H ( 1 ) , ’Color ’ , [ 1 0 0 ] ) ;
set ( s e r i e s H ( 2 ) , ’Color ’ , [ 0 . 5 0 . 5 0 . 5 ] ) ;
set ( s e r i e s H ( 3 ) , ’Color ’ , [ 0 1 0 ] ) ;
ylim ( axisH , [ 0 1 . 1 ] ) ;
ylabel ( axisH , ’y’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
144
xlabel ( axisH , ’\ phi_1 ’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi...
’) ;
t i t l e ( axisH , ’y=d\ phi_1 /dt , \tau_ {1/2}=3 , T_1 =0 ’ , ’...
Interpreter ’ , ’tex ’ , ’FontSize ’ , 1 4 , ’FontWeight ’ , ’demi ’ ) ;
legend ( axisH , ’t\leq0 ’ , ’t=3 ’ , ’t\geq6 ’ , ’Location ’ , ’...
southeast ’ ) ;
set ( axisH , ’XMinorTIck ’ , ’on ’ , ’YMinorTIck ’ , ’on ’ , ’...
FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
f i g u r e H= f i g u r e ( 3 ) ; c l f ( f i g u r e H ) ;
set ( f i g u r e H , ’PaperSize ’ , [ 6 3 ] , ’PaperType ’ , ’<custom >’ , ’...
Units ’ , ’inches ’ ) ;
p o s i t i o n= get ( f i g u r e H , ’Position ’ ) ;
p o s i t i o n ( 3 : 4 )= [ 6 3 ] ;
set ( f i g u r e H , ’Position ’ , p o s i t i o n ) ;
axisH= subplot ( 1 , 1 , 1 ) ;
s e r i e s H= plot ( axisH , s o l . x , s o l . y ( 1 : 6 , : ) , ’LineWidth ’ , 2 ) ;
ylabel ( axisH , ’y’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
xlim ( axisH , [ 0 8 ] ) ;
xlabel ( axisH , ’t’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
t i t l e ( axisH , ’y=\ phi_k (t), \tau_ {1/2}=3 , T_k =(k -1) /6, k...
=1 ,... ,6 ’ , ’FontSize ’ , 1 4 , ’FontWeight ’ , ’demi ’ ) ;
legend ( axisH , ’\ phi_1 (t)’ , ’\ phi_2 (t)’ , ’\ phi_3 (t)’ , ’\...
phi_4 (t)’ , ’\ phi_5 (t)’ , ’\ phi_6 (t)’ , ’Location ’ , ’NorthWest ’ ) ;
set ( axisH , ’XMinorTIck ’ , ’on ’ , ’YMinorTIck ’ , ’on ’ , ’...
FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
145
f i g u r e H= f i g u r e ( 3 ) ; c l f ( f i g u r e H ) ;
set ( f i g u r e H , ’PaperSize ’ , [ 6 3 ] , ’PaperType ’ , ’<custom >’ , ’...
Units ’ , ’inches ’ ) ;
p o s i t i o n= get ( f i g u r e H , ’Position ’ ) ;
p o s i t i o n ( 3 : 4 )= [ 6 3 ] ;
set ( f i g u r e H , ’Position ’ , p o s i t i o n ) ;
axisH= subplot ( 1 , 1 , 1 ) ;
s e r i e s H= plot ( axisH , s o l . x , s o l . y ( 1 : 6 , : ) , ’LineWidth ’ , 2 ) ;
ylabel ( axisH , ’y’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
xlim ( axisH , [ 0 8 ] ) ;
xlabel ( axisH , ’t ( unitless time)’ , ’FontSize ’ , 1 2 , ’...
FontWeight ’ , ’demi ’ ) ;
t i t l e ( axisH , ’y=\ phi_k (t), \tau_ {1/2}=3 , T_k =(k -1) /6, k...
=1 ,... ,6 ’ , ’FontSize ’ , 1 4 , ’FontWeight ’ , ’demi ’ ) ;
legend ( axisH , ’\ phi_1 (t)’ , ’\ phi_2 (t)’ , ’\ phi_3 (t)’ , ’\...
phi_4 (t)’ , ’\ phi_5 (t)’ , ’\ phi_6 (t)’ , ’Location ’ , ’northwest ’ ) ;
set ( axisH , ’XMinorTIck ’ , ’on ’ , ’YMinorTIck ’ , ’on ’ , ’...
FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
f i g u r e H= f i g u r e ( 4 ) ; c l f ( f i g u r e H ) ;
set ( f i g u r e H , ’PaperSize ’ , [ 6 3 ] , ’PaperType ’ , ’<custom >’ , ’...
Units ’ , ’inches ’ ) ;
p o s i t i o n= get ( f i g u r e H , ’Position ’ ) ;
p o s i t i o n ( 3 : 4 )= [ 6 3 ] ;
set ( f i g u r e H , ’Position ’ , p o s i t i o n ) ;
146
axisH= subplot ( 1 , 1 , 1 ) ;
s e r i e s H= plot ( axisH , ( ( 0 : K−1) + 0 . 5 ) / c e l l s P e r S o m i t e , s o l . y...
( : , end ) , ’.-’ , ’LineWidth ’ , 0 . 5 ) ;
ylabel ( axisH , ’y’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
xlim ( axisH , [ 0 K/ c e l l s P e r S o m i t e ] ) ;
xlabel ( axisH , ’x ( somite #) ’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ...
’demi ’ ) ;
t i t l e ( axisH , [ ’y=\ phi_k (’ num2str ( s o l . x ( end ) , ’%1.2f’ ) ’)...
, k=1 ,2 ,... ,60 ’ ] , ’FontSize ’ , 1 4 , ’FontWeight ’ , ’demi ’ ) ;
set ( axisH , ’XMinorTIck ’ , ’on ’ , ’YMinorTIck ’ , ’on ’ , ’...
FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
f i g u r e H= f i g u r e ( 5 ) ; c l f ( f i g u r e H ) ;
colormap ( [ 1 0 0 ; 0 . 5 0 . 5 0 . 5 ; 0 1 0 ] ) ;
set ( f i g u r e H , ’PaperSize ’ , [ 6 8 ] , ’PaperType ’ , ’<custom >’ , ’...
Units ’ , ’inches ’ ) ;
p o s i t i o n= get ( f i g u r e H , ’Position ’ ) ;
p o s i t i o n ( 3 : 4 )= [ 6 8 ] ;
set ( f i g u r e H , ’Position ’ , p o s i t i o n ) ;
t G r i d= 6 + 0 . 2 5 ∗ ( ( 1 : 8 ) −1) ;
l a b e l s= { ’a’ , ’b’ , ’c’ , ’d’ , ’e’ , ’f’ , ’g’ , ’h’ } ;
y P o s i t i o n s= [ 0 . 7 8 0 . 5 4 0 . 3 0 . 0 6 0 . 7 8 0 . 5 4 0 . 3 0 . 0 6 ] ;
f o r k= 1 : 8
c o l o r s= o n e s (K, 1 ) ; % D e f a u l t : 1 <=> Immature c o l o r i n d e x
c o l o r s (T <= t G r i d ( k ) )= 2 ; % 2 <=> Maturing c o l o r i n d e x
147
c o l o r s (T + 2∗ tau <= t G r i d ( k ) )= 3 ; % 3 <=> Mature c o l o r ...
index
i f k <= 4
axisH= subplot ( 4 , 2 , 2 ∗ k−1) ;
p o s i t i o n= get ( axisH , ’Position ’ ) ;
p o s i t i o n ( 1 )= 0 . 1 2 5 ;
else
axisH= subplot ( 4 , 2 , 2 ∗ k−8) ;
p o s i t i o n= get ( axisH , ’Position ’ ) ;
p o s i t i o n ( 1 )= 0 . 6 2 5 ;
end
p o s i t i o n ( 2 )= y P o s i t i o n s ( k ) ;
p o s i t i o n ( 3 )= 1 / 3 ;
p o s i t i o n ( 4 )= 0 . 1 5 ;
barH= bar ( axisH , ( ( 0 : K−1) + 0 . 5 ) / c e l l s P e r S o m i t e , 0 . 5 ∗ ( 1 + ...
sin ( 2 ∗ pi ∗ d e v a l ( s o l , t G r i d ( k ) ) ) ) ) ;
set ( axisH , ’Position ’ , p o s i t i o n ) ;
c h i l d r e n H= get ( barH , ’children ’ ) ;
set ( c h i l d r e n H , ’facevertexcdata ’ , c o l o r s ) ; % S e t c o l o r o f ...
bars
set ( c h i l d r e n H , ’CDataMapping ’ , ’direct ’ ) ;
xlim ( axisH , [ 0 K/ c e l l s P e r S o m i t e ] ) ;
i f rem( k , 4 )==0
xlabel ( axisH , ’x ( somite #) ’ , ’FontSize ’ , 1 2 , ’...
FontWeight ’ , ’demi ’ ) ;
148
end
ylim ( axisH , [ −0.1 1 . 1 ] ) ;
i f k<=4
ylabel ( axisH , ’y’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
end
t i t l e ( axisH , [ ’y=p_k(’ num2str ( t G r i d ( k ) , ’%1.2f’ ) ’)’ ] , ’...
FontSize ’ , 1 4 , ’FontWeight ’ , ’demi ’ ) ;
set ( axisH , ’XMinorTIck ’ , ’on ’ , ’YMinorTIck ’ , ’on ’ , ’...
FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
text ( −2.75 , 1 . 3 7 5 , [ ’(’ l a b e l s {k} ’)’ ] , ’FontSize ’ , 1 6 , ’...
FontWeight ’ , ’demi ’ ) ;
%{
f i g u r e ( 6 ) ; c l f ( gcf ) ;
colormap ( [ 1 0 0 ; 0 . 5 0 . 5 0 . 5 ; 0 1 0 ] ) ;
set ( gcf , ’PaperSize ’ , [ 6 3 ] , ’PaperType ’ , ’<custom >’ , ’...
Units ’ , ’inches ’ ) ;
p o s i t i o n= get ( gcf , ’Position ’ ) ;
p o s i t i o n ( 3 : 4 )= [ 6 3 ] ;
set ( gcf , ’Position ’ , p o s i t i o n ) ;
axisH= subplot ( 1 , 1 , 1 ) ;
barH= bar ( axisH , ( ( 0 : K−1) + 0 . 5 ) / c e l l s P e r S o m i t e , ...
0 . 5 ∗ ( 1 + sin ( 2 ∗ pi ∗ d e v a l ( s o l , t G r i d ( k ) ) ) ) ) ;
c h i l d r e n H= get ( barH , ’children ’ ) ;
set ( c h i l d r e n H , ’facevertexcdata ’ , c o l o r s ) ; % S e t c o l o r ...
of bars
set ( c h i l d r e n H , ’CDataMapping ’ , ’direct ’ ) ;
xlim ( axisH , [ 0 K/ c e l l s P e r S o m i t e ] ) ;
149
xlabel ( axisH , ’x ( somite #) ’ , ’FontSize ’ , 1 2 , ’...
FontWeight ’ , ’demi ’ ) ;
ylim ( axisH , [ −0.1 1 . 1 ] ) ;
ylabel ( axisH , ’y’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ...
;
t i t l e ( axisH , [ ’y=p_k(’ num2str ( t G r i d ( k ) , ’%1.2f’ ) ’)’...
] , ’FontSize ’ , 1 4 , ’FontWeight ’ , ’demi ’ ) ;
set ( axisH , ’XMinorTIck ’ , ’on ’ , ’YMinorTIck ’ , ’on ’ , ’...
FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
pause ;
%}
end
f i g u r e H= f i g u r e ( 6 ) ; c l f ( f i g u r e H ) ;
colormap ( [ 1 0 0 ; 0 . 5 0 . 5 0 . 5 ; 0 1 0 ] ) ;
set ( f i g u r e H , ’PaperSize ’ , [ 6 3 ] , ’PaperType ’ , ’<custom >’ , ’...
Units ’ , ’inches ’ ) ;
p o s i t i o n= get ( f i g u r e H , ’Position ’ ) ;
p o s i t i o n ( 3 : 4 )= [ 6 3 ] ;
set ( f i g u r e H , ’Position ’ , p o s i t i o n ) ;
c o l o r s= o n e s (K, 1 ) ; % D e f a u l t : 1 <=> Immature c o l o r i n d e x
c o l o r s (T <= s o l . x ( end ) )= 2 ; % 2 <=> Maturing c o l o r i n d e x
c o l o r s (T + 2∗ tau <= s o l . x ( end ) )= 3 ; % 3 <=> Mature c o l o r ...
index
axisH= subplot ( 1 , 1 , 1 ) ;
150
barH= bar ( axisH , ( ( 0 : K−1) + 0 . 5 ) / c e l l s P e r S o m i t e , 0 . 5 ∗ ( 1 + ...
sin ( 2 ∗ pi ∗ s o l . y ( : , end ) ) ) ) ;
c h i l d r e n H= get ( barH , ’children ’ ) ;
set ( c h i l d r e n H , ’facevertexcdata ’ , c o l o r s ) ; % S e t c o l o r o f ...
bars
set ( c h i l d r e n H , ’CDataMapping ’ , ’direct ’ ) ;
xlim ( axisH , [ 0 K/ c e l l s P e r S o m i t e ] ) ;
xlabel ( axisH , ’x ( somite #) ’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ...
’demi ’ ) ;
ylim ( axisH , [ −0.1 1 . 1 ] ) ;
ylabel ( axisH , ’y’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
t i t l e ( axisH , [ ’y=p_k(’ num2str ( s o l . x ( end ) , ’%1.2f’ ) ’)’ ] , ...
’FontSize ’ , 1 4 , ’FontWeight ’ , ’demi ’ ) ;
set ( axisH , ’XMinorTIck ’ , ’on ’ , ’YMinorTIck ’ , ’on ’ , ’...
FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
f i g u r e H= f i g u r e ( 7 ) ; c l f ( f i g u r e H ) ;
set ( f i g u r e H , ’PaperSize ’ , [ 6 3 ] , ’PaperType ’ , ’<custom >’ , ’...
Units ’ , ’inches ’ ) ;
p o s i t i o n= get ( f i g u r e H , ’Position ’ ) ;
p o s i t i o n ( 3 : 4 )= [ 6 3 ] ;
set ( f i g u r e H , ’Position ’ , p o s i t i o n ) ;
axisH= subplot ( 1 , 1 , 1 ) ;
xGrid= ( −60:0) / 6 ;
s e r i e s H= plot ( axisH , xGrid , log ( exp(−xGrid ) + 2∗exp(−xGrid...
/ 2 ) + 1 ) , ’.-’ , ’LineWidth ’ , 0 . 5 ) ;
151
xlim ( axisH , [ −10 0 ] ) ;
xlabel ( axisH , ’x ( somite #) ’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ...
’demi ’ ) ;
ylim ( axisH , [ 0 1 2 ] ) ;
ylabel ( axisH , ’y’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
t i t l e ( axisH , ’y=\ phi_\ infty (x), Lewis Phase Oscillator ’ , ...
’FontSize ’ , 1 4 , ’FontWeight ’ , ’demi ’ ) ;
set ( axisH , ’XMinorTIck ’ , ’on ’ , ’YMinorTIck ’ , ’on ’ , ’...
FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
f i g u r e H= f i g u r e ( 8 ) ; c l f ( f i g u r e H ) ;
colormap ( [ 0 1 0 ] ) ;
set ( f i g u r e H , ’PaperSize ’ , [ 6 3 ] , ’PaperType ’ , ’<custom >’ , ’...
Units ’ , ’inches ’ ) ;
p o s i t i o n= get ( f i g u r e H , ’Position ’ ) ;
p o s i t i o n ( 3 : 4 )= [ 6 3 ] ;
set ( f i g u r e H , ’Position ’ , p o s i t i o n ) ;
axisH= subplot ( 1 , 1 , 1 ) ;
xGrid= ( −60:1) / 6 ;
barH= bar ( axisH , xGrid , 1 . / ( 1 + exp ( 1 0 ∗ cos ( 2 ∗ pi ∗ log ( exp(−...
xGrid ) + 2∗exp(−xGrid / 2 ) + 1 ) ) ) ) ) ;
xlim ( axisH , [ −10 0 ] ) ;
xlabel ( axisH , ’x ( somite #) ’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ...
’demi ’ ) ;
ylim ( axisH , [ −0.1 1 . 1 ] ) ;
ylabel ( axisH , ’y’ , ’FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
152
t i t l e ( axisH , ’y=w(z(\ phi_\ infty (x))), Lewis Phase ...
Oscillator ’ , ’FontSize ’ , 1 4 , ’FontWeight ’ , ’demi ’ ) ;
set ( axisH , ’XMinorTIck ’ , ’on ’ , ’YMinorTIck ’ , ’on ’ , ’...
FontSize ’ , 1 2 , ’FontWeight ’ , ’demi ’ ) ;
% Nested f u n c t i o n s
function z= s ( t , tau )
% S u s c e p t i b i l i t y / maturity function with half −l i f e tau .
z= zeros ( s i z e ( t ) ) ;
tMask= ( 0 < t ) & ( t <= tau ) ;
z ( tMask )= 0 . 5 ∗ exp ( 2 ∗ ( t ( tMask )−tau ) . / t ( tMask ) ) ;
tMask= ( tau < t ) & ( t < 2∗ tau ) ;
z ( tMask )= 1 − 0 . 5 ∗ exp ( 2 ∗ ( t ( tMask )−tau ) . / ( t ( tMask ) −2∗tau ) ) ;
tMask= 2∗ tau <= t ;
z ( tMask )= 1 ;
end
function p h i D e r i v= r h s ( t , p h i )
% RHS o f system o f ODE’ s .
p h i D e r i v= 1 − s ( t − T, tau ) . ∗ ( sin ( 2 ∗ pi ∗ p h i ) ) . ˆ 2 ;
end
end
153
Posterior Clock Wave (PCW) Model Code
Simulation Related Code
Params.m: This is the class definition for the parameters object that holds
the system parameters and other configuration information for model simulation.
Objects of this class can provide either deterministic parameter values or random
values drawn from a specified normal or uniform distribution. The code is lengthy
and uninteresting, but it ensures accurate and repeatable parameter specification.
c l a s s d e f Params
% C l a s s d e f i n i t i o n f o r managing p a r a m e t e r s and s e t t i n g s f o r t h e
%
s o m i t o g e n e s i s .m model .
% The parameter mean i s t h e nominal v a l u e o f t h e parameter . I f
%
n o i s y p a r a m e t e r s a r e t u r n e d o f f , t h e n t h e nominal ( s c a l a r )
%
value i s returned , o t h e r w i s e the parameters returned are
%
random v e c t o r s drawn from a s p e c i f i e d d i s t r i b u t i o n , one
%
parameter f o r each c e l l . The same d e f a u l t n o i s e s e e d i s used
%
u n l e s s a d i f f e r e n t s e e d i s s e l e c t e d . This a l l o w s r e p e a t a b l e
%
sampling .
p r o p e r t i e s ( Constant= t r u e )
mRNAProductionRate= 3 3 ;
p e r i o d T o l= 1 e −2;
end
p r o p e r t i e s % D e f a u l t s o n l y used f o r s i m p l e r C o n s t r u c t o r l o g i c
% Simulation / Solver parameters
154
n o i s e B o o l= f a l s e ; noiseType ; n o i s e S e e d ;
nois e Propo rtio nOfMea n ; t i m e I n t e r v a l ; s e t t l i n g T i m e ; h i s t o r y ;
ddeOptions ; fastModeBool ;
% Model p a r a m e t e r s
% C e l l /PSM p a r a m e t e r s
c e l l s P e r S o m i t e ; somitesInPSM ; t a i l b u d P e r i o d ;
c e l l A r r a y S i z e= [ 2 1 ] ; numGenes= 2 ;
% C e l l background parameters
rhoMean ; kH13H13Mean ; kH7H7Mean ; kH7H13Mean ;
% HER7 Parameters
numBindingSites ; tau h7Mean= 4 . 2 5 ; tau H7Mean= 1 . 7 ; aH7Mean ;
bH7Mean ; bH7H7Mean ; bH7H13Mean ; dH7Mean ; rH7H7Mean ;
wH7H7 H7H7Mean ;
% DELTAC/NOTCH Parameters
c o u p l i n g B o o l ; tau dMean= 1 2 . 2 ; tau DMean= 2 0 ; aDMean ; bDMean...
;
dDMean ; rNMean ; wNMean ; wN H7H7Mean ; wN H7H7 H7H7Mean ;
% Her13 . 2 / g r a d i e n t p a r a m e t e r s
g r a d i e n t F u n c ; H13TotalMaxMean ; H13TotalMinMean= 0 ;
H13TotalHalfLifeMean ;
end
p r o p e r t i e s ( Dependent= t r u e , S e t A c c e s s= p r i v a t e )
timeLags ; somiteGrowthRate ; c e l l B i r t h R a t e ; b i r t h T i m e s ;
numCells ; e q u a t i o n s P e r C e l l ; numEquations ; gH7Mean ; gDMean ;
couplingMatrix ;
% Indexing vectors
155
H7TotalIndices ; h7Indices ; DIndices ; dIndices ;
H7TotalDelayedh7Indices ; h7DelayedH7Indices ;
DDelayedh7Indices ; H7TotalDelayeddIndices ; dDelayedDIndices ;
% Computed p a r a m e t e r s
rho ; kH13H13 ; kH7H7 ; kH7H13 ; t a u h 7 ; tau H7 ; aH7 ; bH7 ; bH7H7...
;
bH7H13 ; gH7 ; rH7H7 ; wH7H7 H7H7 ; dH7 ; t a u d ; tau D ; aD ; bD ;
gD ; rN ; wN; wN H7H7 ; wN H7H7 H7H7 ; dD ; H13TotalMax ;
H13TotalMin ; H 1 3 T o t a l H a l f L i f e ;
end
p r o p e r t i e s ( SetAccess = private )
% Random number s t r e a m s f o r r e p r o d u c i b i l i t y p u r p o s e s
rhoStream ; kH13H13Stream ; kH7H7Stream ; kH7H13Stream ;
tau h7Stream ; tau H7Stream ; aH7Stream ; bH7Stream ;
bH7H7Stream ; bH7H13Stream ; gH7Stream ; rH7H7Stream ;
wH7H7 H7H7Stream ; dH7Stream ; tau dStream ; tau DStream ;
aDStream ; bDStream ; gDStream ; rNStream ; wNStream ;
wN H7H7Stream ; wN H7H7 H7H7Stream ; dDStream ;
H13TotalMaxStream ; H13TotalMinStream ; H 1 3 T o t a l H a l f L i f e S t r e a m ...
;
end
methods % CONSTRUCTOR
function params= Params ( n o i s e B o o l )
i f ( nargin > 0 )
params . n o i s e B o o l= n o i s e B o o l ;
156
else
params . n o i s e B o o l= params . n o i s e B o o l ;
end
params . noiseType= ’normal ’ ;
params . n o i s e S e e d= 0 ;
params . nois e Propo rtionOfMea n= 0 . 0 1 ;
params= i n i t S t r e a m s ( params ) ;
params . t i m e I n t e r v a l= [ 0 5 0 0 ] ;
params . s e t t l i n g T i m e= 5 0 0 ;
params . h i s t o r y= 0 ; % Becomes c o n s t a n t z e r o v e c t o r
params . ddeOptions= d d e s e t ( ’AbsTol ’ , 1 e −15 , ’RelTol ’ , 1 e −4, ...
’MaxStep ’ , 5 , ’Events ’ , @ z e r o C r o s s i n g E v e n t ) ;
params . fastModeBool= f a l s e ;
params . c e l l s P e r S o m i t e= 5 ;
params . somitesInPSM= 7 ;
params . t a i l b u d P e r i o d= 3 0 ; % May be u p d a t e d a f t e r ...
s e t t l i n g T i m e t o a computed p e r i o d
params . c e l l A r r a y S i z e= params . c e l l A r r a y S i z e ; % [ ax l a t ]
params . numGenes= params . numGenes ; % 1 or 2 genes , HER7 w/...
p o s s i b l y DELTAC
params . rhoMean= 1 ; % rRNAP∗RNAP, u n i t l e s s
params . kH13H13Mean= 3 0 0 ; % c o p i e s
params . kH7H7Mean= 3 0 0 ; % c o p i e s
params . kH7H13Mean= 3 0 0 ; % c o p i e s
params . numBindingSites= 2 ; % 1 or 2 f o r Her7 dimer
params . tau h7Mean= params . tau h7Mean ; % min
params . tau H7Mean= params . tau H7Mean ; % min
157
params . aH7Mean= 4 . 5 ; % Her7 p r o t e i n c o p i e s /min
params . bH7Mean= 0 . 4 5 ; % minˆ( −1)
params . bH7H7Mean= 0 ; % minˆ( −1)
params . bH7H13Mean= 0 ; % minˆ( −1)
params . rH7H7Mean= 0 . 1 ; % c o p i e s ˆ( −1)
params . wH7H7 H7H7Mean= 2 5 ; % u n i t l e s s
params . dH7Mean= 1 / ( 0 . 6 ∗ 8 . 1 ) ; % minˆ( −1)
params . c o u p l i n g B o o l= t r u e ;
params . tau dMean= params . tau dMean ; % min
params . tau DMean= params . tau DMean ; % min
params . aDMean= 4 . 5 ; % DeltaC p r o t e i n c o p i e s /min
params . bDMean= 0 . 2 3 ; % minˆ( −1)
params . rNMean= params . rH7H7Mean / 1 0 ; % c o p i e s ˆ( −1)
params . wNMean= 2 5 ; % u n i t l e s s
params . wN H7H7Mean= 1 ; % u n i t l e s s
params . wN H7H7 H7H7Mean= 1 ; % u n i t l e s s
params . dDMean= 1 / ( 0 . 6 ∗ 6 . 1 ) ; % minˆ( −1)
params . g r a d i e n t F u n c= ’sigmoidalGradient ’ ;
params . H13TotalMaxMean= 0 ; % Copies ( i n n u c l e u s )
params . H13TotalMinMean= params . H13TotalMinMean ; % Copies (...
in nucleus )
params . H13TotalHalfLifeMean= 3 0 ; % min
end % Params C o n s t r u c t o r
end % CONSTRUCTOR
methods % P u b l i c SET methods
function params= set . n o i s e B o o l ( params , n o i s e B o o l )
158
i f ˜ i s l o g i c a l ( noiseBool )
disp ( ’noiseBool not set , true/ false noiseBool required...
.’ )
else
params . n o i s e B o o l= n o i s e B o o l ;
end
end % n o i s e B o o l s e t f u n c t i o n
function params= set . noiseType ( params , noiseType )
i f ˜ i s c h a r ( noiseType ) | | ˜ ( s t r c m p i ( noiseType , ’uniform ’ ) ...
| | s t r c m p i ( noiseType , ’normal ’ ) )
disp ( ’noiseType not set , uniform OR normal noiseType ...
required .’ )
else
params . noiseType= noiseType ;
end
end % noiseType s e t f u n c t i o n
function params= set . n o i s e S e e d ( params , n o i s e S e e d )
i f ˜ i s s c a l a r ( noiseSeed ) | | ( noiseSeed <
0)
disp ( ’noiseSeed not set , 0 <= noiseSeed required .’ )
else
params . n o i s e S e e d= n o i s e S e e d ;
params= i n i t S t r e a m s ( params ) ;
end
end % n o i s e S e e d s e t f u n c t i o n
function params= set . nois e Propo rtio nOfMea n ( params , ...
nois e Propo rtio nOfMea n )
159
i f ˜ i s s c a l a r ( noise ProportionOfMea n ) | | ( ...
nois e Propo rtio nOfMea n <
0)
disp ( ’noiseProportionOfMean not set , 0 <= ...
noiseProportionOfMean required .’ )
else
params . nois e Propo rtio nOfMea n= nois e Propo rtio nOfMea n ;
end
end % noiseProportionOfMean s e t f u n c t i o n
function params= set . t i m e I n t e r v a l ( params , t i m e I n t e r v a l )
i f ˜ i s v e c t o r ( t i m e I n t e r v a l ) | | any ( s i z e ( t i m e I n t e r v a l ) ˜= [ 1 ...
2]) | | ( timeInterval (1) > timeInterval (2) )
disp ( ’timeInterval not set , timeInterval (1) <= ...
timeInterval (2) required .’ ) ;
else
params . t i m e I n t e r v a l ( 1 , 1 )= t i m e I n t e r v a l ( 1 ) ;
params . t i m e I n t e r v a l ( 1 , 2 )= t i m e I n t e r v a l ( 2 ) ;
end
end % t i m e I n t e r v a l s e t f u n c t i o n
function params= set . s e t t l i n g T i m e ( params , s e t t l i n g T i m e )
i f ˜ i s s c a l a r ( settlingTime ) | | ( settlingTime < 0)
disp ( ’settlingTime not set , 0 <= settlingTime required ....
’) ;
else
params . s e t t l i n g T i m e= s e t t l i n g T i m e ;
end
end % s e t t l i n g T i m e s e t f u n c t i o n
function params= set . h i s t o r y ( params , h i s t o r y )
160
if isstruct ( history )
i f i s f i e l d ( h i s t o r y , ’solver ’ ) && ( strcmp ( h i s t o r y . s o l v e r , ...
’dde23 ’ ) | | strcmp ( h i s t o r y . s o l v e r , ’ddesd ’ ) )
params . h i s t o r y= h i s t o r y ;
else
disp ( ’history not set , unrecognized previous solution...
.’ ) ;
end
e l s e i f i s a ( h i s t o r y , ’function_handle ’ )
params . h i s t o r y= h i s t o r y ;
elseif isscalar ( history )
if history < 0
disp ( ’history not set , 0 <= history required .’ ) ;
else
params . h i s t o r y= h i s t o r y ∗ o n e s ( 1 , params . numEquations ) ;
end
elseif isvector ( history )
i f length ( h i s t o r y ) ˜= params . numEquations
disp ( ’history not set , length ( history ) == ...
numEquations required .’ ) ;
else
params . h i s t o r y= h i s t o r y ;
end
else
disp ( ’history not set , unrecognized history type.’ ) ;
end
end % h i s t o r y s e t f u n c t i o n
161
function params= set . ddeOptions ( params , ddeOptions )
i f ˜ i s s t r u c t ( ddeOptions )
disp ( ’ddeOptions not set , use ddeset () to get the ...
appropriate struct .’ ) ;
else
params . ddeOptions= ddeOptions ;
end
end % d d e O p t i o n s s e t f u n c t i o n
function params= set . fastModeBool ( params , fastModeBool )
i f ˜ i s l o g i c a l ( fastModeBool )
disp ( ’fastModeBool not set , true/ false fastModeBool ...
required .’ ) ;
else
params . fastModeBool= fastModeBool ;
end
end % fastModeBool s e t f u n c t i o n
function params= set . c e l l s P e r S o m i t e ( params , c e l l s P e r S o m i t e )
i f ˜ i s s c a l a r ( c e l l s P e r S o m i t e ) | | ( c e l l s P e r S o m i t e <= 0 )
disp ( ’cellsPerSomite not set , 0 < cellsPerSomite ...
required .’ ) ;
else
params . c e l l s P e r S o m i t e= c e l l s P e r S o m i t e ;
end
end % c e l l s P e r S o m i t e s e t f u n c t i o n
function params= set . somitesInPSM ( params , somitesInPSM )
i f ˜ i s s c a l a r ( somitesInPSM ) | | ( somitesInPSM <= 0 )
162
disp ( ’somitesInPSM not set , 0 < somitesInPSM required .’ ) ...
;
else
params . somitesInPSM= somitesInPSM ;
end
end % somitesInPSM s e t f u n c t i o n
function params= set . t a i l b u d P e r i o d ( params , t a i l b u d P e r i o d )
i f ˜ i s s c a l a r ( t a i l b u d P e r i o d ) | | ( t a i l b u d P e r i o d <= 0 )
disp ( ’tailbudPeriod not set , 0 < tailbudPeriod required...
.’ ) ;
else
params . t a i l b u d P e r i o d= t a i l b u d P e r i o d ;
end
end % t a i l b u d P e r i o d s e t f u n c t i o n
function params= set . c e l l A r r a y S i z e ( params , c e l l A r r a y S i z e )
i f ˜ i s v e c t o r ( c e l l A r r a y S i z e ) | | any ( s i z e ( c e l l A r r a y S i z e ) ˜= ...
[ 1 2 ] ) | | (rem( c e l l A r r a y S i z e ( 1 ) , 1 ) ˜= 0 ) | | ( c e l l A r r a y S i z e ( 1 ) < ...
0 ) | | (rem( c e l l A r r a y S i z e ( 2 ) , 1 ) ˜= 0 ) | | ( c e l l A r r a y S i z e ( 2 ) < 0 )
disp ( ’cellArraySize not set , cellArraySize = [M N], M ...
and N positive integers , required .’ ) ;
e l s e i f any ( params . c e l l A r r a y S i z e ˜= c e l l A r r a y S i z e )
i f params . numCells > 1 % E x i s t i n g v a l u e
params . c e l l A r r a y S i z e= c e l l A r r a y S i z e ;
i f params . numCells == 1 % New v a l u e
params . c o u p l i n g B o o l= f a l s e ;
e l s e % params . numCells > 1
params . c o u p l i n g B o o l= params . c o u p l i n g B o o l ;
163
end
e l s e % E x i s t i n g params . numCells == 1
params . c e l l A r r a y S i z e= c e l l A r r a y S i z e ;
i f params . numCells > 1
params . c o u p l i n g B o o l= t r u e ;
end
end
i f i s v e c t o r ( params . h i s t o r y )
disp ( ’Notice : history reset to default constant zero ...
vector .’ )
params . h i s t o r y= 0 ;
else
disp ( ’Notice : Existing history may be invalid .’ ) ;
end
end
end % c e l l A r r a y S i z e s e t f u n c t i o n
function params= set . numGenes ( params , numGenes )
i f ˜ i s s c a l a r ( numGenes ) | | ( ( numGenes ˜= 1 ) && ( numGenes ˜=...
2) )
disp ( ’numGenes not set , numGenes = 1 or 2 required .’ ) ;
e l s e i f params . numGenes ˜= numGenes
params . numGenes= numGenes ;
i f i s v e c t o r ( params . h i s t o r y )
disp ( ’Notice : history reset to default constant zero ...
vector .’ )
params . h i s t o r y= 0 ;
else
164
disp ( ’Notice : Existing history may be invalid .’ )
end
i f params . numGenes == 1
params . c o u p l i n g B o o l= f a l s e ; % Turn o f f c o u p l i n g
e l s e i f params . numGenes == 2
i f params . numCells > 1
i f ˜ params . c o u p l i n g B o o l
disp ( ’Notice : couplingBool reset to default true....
’) ;
params . c o u p l i n g B o o l= t r u e ; % C o u p l i n g on by ...
default
end
e l s e % params . numCells == 1
params . c o u p l i n g B o o l= f a l s e ;
end
end
end
end % numGenes s e t f u n c t i o n
function params= set . rhoMean ( params , rhoMean )
i f ˜ i s s c a l a r ( rhoMean ) | | ( rhoMean < 0 )
disp ( ’rhoMean not set , 0 <= rhoMean required .’ ) ;
else
params . rhoMean= rhoMean ;
end
end % rhoMean s e t f u n c t i o n
function params= set . kH13H13Mean ( params , kH13H13Mean )
i f ˜ i s s c a l a r ( kH13H13Mean ) | | ( kH13H13Mean <= 0 )
165
disp ( ’kH13H13Mean not set , 0 < kH13H13Mean required .’ ) ;
else
params . kH13H13Mean= kH13H13Mean ;
end
end % kH13H13Mean s e t f u n c t i o n
function params= set . kH7H7Mean ( params , kH7H7Mean )
i f ˜ i s s c a l a r ( kH7H7Mean ) | | ( kH7H7Mean <= 0 )
disp ( ’kH7H7Mean not set , 0 < kH7H7Mean required .’ ) ;
else
params . kH7H7Mean= kH7H7Mean ;
end
end % kH7H7Mean s e t f u n c t i o n
function params= set . kH7H13Mean ( params , kH7H13Mean )
i f ˜ i s s c a l a r ( kH7H13Mean ) | | ( kH7H13Mean <= 0 )
disp ( ’kH7H13Mean not set , 0 < kH7H13Mean required .’ ) ;
else
params . kH7H13Mean= kH7H13Mean ;
end
end % kH7H13Mean s e t f u n c t i o n
function params= set . numBindingSites ( params , numBindingSites...
)
i f ˜ i s s c a l a r ( numBindingSites ) | | ( ( numBindingSites ˜= 1 ) ...
&& ( numBindingSites ˜= 2 ) )
disp ( ’numBindingSites not set , numBindingSites = 1 or 2 ...
required .’ ) ;
else
params . numBindingSites= numBindingSites ;
166
end
end % n u m B in d i n gS i t e s s e t f u n c t i o n
function params= set . tau h7Mean ( params , tau h7Mean )
i f ˜ i s s c a l a r ( tau h7Mean ) | | ( tau h7Mean <= 0 )
disp ( ’tau_h7Mean not set , 0 < tau_h7Mean required .’ ) ;
else
params . tau h7Mean= tau h7Mean ;
end
end % tau h7Mean s e t f u n c t i o n
function params= set . tau H7Mean ( params , tau H7Mean )
i f ˜ i s s c a l a r ( tau H7Mean ) | | ( tau H7Mean <= 0 )
disp ( ’tau_H7Mean not set , 0 < tau_H7Mean required .’ ) ;
else
params . tau H7Mean= tau H7Mean ;
end
end % tau H7Mean s e t f u n c t i o n
function params= set . aH7Mean ( params , aH7Mean )
i f ˜ i s s c a l a r ( aH7Mean ) | | ( aH7Mean < 0 )
disp ( ’aH7Mean not set , 0 <= aH7Mean required .’ ) ;
else
params . aH7Mean= aH7Mean ;
end
end % aH7Mean s e t f u n c t i o n
function params= set . bH7Mean ( params , bH7Mean )
i f ˜ i s s c a l a r ( bH7Mean ) | | ( bH7Mean < 0 )
disp ( ’bH7Mean not set , 0 <= bH7Mean required .’ ) ;
else
167
params . bH7Mean= bH7Mean ;
end
end % bH7Mean s e t f u n c t i o n
function params= set . bH7H7Mean( params , bH7H7Mean)
i f ˜ i s s c a l a r (bH7H7Mean) | | (bH7H7Mean < 0 )
disp ( ’bH7H7Mean not set , 0 <= bH7H7Mean required .’ ) ;
else
params . bH7H7Mean= bH7H7Mean ;
end
end % bH7H7Mean s e t f u n c t i o n
function params= set . bH7H13Mean ( params , bH7H13Mean )
i f ˜ i s s c a l a r ( bH7H13Mean ) | | ( bH7H13Mean < 0 )
disp ( ’bH7H13Mean not set , 0 <= bH7H13Mean required .’ ) ;
else
params . bH7H13Mean= bH7H13Mean ;
end
end % bH7H13Mean s e t f u n c t i o n
function params= set . rH7H7Mean ( params , rH7H7Mean )
i f ˜ i s s c a l a r ( rH7H7Mean ) | | ( rH7H7Mean < 0 )
disp ( ’rH7H7Mean not set , 0 <= rH7H7Mean required .’ ) ;
else
params . rH7H7Mean= rH7H7Mean ;
end
end % rH7H7Mean s e t f u n c t i o n
function params= set . wH7H7 H7H7Mean( params , wH7H7 H7H7Mean)
i f ˜ i s s c a l a r (wH7H7 H7H7Mean) | | (wH7H7 H7H7Mean < 0 )
168
disp ( ’wH7H7_H7H7Mean not set , 0 <= wH7H7_H7H7Mean ...
required .’ ) ;
else
params . wH7H7 H7H7Mean= wH7H7 H7H7Mean ;
end
end % wH7H7 H7H7Mean s e t f u n c t i o n
function params= set . dH7Mean ( params , dH7Mean )
i f ˜ i s s c a l a r ( dH7Mean ) | | ( dH7Mean < 0 )
disp ( ’dH7Mean not set , 0 <= dH7Mean required .’ ) ;
else
params . dH7Mean= dH7Mean ;
end
end % dH7Mean s e t f u n c t i o n
function params= set . c o u p l i n g B o o l ( params , c o u p l i n g B o o l )
i f ˜ i s l o g i c a l ( couplingBool )
disp ( ’couplingBool not set , true/ false couplingBool ...
required .’ ) ;
e l s e i f c o u p l i n g B o o l && ( ( params . numGenes ˜= 2 ) | | ( params . ...
numCells == 1 ) )
disp ( ’couplingBool not set , numGenes == 2 and numCells ...
>= 2 required .’ ) ;
else
params . c o u p l i n g B o o l= c o u p l i n g B o o l ;
end
end % c o u p l i n g B o o l s e t f u n c t i o n
function params= set . tau dMean ( params , tau dMean )
i f ˜ i s s c a l a r ( tau dMean ) | | ( tau dMean <= 0 )
169
disp ( ’tau_dMean not set , 0 < tau_dMean required .’ ) ;
else
params . tau dMean= tau dMean ;
end
end % tau dMean s e t f u n c t i o n
function params= set . tau DMean ( params , tau DMean )
i f ˜ i s s c a l a r ( tau DMean ) | | ( tau DMean <= 0 )
disp ( ’tau_DMean not set , 0 < tau_DMean required .’ ) ;
else
params . tau DMean= tau DMean ;
end
end % tau DMean s e t f u n c t i o n
function params= set . aDMean( params , aDMean)
i f ˜ i s s c a l a r (aDMean) | | (aDMean < 0 )
disp ( ’aDMean not set , 0 <= aDMean required .’ ) ;
else
params . aDMean= aDMean ;
end
end % aDMean s e t f u n c t i o n
function params= set . bDMean( params , bDMean)
i f ˜ i s s c a l a r (bDMean) | | (bDMean < 0 )
disp ( ’bDMean not set , 0 <= bDMean required .’ ) ;
else
params . bDMean= bDMean ;
end
end % bDMean s e t f u n c t i o n
function params= set . rNMean ( params , rNMean )
170
i f ˜ i s s c a l a r ( rNMean ) | | ( rNMean < 0 )
disp ( ’rNMean not set , 0 <= rNMean required .’ ) ;
else
params . rNMean= rNMean ;
end
end % rNMean s e t f u n c t i o n
function params= set . wNMean( params , wNMean)
i f ˜ i s s c a l a r (wNMean) | | (wNMean < 0 )
disp ( ’wNMean not set , 0 <= wNMean required .’ ) ;
else
params . wNMean= wNMean ;
end
end % wNMean s e t f u n c t i o n
function params= set . wN H7H7Mean( params , wN H7H7Mean)
i f ˜ i s s c a l a r (wN H7H7Mean) | | (wN H7H7Mean < 0 )
disp ( ’wN_H7H7Mean not set , 0 <= wN_H7H7Mean required .’ ) ;
else
params . wN H7H7Mean= wN H7H7Mean ;
end
end % wN H7H7Mean s e t f u n c t i o n
function params= set . wN H7H7 H7H7Mean ( params , ...
wN H7H7 H7H7Mean )
i f ˜ i s s c a l a r ( wN H7H7 H7H7Mean ) | | ( wN H7H7 H7H7Mean < 0 )
disp ( ’wN_H7H7_H7H7Mean not set , 0 <= wN_H7H7_H7H7Mean ...
required .’ ) ;
else
params . wN H7H7 H7H7Mean= wN H7H7 H7H7Mean ;
171
end
end % wN H7H7 H7H7Mean s e t f u n c t i o n
function params= set . dDMean( params , dDMean)
i f ˜ i s s c a l a r (dDMean) | | (dDMean < 0 )
disp ( ’dDMean not set , 0 <= dDMean required .’ ) ;
else
params . dDMean= dDMean ;
end
end % dDMean s e t f u n c t i o n
function params= set . H13TotalMaxMean ( params , H13TotalMaxMean...
)
i f ˜ i s s c a l a r ( H13TotalMaxMean ) | | ( H13TotalMaxMean < 0 ) | | ...
( H13TotalMaxMean < params . H13TotalMinMean )
disp ( ’H13TotalMaxMean not set , 0 <= H13TotalMinMean <= ...
H13TotalMaxMean required .’ ) ;
else
params . H13TotalMaxMean= H13TotalMaxMean ;
end
end % H13TotalMaxMean s e t f u n c t i o n
function params= set . H13TotalMinMean ( params , H13TotalMinMean...
)
i f ˜ i s s c a l a r ( H13TotalMinMean ) | | ( H13TotalMinMean < 0 ) | | ...
( params . H13TotalMaxMean < H13TotalMinMean )
disp ( ’H13TotalMinMean not set , 0 <= H13TotalMinMean <= ...
H13TotalMaxMean required .’ ) ;
else
params . H13TotalMinMean= H13TotalMinMean ;
172
end
end % H13TotalMinMean s e t f u n c t i o n
function params= set . H13TotalHalfLifeMean ( params , ...
H13TotalHalfLifeMean )
i f ˜ i s s c a l a r ( H13TotalHalfLifeMean ) | | ( ...
H13TotalHalfLifeMean <= 0 )
disp ( ’H13TotalHalfLifeMean not set , 0 < ...
H13TotalHalfLifeMean required .’ ) ;
else
params . H13TotalHalfLifeMean= H13TotalHalfLifeMean ;
end
end % H13TotalHalfLifeMean s e t f u n c t i o n
function params= set . g r a d i e n t F u n c ( params , g r a d i e n t F u n c )
i f ˜ i s c h a r ( gradientFunc )
disp ( ’gradientFunc not set , a string is required .’ ) ;
else
params . g r a d i e n t F u n c= g r a d i e n t F u n c ;
end
end % g r a d i e n t F u n c s e t f u n c t i o n
end % P u b l i c SET methods
methods % P u b l i c GET methods
function timeLags= get . timeLags ( params )
i f params . numGenes == 1
timeLags= [ params . tau h7 ’ ; params . tau H7 ’ ] ;
timeLags= timeLags ( : ) ’ ; % Reorder by c e l l
e l s e i f params . numGenes == 2
173
timeLags= [ params . tau h7 ’ ; params . tau H7 ’ ; params . tau d ...
’ ; params . tau D ’ ] ;
timeLags= timeLags ( : ) ’ ; % Reorder by c e l l
end
end
function somiteGrowthRate= get . somiteGrowthRate ( params )
somiteGrowthRate= 1/ params . t a i l b u d P e r i o d ;
end % g e t somiteGrowthRate
function c e l l B i r t h R a t e= get . c e l l B i r t h R a t e ( params )
c e l l B i r t h R a t e= params . c e l l s P e r S o m i t e ∗ params . ...
somiteGrowthRate ;
end % g e t somiteGrowthRate
function b i r t h T i m e s= get . b i r t h T i m e s ( params )
b i r t h T i m e s= repmat ( params . t i m e I n t e r v a l ( 1 ) + ( 0 : params . ...
c e l l A r r a y S i z e ( 1 ) − 1 ) ’ / params . c e l l B i r t h R a t e , params . ...
c e l l A r r a y S i z e (2) , 1) ;
end
function numCells= get . numCells ( params )
numCells= prod ( params . c e l l A r r a y S i z e ) ;
end
function e q u a t i o n s P e r C e l l= get . e q u a t i o n s P e r C e l l ( params )
e q u a t i o n s P e r C e l l= 2∗ params . numGenes ;
end
function numEquations= get . numEquations ( params )
numEquations= params . numCells ∗ params . e q u a t i o n s P e r C e l l ;
end
function gH7Mean= get . gH7Mean ( params )
174
gH7Mean= params . mRNAProductionRate ∗ ( 1 + params . rhoMean ) /...
params . rhoMean ;
end
function gDMean= get . gDMean( params )
gDMean= params . mRNAProductionRate ∗ ( 1 + params . rhoMean ) /...
params . rhoMean ;
end
function c o u p l i n g M a t r i x= get . c o u p l i n g M a t r i x ( params )
i f params . c o u p l i n g B o o l
% A d d i t i v e n e a r e s t −n e i g h b o r ( i n c l u d e s c o r n e r i n g c e l l s )
c o u p l i n g M a t r i x= Params . ...
c r e a t e N e a r e s t N e i g h b o r C o u p l i n g M a t r i x ( params . c e l l A r r a y S i z e ) ;
% A l t e r n a t i v e : A d d i t i v e , a x i a l −only , n e a r e s t −n e i g h b o r
%
c o u p l i n g M a t r i x= Params . c r e a t e A x i a l C o u p l i n g M a t r i x ( params...
. cellArraySize ) ;
% Option : n e i g h b o r −a v e r a g e d c o u p l i n g
%
c o u p l i n g M a t r i x= c o u p l i n g M a t r i x . / repmat ( sum (...
c o u p l i n g M a t r i x , 2) , 1 , params . numCells ) ;
e l s e i f params . numGenes == 2
c o u p l i n g M a t r i x= zeros ( params . numCells ) ;
else
c o u p l i n g M a t r i x= [ ] ;
end
end
% Indexing vectors
function H 7 T o t a l I n d i c e s= get . H 7 T o t a l I n d i c e s ( params )
175
H 7 T o t a l I n d i c e s= 1 : params . e q u a t i o n s P e r C e l l : params . ...
numEquations ;
end
function h 7 I n d i c e s= get . h 7 I n d i c e s ( params )
h 7 I n d i c e s= 2 : params . e q u a t i o n s P e r C e l l : params . numEquations ;
end
function D I n d i c e s= get . D I n d i c e s ( params )
i f params . numGenes == 1
D I n d i c e s= [ ] ;
e l s e i f params . numGenes == 2
D I n d i c e s= 3 : params . e q u a t i o n s P e r C e l l : params . numEquations ;
end
end
function d I n d i c e s= get . d I n d i c e s ( params )
i f params . numGenes == 1
d I n d i c e s= [ ] ;
e l s e i f params . numGenes == 2
d I n d i c e s= 4 : params . e q u a t i o n s P e r C e l l : params . numEquations ;
end
end
function H 7 T o t a l D e l a y e d h 7 I n d i c e s= get . ...
H 7 T o t a l D e l a y e d h 7 I n d i c e s ( params )
i f params . n o i s e B o o l
i f params . numGenes == 1
H 7 T o t a l D e l a y e d h 7 I n d i c e s= diag ( repmat ( [ t r u e f a l s e ] , 1 , ...
params . numCells ) ) ;
e l s e i f params . numGenes == 2
176
H 7 T o t a l D e l a y e d h 7 I n d i c e s= diag ( repmat ( [ t r u e f a l s e f a l s e ...
f a l s e ] , 1 , params . numCells ) ) ;
end
e l s e % No n o i s e
i f params . numGenes == 1
H 7 T o t a l D e l a y e d h 7 I n d i c e s= repmat ( [ t r u e f a l s e ; f a l s e ...
f a l s e ] , params . numCells , 1 ) ;
e l s e i f params . numGenes == 2
H 7 T o t a l D e l a y e d h 7 I n d i c e s= repmat ( [ t r u e f a l s e f a l s e ...
f a l s e ; f a l s e f a l s e f a l s e f a l s e ; f a l s e f a l s e f a l s e f a l s e ; f a l s e ...
f a l s e f a l s e f a l s e ] , params . numCells , 1 ) ;
end
end
end
function h 7 D e l a y e d H 7 I n d i c e s= get . h 7 D e l a y e d H 7 I n d i c e s ( params )
i f params . n o i s e B o o l
i f params . numGenes == 1
h 7 D e l a y e d H 7 I n d i c e s= diag ( repmat ( [ f a l s e t r u e ] , 1 , ...
params . numCells ) ) ;
e l s e i f params . numGenes == 2
h 7 D e l a y e d H 7 I n d i c e s= diag ( repmat ( [ f a l s e t r u e f a l s e ...
f a l s e ] , 1 , params . numCells ) ) ;
end
e l s e % No n o i s e
i f params . numGenes == 1
h 7 D e l a y e d H 7 I n d i c e s= repmat ( [ f a l s e f a l s e ; f a l s e t r u e ] , ...
params . numCells , 1 ) ;
177
e l s e i f params . numGenes == 2
h 7 D e l a y e d H 7 I n d i c e s= repmat ( [ f a l s e f a l s e f a l s e f a l s e ; ...
f a l s e t r u e f a l s e f a l s e ; f a l s e f a l s e f a l s e f a l s e ; f a l s e f a l s e ...
f a l s e f a l s e ] , params . numCells , 1 ) ;
end
end
end
function D D e l a y e d h 7 I n d i c e s= get . D D e l a y e d h 7 I n d i c e s ( params )
i f params . n o i s e B o o l
i f params . numGenes == 1
D D e l a y e d h 7 I n d i c e s= [ ] ;
e l s e i f params . numGenes == 2
D D e l a y e d h 7 I n d i c e s= diag ( [ repmat ( [ t r u e f a l s e f a l s e ...
f a l s e ] , 1 , params . numCells − 1 ) , t r u e , f a l s e ] , −2) ;
end
else
i f params . numGenes == 1
D D e l a y e d h 7 I n d i c e s= [ ] ;
e l s e i f params . numGenes == 2
D D e l a y e d h 7 I n d i c e s= repmat ( [ f a l s e f a l s e f a l s e f a l s e ; ...
f a l s e f a l s e f a l s e f a l s e ; t r u e f a l s e f a l s e f a l s e ; f a l s e f a l s e ...
f a l s e f a l s e ] , params . numCells , 1 ) ;
end
end
end
function H 7 T o t a l D e l a y e d d I n d i c e s= get . H 7 T o t a l D e l a y e d d I n d i c e s ( ...
params )
178
i f params . n o i s e B o o l
i f params . numGenes == 1
H 7 T o t a l D e l a y e d d I n d i c e s= [ ] ;
e l s e i f params . numGenes == 2
H 7 T o t a l D e l a y e d d I n d i c e s= diag ( [ repmat ( [ t r u e f a l s e f a l s e ...
f a l s e ] , 1 , params . numCells − 1 ) , t r u e , f a l s e ] , 2 ) ;
end
else
i f params . numGenes == 1
H 7 T o t a l D e l a y e d d I n d i c e s= [ ] ;
e l s e i f params . numGenes == 2
H 7 T o t a l D e l a y e d d I n d i c e s= repmat ( [ f a l s e f a l s e t r u e f a l s e ...
; f a l s e f a l s e f a l s e f a l s e ; f a l s e f a l s e f a l s e f a l s e ; f a l s e f a l s e ...
f a l s e f a l s e ] , params . numCells , 1 ) ;
end
end
end
function d D e l a y e d D I n d i c e s= get . d D e l a y e d D I n d i c e s ( params )
i f params . n o i s e B o o l
i f params . numGenes == 1
d D e l a y e d D I n d i c e s= [ ] ;
e l s e i f params . numGenes == 2
d D e l a y e d D I n d i c e s= diag ( repmat ( [ f a l s e f a l s e f a l s e t r u e ...
] , 1 , params . numCells ) ) ;
end
else
i f params . numGenes == 1
179
d D e l a y e d D I n d i c e s= [ ] ;
e l s e i f params . numGenes == 2
d D e l a y e d D I n d i c e s= repmat ( [ f a l s e f a l s e f a l s e f a l s e ; ...
f a l s e f a l s e f a l s e f a l s e ; f a l s e f a l s e f a l s e f a l s e ; f a l s e f a l s e ...
f a l s e t r u e ] , params . numCells , 1 ) ;
end
end
end
% Computed Parameters
function rho= get . rho ( params )
i f params . n o i s e B o o l
rho= addNoise ( params , params . rhoMean , params . rhoStream ) ;
else
rho= params . rhoMean ;
end
end
function kH13H13= get . kH13H13 ( params )
i f params . n o i s e B o o l
kH13H13= addNoise ( params , params . kH13H13Mean , params . ...
kH13H13Stream ) ;
else
kH13H13= params . kH13H13Mean ;
end
end
function kH7H7= get . kH7H7 ( params )
i f params . n o i s e B o o l
180
kH7H7= addNoise ( params , params . kH7H7Mean , params . ...
kH7H7Stream ) ;
else
kH7H7= params . kH7H7Mean ;
end
end
function kH7H13= get . kH7H13 ( params )
i f params . n o i s e B o o l
kH7H13= addNoise ( params , params . kH7H13Mean , params . ...
kH7H13Stream ) ;
else
kH7H13= params . kH7H13Mean ;
end
end
function t a u h 7= get . t a u h 7 ( params )
i f params . n o i s e B o o l
t a u h 7= addNoise ( params , params . tau h7Mean , params . ...
tau h7Stream ) ;
else
t a u h 7= params . tau h7Mean ;
end
end
function tau H7= get . tau H7 ( params )
i f params . n o i s e B o o l
tau H7= addNoise ( params , params . tau H7Mean , params . ...
tau H7Stream ) ;
else
181
tau H7= params . tau H7Mean ;
end
end
function aH7= get . aH7 ( params )
i f params . n o i s e B o o l
aH7= addNoise ( params , params . aH7Mean , params . aH7Stream ) ;
else
aH7= params . aH7Mean ;
end
end
function bH7= get . bH7 ( params )
i f params . n o i s e B o o l
bH7= addNoise ( params , params . bH7Mean , params . bH7Stream ) ;
else
bH7= params . bH7Mean ;
end
end
function bH7H7= get . bH7H7( params )
i f params . n o i s e B o o l
bH7H7= addNoise ( params , params . bH7H7Mean , params . ...
bH7H7Stream ) ;
else
bH7H7= params . bH7H7Mean ;
end
end
function bH7H13= get . bH7H13 ( params )
i f params . n o i s e B o o l
182
bH7H13= addNoise ( params , params . bH7H13Mean , params . ...
bH7H13Stream ) ;
else
bH7H13= params . bH7H13Mean ;
end
end
function gH7= get . gH7 ( params )
i f params . n o i s e B o o l
gH7= addNoise ( params , params . gH7Mean , params . gH7Stream ) ;
else
gH7= params . gH7Mean ;
end
end
function dH7= get . dH7 ( params )
i f params . n o i s e B o o l
dH7= addNoise ( params , params . dH7Mean , params . dH7Stream ) ;
else
dH7= params . dH7Mean ;
end
end
function rH7H7= get . rH7H7 ( params )
i f params . n o i s e B o o l
rH7H7= addNoise ( params , params . rH7H7Mean , params . ...
rH7H7Stream ) ;
else
rH7H7= params . rH7H7Mean ;
end
183
end
function wH7H7 H7H7= get . wH7H7 H7H7( params )
i f params . n o i s e B o o l
wH7H7 H7H7= addNoise ( params , params . wH7H7 H7H7Mean , ...
params . wH7H7 H7H7Stream ) ;
else
wH7H7 H7H7= params . wH7H7 H7H7Mean ;
end
end
function t a u d= get . t a u d ( params )
i f params . n o i s e B o o l
t a u d= addNoise ( params , params . tau dMean , params . ...
tau dStream ) ;
else
t a u d= params . tau dMean ;
end
end
function tau D= get . tau D ( params )
i f params . n o i s e B o o l
tau D= addNoise ( params , params . tau DMean , params . ...
tau DStream ) ;
else
tau D= params . tau DMean ;
end
end
function aD= get . aD( params )
i f params . n o i s e B o o l
184
aD= addNoise ( params , params . aDMean , params . aDStream ) ;
else
aD= params . aDMean ;
end
end
function bD= get . bD( params )
i f params . n o i s e B o o l
bD= addNoise ( params , params . bDMean , params . bDStream ) ;
else
bD= params . bDMean ;
end
end
function gD= get . gD( params )
i f params . n o i s e B o o l
gD= addNoise ( params , params . gDMean , params . gDStream ) ;
else
gD= params . gDMean ;
end
end
function dD= get . dD( params )
i f params . n o i s e B o o l
dD= addNoise ( params , params . dDMean , params . dDStream ) ;
else
dD= params . dDMean ;
end
end
function rN= get . rN ( params )
185
i f params . n o i s e B o o l
rN= addNoise ( params , params . rNMean , params . rNStream ) ;
else
rN= params . rNMean ;
end
end
function wN= get .wN( params )
i f params . n o i s e B o o l
wN= addNoise ( params , params . wNMean, params . wNStream ) ;
else
wN= params . wNMean ;
end
end
function wN H7H7= get . wN H7H7( params )
i f params . n o i s e B o o l
wN H7H7= addNoise ( params , params . wN H7H7Mean , params . ...
wN H7H7Stream ) ;
else
wN H7H7= params . wN H7H7Mean ;
end
end
function wN H7H7 H7H7= get . wN H7H7 H7H7 ( params )
i f params . n o i s e B o o l
wN H7H7 H7H7= addNoise ( params , params . wN H7H7 H7H7Mean , ...
params . wN H7H7 H7H7Stream ) ;
else
wN H7H7 H7H7= params . wN H7H7 H7H7Mean ;
186
end
end
function H13TotalMax= get . H13TotalMax ( params )
i f params . n o i s e B o o l
H13TotalMax= addNoise ( params , params . H13TotalMaxMean , ...
params . H13TotalMaxStream ) ;
else
H13TotalMax= params . H13TotalMaxMean ;
end
end
function H13TotalMin= get . H13TotalMin ( params )
i f params . n o i s e B o o l
H13TotalMin= addNoise ( params , params . H13TotalMinMean , ...
params . H13TotalMinStream ) ;
else
H13TotalMin= params . H13TotalMinMean ;
end
end
function H 1 3 T o t a l H a l f L i f e= get . H 1 3 T o t a l H a l f L i f e ( params )
i f params . n o i s e B o o l
H 1 3 T o t a l H a l f L i f e= addNoise ( params , params . ...
H13TotalHalfLifeMean , params . H 1 3 T o t a l H a l f L i f e S t r e a m ) ;
else
H 1 3 T o t a l H a l f L i f e= params . H13TotalHalfLifeMean ;
end
end
187
% addNoise method
function paramVec= addNoise ( params , paramMean , paramStream )
paramVec= zeros ( params . numCells , 1 ) ;
i f 0 < paramMean % Only add n o i s e t o ( s t r i c t l y ) p o s i t i v e ...
parameters
paramStream . reset ; % Makes n o i s e r e p e a t a b l e
i f s t r c m p i ( params . noiseType , ’uniform ’ )
paramVec= paramMean ∗ ( 1 + (−params . ...
nois e Propo rtio nOfMea n + 2∗ params . nois e Propo rtio nOfMea n ∗ ( rand ( ...
paramStream , s i z e ( paramVec ) ) ) ) ) ;
e l s e % ” p o s i t i v e ” normal
u n c o n v e r g e d I n d i c e s= ( paramVec <= 0 ) ; % Non−n e g a t i v i t y ...
constraint
while any ( u n c o n v e r g e d I n d i c e s ( : ) )
% Note : 99.7% w i t h i n +/− noiseProportionOfMean o f ...
mean v a l u e
paramVec ( u n c o n v e r g e d I n d i c e s )= paramMean ∗ ( 1 + params . ...
nois e Propo rtio nOfMea n /3∗randn ( paramStream , s i z e ( find ( ...
unconvergedIndices ) ) ) ) ;
u n c o n v e r g e d I n d i c e s= ( paramVec <= 0 ) ;
end
end
end
end % addNoise method
% DISP method
function disp ( params )
188
i f params . n o i s e B o o l
disp ( ’noiseBool : true ’ ) ;
else
disp ( ’noiseBool : false ’ ) ;
end
disp ( [ ’noiseType : ’ params . noiseType ] ) ;
disp ( [ ’noiseSeed : ’ num2str ( params . n o i s e S e e d ) ] ) ;
disp ( [ ’noiseProportionOfMean : ’ num2str ( params . ...
nois e Propo rtio nOfMea n ) ] ) ;
disp ( [ ’timeInterval : ’ m a t 2 s t r ( params . t i m e I n t e r v a l ) ] ) ;
disp ( [ ’settlingTime : ’ num2str ( params . s e t t l i n g T i m e ) ] ) ;
disp ( [ ’timeLags : ’ m a t 2 s t r ( params . timeLags ) ] ) ;
i f i s s t r u c t ( params . h i s t o r y )
disp ( ’history : ’ ) ; disp ( params . h i s t o r y ) ;
e l s e i f i s a ( params . h i s t o r y , ’function_handle ’ )
disp ( [ ’history : @’ f u n c 2 s t r ( params . h i s t o r y ) ] ) ;
e l s e i f i s v e c t o r ( params . h i s t o r y )
disp ( [ ’history : ’ m a t 2 s t r ( params . h i s t o r y ) ] ) ;
end
i f isempty ( params . ddeOptions )
disp ( ’ddeOptions : [] ’ ) ;
else
disp ( ’ddeOptions : ’ ) ; disp ( params . ddeOptions ) ;
end
i f ( params . fastModeBool )
disp ( ’fastMode : true ’ ) ;
else
189
disp ( ’fastMode : false ’ ) ;
end
disp ( [ ’cellsPerSomite : ’ num2str ( params . c e l l s P e r S o m i t e ) ...
]) ;
disp ( [ ’somitesInPSM : ’ num2str ( params . somitesInPSM ) ] ) ;
disp ( [ ’tailbudPeriod : ’ num2str ( params . t a i l b u d P e r i o d ) ] ) ;
disp ( [ ’somiteGrowthRate : ’ num2str ( params . ...
somiteGrowthRate ) ] ) ;
disp ( [ ’cellBirthRate : ’ num2str ( params . c e l l B i r t h R a t e ) ] ) ;
disp ( [ ’birthTimes : ’ m a t 2 s t r ( params . birthTimes ’ ) ] ) ;
disp ( [ ’cellArraySize : ’ m a t 2 s t r ( params . c e l l A r r a y S i z e ) ] ) ;
disp ( [ ’numCells : ’ num2str ( params . numCells ) ] ) ;
disp ( [ ’numGenes : ’ num2str ( params . numGenes ) ] ) ;
disp ( [ ’equationsPerCell : ’ num2str ( params . ...
equationsPerCell ) ] ) ;
disp ( [ ’numEquations : ’ num2str ( params . numEquations ) ] ) ;
disp ( [ ’gradientFunc : ’ params . g r a d i e n t F u n c ] ) ;
disp ( [ ’H13TotalMaxMean : ’ num2str ( params . H13TotalMaxMean ) ...
]) ;
disp ( [ ’H13TotalMinMean : ’ num2str ( params . H13TotalMinMean ) ...
]) ;
disp ( [ ’H13TotalHalfLifeMean : ’ num2str ( params . ...
H13TotalHalfLifeMean ) ] ) ;
disp ( [ ’rhoMean : ’ num2str ( params . rhoMean ) ] ) ;
disp ( [ ’numBindingSites : ’ num2str ( params . numBindingSites ) ...
]) ;
disp ( [ ’tau_h7Mean : ’ num2str ( params . tau h7Mean ) ] ) ;
190
disp ( [ ’tau_H7Mean : ’ num2str ( params . tau H7Mean ) ] ) ;
disp ( [ ’aH7Mean : ’ num2str ( params . aH7Mean ) ] ) ;
disp ( [ ’bH7Mean : ’ num2str ( params . bH7Mean ) ] ) ;
disp ( [ ’bH7H7Mean : ’ num2str ( params . bH7H7Mean) ] ) ;
disp ( [ ’bH7H13Mean : ’ num2str ( params . bH7H13Mean ) ] ) ;
disp ( [ ’gH7Mean : ’ num2str ( params . gH7Mean ) ] ) ;
disp ( [ ’dH7Mean : ’ num2str ( params . dH7Mean ) ] ) ;
disp ( [ ’rH7H7Mean : ’ num2str ( params . rH7H7Mean ) ] ) ;
disp ( [ ’wH7H7_H7H7Mean : ’ num2str ( params . wH7H7 H7H7Mean) ...
]) ;
disp ( [ ’kH13H13Mean : ’ num2str ( params . kH13H13Mean ) ] ) ;
disp ( [ ’kH7H7Mean : ’ num2str ( params . kH7H7Mean ) ] ) ;
disp ( [ ’kH7H13Mean : ’ num2str ( params . kH7H13Mean ) ] ) ;
disp ( [ ’tau_dMean : ’ num2str ( params . tau dMean ) ] ) ;
disp ( [ ’tau_DMean : ’ num2str ( params . tau DMean ) ] ) ;
disp ( [ ’aDMean : ’ num2str ( params . aDMean) ] ) ;
disp ( [ ’bDMean : ’ num2str ( params . bDMean) ] ) ;
disp ( [ ’gDMean : ’ num2str ( params . gDMean) ] ) ;
disp ( [ ’dDMean : ’ num2str ( params . dDMean) ] ) ;
disp ( [ ’rNMean : ’ num2str ( params . rNMean ) ] ) ;
disp ( [ ’wNMean : ’ num2str ( params . wNMean) ] ) ;
disp ( [ ’wN_H7H7Mean : ’ num2str ( params . wN H7H7Mean) ] ) ;
disp ( [ ’wN_H7H7_H7H7Mean : ’ num2str ( params . ...
wN H7H7 H7H7Mean ) ] ) ;
i f ( params . c o u p l i n g B o o l )
disp ( ’couplingBool : true ’ ) ;
else
191
disp ( ’couplingBool : false ’ ) ;
end
i f isempty ( params . c o u p l i n g M a t r i x )
disp ( ’couplingMatrix : [] ’ ) ;
else
[M N]= s i z e ( params . c o u p l i n g M a t r i x ) ;
i f (M > 2 ) | | (N > 2 )
disp ( [ ’couplingMatrix : ’ num2str (M) ’ x ’ num2str (N) ...
’ matrix ’ ] ) ;
else
disp ( ’couplingMatrix : ’ ) ; disp ( params . c o u p l i n g M a t r i x ) ;
end
end
disp ( [ ’H7TotalIndices : ’ m a t 2 s t r ( params . H 7 T o t a l I n d i c e s ) ...
]) ;
disp ( [ ’h7Indices : ’ m a t 2 s t r ( params . h 7 I n d i c e s ) ] ) ;
disp ( [ ’DIndices : ’ m a t 2 s t r ( params . D I n d i c e s ) ] ) ;
disp ( [ ’dIndices : ’ m a t 2 s t r ( params . d I n d i c e s ) ] ) ;
[M N]= s i z e ( params . H 7 T o t a l D e l a y e d h 7 I n d i c e s ) ;
i f (M == 0 ) && (N == 0 )
disp ( ’H7TotalDelayedh7Indices : [] ’ ) ;
disp ( ’h7DelayedH7Indices : [] ’ ) ;
disp ( ’DDelayedh7Indices : [] ’ ) ;
disp ( ’H7TotalDelayeddIndices : [] ’ ) ;
disp ( ’dDelayedDIndices : [] ’ ) ;
e l s e i f (M > 8 ) | | (N > 8 )
192
disp ( [ ’H7TotalDelayedh7Indices : ’ num2str (M) ’ x ’ ...
num2str (N) ’ matrix ’ ] ) ;
disp ( [ ’h7DelayedH7Indices : ’ num2str (M) ’ x ’ num2str (N...
) ’ matrix ’ ] ) ;
disp ( [ ’DDelayedh7Indices : ’ num2str (M) ’ x ’ num2str (N) ...
’ matrix ’ ] ) ;
disp ( [ ’H7TotalDelayeddIndices : ’ num2str (M) ’ x ’ ...
num2str (N) ’ matrix ’ ] ) ;
disp ( [ ’dDelayedDIndices : ’ num2str (M) ’ x ’ num2str (N) ...
’ matrix ’ ] ) ;
else
disp ( ’H7TotalDelayedh7Indices : ’ ) ; disp ( params . ...
H7TotalDelayedh7Indices ) ;
disp ( ’h7DelayedH7Indices : ’ ) ; disp ( params . ...
h7DelayedH7Indices ) ;
disp ( ’DDelayedh7Indices : ’ ) ; disp ( params . ...
DDelayedh7Indices ) ;
disp ( ’H7TotalDelayeddIndices : ’ ) ; disp ( params . ...
H7TotalDelayeddIndices ) ;
disp ( ’dDelayedDIndices : ’ ) ; disp ( params . d D e l a y e d D I n d i c e s ...
);
end
end % d i s p
end % P u b l i c GET methods
methods ( A c c e s s = ’private ’ ) % Access by c l a s s members o n l y
function params= i n i t S t r e a m s ( params )
193
[ params . rhoStream , params . kH13H13Stream , params . ...
kH7H7Stream , params . kH7H13Stream , params . tau h7Stream , params . ...
tau H7Stream , params . gH7Stream , params . aH7Stream , params . ...
bH7Stream , params . bH7H7Stream , params . bH7H13Stream , params . ...
dH7Stream , params . rH7H7Stream , params . wH7H7 H7H7Stream , params . ...
tau dStream , params . tau DStream , params . gDStream , params . ...
aDStream , params . bDStream , params . dDStream , params . rNStream , ...
params . wNStream , params . wN H7H7Stream , params . wN H7H7 H7H7Stream...
, params . H13TotalMaxStream , params . H13TotalMinStream , params . ...
H 1 3 T o t a l H a l f L i f e S t r e a m ]= RandStream . c r e a t e ( ’mrg32k3a ’ , ’...
NumStreams ’ , 2 7 , ’Seed ’ , params . n o i s e S e e d ) ;
end
end % P r i v a t e methods
methods ( S t a t i c )
function c o u p l i n g M a t r i x= c r e a t e N e a r e s t N e i g h b o r C o u p l i n g M a t r i x ...
( cellArraySize )
% Note : Works f o r 1D v e c t o r s and 2D a r r a y s .
% Note : DeltaC assumed t o be e v e n l y d i s t r i b u t e d t h r o u g h c e l l ...
membrane
%
t o a d j a c e n t c e l l s w i t h o u t Notch r e c e p t o r s a t u r a t i o n . This...
scaling
%
i s lumped i n t o t h e rN parameter .
M= c e l l A r r a y S i z e ( 1 ) ;
N= c e l l A r r a y S i z e ( 2 ) ;
K= M∗N;
194
c o u p l i n g M a t r i x= zeros (K) ;
f o r k= 1 :K
i f k == 1 % Top− l e f t c o r n e r
if M > 1
c o u p l i n g M a t r i x ( 1 , 2 )= 1 ;
if N > 1
c o u p l i n g M a t r i x ( 1 ,M+1:M+2)= 1 ;
end
elseif N > 1
c o u p l i n g M a t r i x ( 1 ,M+1)= 1 ;
end
e l s e i f ( 2 <= k ) && ( k <= M−1) % L e f t s i d e
c o u p l i n g M a t r i x ( k , [ k−1,k +1])= 1 ;
if N > 1
c o u p l i n g M a t r i x ( k , k+M−1:k+M+1)= 1 ;
end
e l s e i f k == M % Bottom− l e f t c o r n e r
c o u p l i n g M a t r i x (M, k−1)= 1 ;
if N > 1
c o u p l i n g M a t r i x (M, k+M−1:k+M)= 1 ;
end
e l s e i f (rem( k , M) == 1 ) && ( k < K−M+1) % Top s i d e
c o u p l i n g M a t r i x ( k , [ k−M: k−M+1, k+1, k+M: k+M+1])= 1 ;
e l s e i f (rem( k , M) == 0 ) && ( k < K) % Bottom s i d e
if M > 1
c o u p l i n g M a t r i x ( k , [ k−M−1:k−M, k−1, k+M−1:k+M] )= 1 ;
195
else
c o u p l i n g M a t r i x ( k , [ k−M, k+M] )= 1 ;
end
e l s e i f k == K−M+1 % Top−r i g h t c o r n e r
if M > 1
c o u p l i n g M a t r i x ( k , [ k−M: k−M+1, k +1])= 1 ;
else
c o u p l i n g M a t r i x ( k , k−M)= 1 ;
end
e l s e i f (K−M+1 < k ) && ( k < K) % R i g h t s i d e
c o u p l i n g M a t r i x ( k , [ k−M−1:k−M+1, k−1, k +1])= 1 ;
e l s e i f k == K % Bottom−r i g h t c o r n e r
c o u p l i n g M a t r i x ( k , [ k−M−1:k−M, k −1])= 1 ;
else % Inside
c o u p l i n g M a t r i x ( k , [ k−M−1:k−M+1, k−1, k+1, k+M−1:k+M+1])...
= 1;
end
end
end % c r e a t e N e a r e s t N e i g h b o r C o u p l i n g M a t r i x ( )
function c o u p l i n g M a t r i x= c r e a t e A x i a l C o u p l i n g M a t r i x ( ...
cellArraySize )
% Note : Works f o r 1D v e c t o r s and 2D a r r a y s .
% Note : DeltaC assumed t o be e v e n l y d i s t r i b u t e d t h r o u g h c e l l ...
membrane
%
t o a d j a c e n t c e l l s w i t h o u t Notch r e c e p t o r s a t u r a t i o n . This...
scaling
196
%
i s lumped i n t o t h e rN parameter .
M= c e l l A r r a y S i z e ( 1 ) ;
N= c e l l A r r a y S i z e ( 2 ) ;
K= M∗N;
c o u p l i n g M a t r i x= zeros (K) ;
f o r k= 1 :K
i f k == 1 % Top− l e f t c o r n e r
if M > 1
c o u p l i n g M a t r i x ( 1 , 2 )= 1 ;
end
e l s e i f ( 2 <= k ) && ( k <= M−1) % L e f t s i d e
c o u p l i n g M a t r i x ( k , [ k−1, k +1])= 1 ;
e l s e i f k == M % Bottom− l e f t c o r n e r
c o u p l i n g M a t r i x (M, k−1)= 1 ;
e l s e i f (rem( k , M) == 1 ) && ( k < K−M+1) % Top s i d e
c o u p l i n g M a t r i x ( k , k+1)= 1 ;
e l s e i f (rem( k , M) == 0 ) && ( k < K) % Bottom s i d e
if M > 1
c o u p l i n g M a t r i x ( k , k−1)= 1 ;
end
e l s e i f k == K−M+1 % Top−r i g h t c o r n e r
if M > 1
c o u p l i n g M a t r i x ( k , k+1)= 1 ;
end
e l s e i f (K−M+1 < k ) && ( k < K) % R i g h t s i d e
197
c o u p l i n g M a t r i x ( k , [ k−1, k +1])= 1 ;
e l s e i f k == K % Bottom−r i g h t c o r n e r
c o u p l i n g M a t r i x ( k , k−1)= 1 ;
else % Inside
c o u p l i n g M a t r i x ( k , [ k−1, k +1])= 1 ;
end
end
end % c r e a t e A x i a l C o u p l i n g M a t r i x ( )
end % S t a t i c methods
end % c l a s s d e f
somitogenesisPCW.m: This is the main simulation function for the PCW
model. This code calls the built-in Matlab DDE solver ddesd by default, but it also
works with dde23.
function s o l= somitogenesisPCW ( params )
% Usage :
%
s o l= somitogenesisPCW ( params )
%
% S i m u l a t e s z e b r a f i s h s o m i t o g e n e s i s i n p o s t e r i o r PSM.
%
S i n g l e c l o c k −gene i s HER7, d e n o t e d H7 ( p r o t e i n ) or h7 (mRNA) .
%
C o n t r o l p r o t e i n i s Her13 . 2 , d e n o t e d H13 .
%
C o o r d i n a t i n g −s i g n a l gene ( o p t i o n a l ) i s DELTAC, d e n o t e d
%
%
D ( p r o t e i n ) or d (mRNA) .
198
% Note : To g e t a d e f a u l t params o b j e c t f o r t h e i n p u t argument ...
use
%
params= Params ( )
%s o m i t o g e n e s i s T i c= t i c ;
% S e l e c t r h s system t o s o l v e ( s e e n e s t e d f u n c t i o n s b e l o w )
i f params . numGenes == 1 % HER7 o n l y
i f params . numBindingSites == 1 % R e p r e s s o r b i n d i n g s i t e s
rhsSystem= @rhs1Gene1Site ;
e l s e i f params . numBindingSites == 2
rhsSystem= @rhs1Gene2Site ;
end
e l s e i f params . numGenes == 2 % HER7 and DELTAC
i f params . numBindingSites == 1 % R e p r e s s o r b i n d i n g s i t e s
rhsSystem= @rhs2Gene1Site ;
e l s e i f params . numBindingSites == 2
rhsSystem= @rhs2Gene2Site ;
end
end
% O p t i m i z a t i o n s t o a v o i d i n e f f i c i e n t params o b j e c t a c c e s s
numEquations= params . numEquations ;
b i r t h T i m e s= params . b i r t h T i m e s ;
H 7 T o t a l I n d i c e s= params . H 7 T o t a l I n d i c e s ;
h 7 I n d i c e s= params . h 7 I n d i c e s ;
D I n d i c e s= params . D I n d i c e s ;
d I n d i c e s= params . d I n d i c e s ;
H 7 T o t a l D e l a y e d h 7 I n d i c e s= params . H 7 T o t a l D e l a y e d h 7 I n d i c e s ;
199
D D e l a y e d h 7 I n d i c e s= params . D D e l a y e d h 7 I n d i c e s ;
h 7 D e l a y e d H 7 I n d i c e s= params . h 7 D e l a y e d H 7 I n d i c e s ;
H 7 T o t a l D e l a y e d d I n d i c e s= params . H 7 T o t a l D e l a y e d d I n d i c e s ;
d D e l a y e d D I n d i c e s= params . d D e l a y e d D I n d i c e s ;
rho= params . rho ;
kH13H13= params . kH13H13 ;
kH7H7= params . kH7H7 ;
kH7H13= params . kH7H13 ;
t a u h 7= params . t a u h 7 ;
%
tau H7= params . tau H7 ;
aH7= params . aH7 ;
bH7= params . bH7 ;
bH7H7= params . bH7H7 ;
bH7H13= params . bH7H13 ;
gH7= params . gH7 ;
rH7H7= params . rH7H7 ;
wH7H7 H7H7= params . wH7H7 H7H7 ;
dH7= params . dH7 ;
t a u d= params . t a u d ;
%
tau D= params . tau D ;
aD= params . aD ;
bD= params . bD ;
gD= params . gD ;
rN= params . rN ;
wN= params .wN;
wN H7H7= params . wN H7H7 ;
wN H7H7 H7H7= params . wN H7H7 H7H7 ;
200
dD= params . dD ;
c o u p l i n g M a t r i x= params . c o u p l i n g M a t r i x ;
gradientFuncH= s t r 2 f u n c ( params . g r a d i e n t F u n c ) ;
H13TotalMax= params . H13TotalMax ;
H13TotalMin= params . H13TotalMin ;
H 1 3 T o t a l H a l f L i f e= params . H 1 3 T o t a l H a l f L i f e ;
i f i s s t r u c t ( params . h i s t o r y ) % Continue e x i s t i n g s o l u t i o n
% Assumes l o n g e r params . t i m e I n t e r v a l f o r c o n t i n u a t i o n
% TBD: How t o t r a c k / r e c o r d c o n t i n u a t i o n s ?
s o l= ddesd ( rhsSystem , params . timeLags , params . h i s t o r y , [ ...
params . h i s t o r y . params . t i m e I n t e r v a l ( 2 ) , params . t i m e I n t e r v a l ( 2 ) ] , ...
params . ddeOptions ) ;
i f ˜isempty ( s o l . xe ) % S o l u t i o n went n e g a t i v e
warning ( ’Zero - crossing in solution during continuation .’ ) ;
end
s o l . params= params ;
s o l . s t a t i s t i c s = s o l . params . h i s t o r y . s t a t i s t i c s ;
e l s e % S t a r t a new s o l u t i o n
i f params . s e t t l i n g T i m e > 0
% Compute ” s e t t l i n g time ” p o r t i o n o f s o l u t i o n
s o l= ddesd ( rhsSystem , params . timeLags , params . h i s t o r y , [ ...
params . t i m e I n t e r v a l ( 1 ) − params . s e t t l i n g T i m e , params . ...
t i m e I n t e r v a l ( 1 ) ] , params . ddeOptions ) ;
i f isempty ( s o l . xe ) % S o l u t i o n d i d not go n e g a t i v e
s o l . params= params ; % Needed f o r calcPerMedAmp ( ) c a l l
201
% C a l c u l a t e p e r i o d s , medians , and a m p l i t u d e s a t end o f ...
settlingTime
[ s t a t i s t i c s . p e r i o d s , s t a t i s t i c s . medians , s t a t i s t i c s . ...
a m p l i t u d e s ]= calcPerMedAmp ( s o l ) ;
% Update d e f a u l t t a i l b u d p e r i o d
i f ˜any ( isnan ( s t a t i s t i c s . p e r i o d s ) ) && ˜any ( s t a t i s t i c s . ...
p e r i o d s <= 0 )
periodMean= mean( s t a t i s t i c s . p e r i o d s ) ;
i f a l l ( abs ( ( s t a t i s t i c s . p e r i o d s − periodMean ) /...
periodMean ) < params . p e r i o d T o l )
params . t a i l b u d P e r i o d= periodMean ;
b i r t h T i m e s= params . b i r t h T i m e s ;
end
end
% Continue w i t h ” time o f i n t e r e s t ” p o r t i o n o f s o l u t i o n
i f params . t i m e I n t e r v a l ( 2 ) − params . t i m e I n t e r v a l ( 1 ) > 0
s o l= ddesd ( rhsSystem , params . timeLags , s o l , params . ...
t i m e I n t e r v a l , params . ddeOptions ) ;
i f ˜isempty ( s o l . xe ) % S o l u t i o n went n e g a t i v e
warning ( ’Zero - crossing in solution during "time of ...
interest ". ’ ) ;
end
end
else
warning ( ’Zero - crossing in solution during " settling ...
time ". ’ ) ;
end
202
e l s e i f params . t i m e I n t e r v a l ( 2 ) − params . t i m e I n t e r v a l ( 1 ) > 0
% Compute s o l u t i o n w i t h o u t any ” s e t t l i n g time ”
s t a t i s t i c s . p e r i o d s= nan ( numEquations , 1 ) ;
s t a t i s t i c s . medians= nan ( numEquations , 1 ) ;
s t a t i s t i c s . a m p l i t u d e s= nan ( numEquations , 1 ) ;
s o l= ddesd ( rhsSystem , params . timeLags , params . h i s t o r y , ...
params . t i m e I n t e r v a l , params . ddeOptions ) ;
i f ˜isempty ( s o l . xe ) % S o l u t i o n went n e g a t i v e
warning ( ’Zero - crossing in solution during "time of ...
interest ". ’ ) ;
end
else
warning ( ’No non - empty time - interval on which to solve the...
system .’ ) ;
s o l . params= params ; % Record p a r a m e t e r s o b j e c t i n s o l u t i o n ...
structure
return ;
end
s o l . params= params ; % Record p a r a m e t e r s o b j e c t i n s o l u t i o n ...
structure
s o l . s t a t i s t i c s = s t a t i s t i c s ; % Record s t a t i s t i c s i n s o l u t i o n ...
structure
end
%t o c ( s o m i t o g e n e s i s T i c )
i f ˜ params . fastModeBool
% C a l c u l a t e Her13 . 2 & Her7 monomer & dimer c o n c e n t r a t i o n s
203
s o l . output . H13Total= gradientFuncH ( repmat ( s o l . x , length ( ...
b i r t h T i m e s ) , 1 ) − repmat ( birthTimes , 1 , length ( s o l . x ) ) , ...
H13TotalMax , H13TotalMin , H 1 3 T o t a l H a l f L i f e ) ;
[ s o l . output . H13 , s o l . output . H7 , s o l . output . H13H13 , s o l . ...
output . H7H7 , s o l . output . H7H13]= c a l c D i m e r i z a t i o n ( s o l . output . ...
H13Total , s o l . y ( H 7 T o t a l I n d i c e s , : ) , kH13H13 , kH7H7 , kH7H13 ) ;
n e g a t i v i t y C h e c k= ( s o l . output . H7 < 0 ) | ( s o l . output . H13 < 0 ) ...
| ( s o l . output . H7H7 < 0 ) | ( s o l . output . H13H13 < 0 ) | ( s o l . output . ...
H7H13 < 0 ) ;
i f any ( n e g a t i v i t y C h e c k ( : ) )
warning ( ’Negative copy number ( possibly near zero) ...
detected in CalcDimerization () computation .’ ) ;
end
plotSolution ( sol ) ; % Plot r e s u l t s
end
% Nested f u n c t i o n s
function dzdt= r h s 1 G e n e 1 S i t e ( t , z , Z )
dzdt= zeros ( numEquations , 1 ) ; % P r e a l l o c a t e
% C a l c u l a t e f a s t d i m e r i z a t i o n ( Her monomer and dimer amounts )
[ H13 , H7 , H13H13 , H7H7 , H7H13]= c a l c D i m e r i z a t i o n ( ...
gradientFuncH ( t − birthTimes , H13TotalMax , H13TotalMin , ...
H 1 3 T o t a l H a l f L i f e ) , z ( H 7 T o t a l I n d i c e s ) , kH13H13 , kH7H7 , kH7H13 ) ;
[ H13Delayedh7 , H7Delayedh7 , H13H13Delayedh7 , H7H7Delayedh7 , ...
H7H13Delayedh7 ]= c a l c D i m e r i z a t i o n ( gradientFuncH ( t − b i r t h T i m e s −...
tau h7 , H13TotalMax , H13TotalMin , H 1 3 T o t a l H a l f L i f e ) , Z ( ...
H 7 T o t a l D e l a y e d h 7 I n d i c e s ) , kH13H13 , kH7H7 , kH7H13 ) ;
204
% System o f d i f f e r e n t i a l e q u a t i o n s
% Her7 t o t a l p r o t e i n
dzdt ( H 7 T o t a l I n d i c e s )= aH7 . ∗ Z ( h 7 D e l a y e d H 7 I n d i c e s ) − bH7 . ∗ H7 −...
2∗bH7H7 . ∗ H7H7 − bH7H13 . ∗ H7H13 ;
% Her7 mRNA
dzdt ( h 7 I n d i c e s )= gH7 . ∗ rho . / ( rho + 1 + rH7H7 . ∗ H7H7Delayedh7 ) ...
− dH7 . ∗ z ( h 7 I n d i c e s ) ;
end
function dzdt= r h s 1 G e n e 2 S i t e ( t , z , Z )
dzdt= zeros ( numEquations , 1 ) ; % P r e a l l o c a t e
% C a l c u l a t e f a s t d i m e r i z a t i o n ( Her monomer and dimer amounts )
[ H13 , H7 , H13H13 , H7H7 , H7H13]= c a l c D i m e r i z a t i o n ( ...
gradientFuncH ( t − birthTimes , H13TotalMax , H13TotalMin , ...
H 1 3 T o t a l H a l f L i f e ) , z ( H 7 T o t a l I n d i c e s ) , kH13H13 , kH7H7 , kH7H13 ) ;
[ H13Delayedh7 , H7Delayedh7 , H13H13Delayedh7 , H7H7Delayedh7 , ...
H7H13Delayedh7 ]= c a l c D i m e r i z a t i o n ( gradientFuncH ( t − b i r t h T i m e s −...
tau h7 , H13TotalMax , H13TotalMin , H 1 3 T o t a l H a l f L i f e ) , Z ( ...
H 7 T o t a l D e l a y e d h 7 I n d i c e s ) , kH13H13 , kH7H7 , kH7H13 ) ;
% System o f d i f f e r e n t i a l e q u a t i o n s
% Her7 t o t a l p r o t e i n
dzdt ( H 7 T o t a l I n d i c e s )= aH7 . ∗ Z ( h 7 D e l a y e d H 7 I n d i c e s ) − bH7 . ∗ H7 −...
2∗bH7H7 . ∗ H7H7 − bH7H13 . ∗ H7H13 ;
% Her7 mRNA
dzdt ( h 7 I n d i c e s )= gH7 . ∗ rho . / ( rho + 1 + 2∗rH7H7 . ∗ H7H7Delayedh7...
+ wH7H7 H7H7 . ∗ ( rH7H7 . ∗ H7H7Delayedh7 ) . ˆ 2 ) − dH7 . ∗ z ( h 7 I n d i c e s ) ;
end
205
function dzdt= r h s 2 G e n e 1 S i t e ( t , z , Z )
dzdt= zeros ( numEquations , 1 ) ; % P r e a l l o c a t e
% C a l c u l a t e f a s t d i m e r i z a t i o n ( Her monomer and dimer amounts )
[ H13 , H7 , H13H13 , H7H7 , H7H13]= c a l c D i m e r i z a t i o n ( ...
gradientFuncH ( t − birthTimes , H13TotalMax , H13TotalMin , ...
H 1 3 T o t a l H a l f L i f e ) , z ( H 7 T o t a l I n d i c e s ) , kH13H13 , kH7H7 , kH7H13 ) ;
[ H13Delayedh7 , H7Delayedh7 , H13H13Delayedh7 , H7H7Delayedh7 , ...
H7H13Delayedh7 ]= c a l c D i m e r i z a t i o n ( gradientFuncH ( t − b i r t h T i m e s −...
tau h7 , H13TotalMax , H13TotalMin , H 1 3 T o t a l H a l f L i f e ) , Z ( ...
H 7 T o t a l D e l a y e d h 7 I n d i c e s ) , kH13H13 , kH7H7 , kH7H13 ) ;
[ H13Delayedd , H7Delayedd , H13H13Delayedd , H7H7Delayedd , ...
H7H13Delayedd ]= c a l c D i m e r i z a t i o n ( gradientFuncH ( t − b i r t h T i m e s − ...
tau d , H13TotalMax , H13TotalMin , H 1 3 T o t a l H a l f L i f e ) , Z ( ...
H 7 T o t a l D e l a y e d d I n d i c e s ) , kH13H13 , kH7H7 , kH7H13 ) ;
% C a l c u l a t e NICD from c o u p l i n g
NDelayedh7= c o u p l i n g M a t r i x ∗Z ( D D e l a y e d h 7 I n d i c e s ) ;
% System o f d i f f e r e n t i a l e q u a t i o n s
% Her7 t o t a l p r o t e i n
dzdt ( H 7 T o t a l I n d i c e s )= aH7∗Z ( h 7 D e l a y e d H 7 I n d i c e s ) − bH7∗H7 − ...
2∗bH7H7∗H7H7 − bH7H13∗H7H13 ;
% Her7 mRNA
dzdt ( h 7 I n d i c e s )= gH7∗ rho ∗ ( 1 + wN∗rN∗ NDelayedh7 ) . / ( rho ∗ ( 1 + ...
wN∗rN∗ NDelayedh7 ) + ( 1 + rH7H7∗ H7H7Delayedh7 ) + rN∗ NDelayedh7...
. ∗ ( 1 + wN H7H7∗rH7H7∗ H7H7Delayedh7 ) ) − dH7∗ z ( h 7 I n d i c e s ) ;
% Delta protein
dzdt ( D I n d i c e s )= aD . ∗ Z ( d D e l a y e d D I n d i c e s ) − bD . ∗ z ( D I n d i c e s ) ;
206
% D e l t a mRNA
dzdt ( d I n d i c e s )= gD . ∗ rho . / ( rho + 1 + rH7H7 . ∗ H7H7Delayedd ) − ...
dD . ∗ z ( d I n d i c e s ) ;
end
function dzdt= r h s 2 G e n e 2 S i t e ( t , z , Z )
dzdt= zeros ( numEquations , 1 ) ; % P r e a l l o c a t e
% C a l c u l a t e f a s t d i m e r i z a t i o n ( Her monomer and dimer amounts )
[ H13 , H7 , H13H13 , H7H7 , H7H13]= c a l c D i m e r i z a t i o n ( ...
gradientFuncH ( t − birthTimes , H13TotalMax , H13TotalMin , ...
H 1 3 T o t a l H a l f L i f e ) , z ( H 7 T o t a l I n d i c e s ) , kH13H13 , kH7H7 , kH7H13 ) ;
[ H13Delayedh7 , H7Delayedh7 , H13H13Delayedh7 , H7H7Delayedh7 , ...
H7H13Delayedh7 ]= c a l c D i m e r i z a t i o n ( gradientFuncH ( t − b i r t h T i m e s −...
tau h7 , H13TotalMax , H13TotalMin , H 1 3 T o t a l H a l f L i f e ) , Z ( ...
H 7 T o t a l D e l a y e d h 7 I n d i c e s ) , kH13H13 , kH7H7 , kH7H13 ) ;
[ H13Delayedd , H7Delayedd , H13H13Delayedd , H7H7Delayedd , ...
H7H13Delayedd ]= c a l c D i m e r i z a t i o n ( gradientFuncH ( t − b i r t h T i m e s − ...
tau d , H13TotalMax , H13TotalMin , H 1 3 T o t a l H a l f L i f e ) , Z ( ...
H 7 T o t a l D e l a y e d d I n d i c e s ) , kH13H13 , kH7H7 , kH7H13 ) ;
% C a l c u l a t e NICD from c o u p l i n g
NDelayedh7= c o u p l i n g M a t r i x ∗Z ( D D e l a y e d h 7 I n d i c e s ) ;
% System o f d i f f e r e n t i a l e q u a t i o n s
% Her7 t o t a l p r o t e i n
dzdt ( H 7 T o t a l I n d i c e s )= aH7 . ∗ Z ( h 7 D e l a y e d H 7 I n d i c e s ) − bH7 . ∗ H7 −...
2∗bH7H7 . ∗ H7H7 − bH7H13 . ∗ H7H13 ;
% Her7 mRNA
207
dzdt ( h 7 I n d i c e s )= gH7 . ∗ rho . ∗ ( 1 + wN. ∗ rN . ∗ NDelayedh7 ) . / ( rho...
. ∗ ( 1 + wN. ∗ rN . ∗ NDelayedh7 ) + ( 1 + 2∗rH7H7 . ∗ H7H7Delayedh7 + ...
wH7H7 H7H7 . ∗ ( rH7H7 . ∗ H7H7Delayedh7 ) . ˆ 2 ) + rN . ∗ NDelayedh7 . ∗ ( 1 + 2∗...
wN H7H7 . ∗ rH7H7 . ∗ H7H7Delayedh7 + wN H7H7 H7H7 . ∗ wH7H7 H7H7 . ∗ ( rH7H7...
. ∗ H7H7Delayedh7 ) . ˆ 2 ) ) − dH7 . ∗ z ( h 7 I n d i c e s ) ;
% Delta protein
dzdt ( D I n d i c e s )= aD . ∗ Z ( d D e l a y e d D I n d i c e s ) − bD . ∗ z ( D I n d i c e s ) ;
% D e l t a mRNA
dzdt ( d I n d i c e s )= gD . ∗ rho . / ( rho + 1 + rH7H7 . ∗ H7H7Delayedd ) − ...
dD . ∗ z ( d I n d i c e s ) ;
end
end
calcDimerization.m: This implements the iterative numerical scheme for
calculating monomer, homodimer, and heterodimer amounts from total clock protein
and total control protein amounts.
function [ H13 , H7 , H13H13 , H7H7 , H7H13]= c a l c D i m e r i z a t i o n ( ...
H13Total , H7Total , kH13H13 , kH7H7 , kH7H13 )
% Usage :
%
[ H13 , H7 , H13H13 , H7H7 , H7H13]= c a l c D i m e r i z a t i o n ( H13Total , ...
H7Total , kH13H13 , kH7H7 , kH7H13 )
%
% Computes ( v i a i t e r a t i o n ) t h e n o n l i n e a r a l g e b r a i c system o f
%
equations for the f a s t dimerization .
% 2D V e c t o r i z e d − assumes H7Total & H13Total have same s i z e and
208
%
a r e non−n e g a t i v e :
%
1) The f i r s t ( row ) dimension i s t h e t o t a l number o f c e l l s .
%
2) The second ( column ) dimension i s t y p i c a l l y time .
% The kappa p a r a m e t e r s may be a l l s c a l a r s or a l l column v e c t o r s
%
w i t h t h e same number o f columns as H7Total & H13Total .
r e s i d u a l T o l= 1 e −6;
promptFlag= t r u e ;
H7= H7Total ; % IC
H13= H13Total ; % IC
H 1 3 T o t a l S i z e= s i z e ( H13Total ) ;
u n c o n v e r g e d I n d i c e s= t r u e ( H 1 3 T o t a l S i z e ) ;
i f i s s c a l a r ( kH13H13 )
k= 0 ; % I t e r a t i o n c o u n t e r
while any ( u n c o n v e r g e d I n d i c e s ( : ) )
k= k + 1 ;
i f promptFlag && ˜rem( k , 1 0 0 0 )
while t r u e % User i n p u t l o o p
r e s p o n s e= menu ( [ ’No convergence to root after ’ ...
num2str ( k ) ’ iterations . Iterate further ? ’ ] , ’Yes ’ , ’Yes , turn...
off prompting ’ , ’No , terminate execution ’ ) ;
switch response
case 1
break ;
case 2
promptFlag= f a l s e ;
209
break ;
case 3
disp ( ’Parameter Dump:’ ) ;
disp ( [ ’H13Total = ’ m a t 2 s t r ( H13Total ) ] ) ;
disp ( [ ’H7Total = ’ m a t 2 s t r ( H7Total ) ] ) ;
disp ( [ ’kH13H13 = ’ m a t 2 s t r ( kH13H13 ) ] ) ;
disp ( [ ’kH7H7 = ’ m a t 2 s t r ( kH7H7 ) ] ) ;
disp ( [ ’kH7H13 = ’ m a t 2 s t r ( kH7H13 ) ] ) ;
error ( ’Failed to converge to root.’ ) ;
otherwise
continue ;
end
end
end
% Only u p d a t e un c on v er ge d i n d i c e s
H7( u n c o n v e r g e d I n d i c e s )= ( sqrt ( ( kH7H7∗ ( 1 + H13 ( ...
u n c o n v e r g e d I n d i c e s ) /kH7H13 ) ) . ˆ 2 + 8∗kH7H7∗ H7Total ( ...
u n c o n v e r g e d I n d i c e s ) ) − kH7H7∗ ( 1 + H13 ( u n c o n v e r g e d I n d i c e s ) /kH7H13...
) ) /4;
H13 ( u n c o n v e r g e d I n d i c e s )= ( sqrt ( ( kH13H13 ∗ ( 1 + H7( ...
u n c o n v e r g e d I n d i c e s ) /kH7H13 ) ) . ˆ 2 + 8∗kH13H13∗ H13Total ( ...
u n c o n v e r g e d I n d i c e s ) ) − kH13H13 ∗ ( 1 + H7( u n c o n v e r g e d I n d i c e s ) /...
kH7H13 ) ) / 4 ;
% Use r e s i d u a l t o compute remainin g un c on v er ge d i n d i c e s
u n c o n v e r g e d I n d i c e s ( u n c o n v e r g e d I n d i c e s )= (H7( ...
u n c o n v e r g e d I n d i c e s ) . ˆ 2 + kH7H7 / 2 . ∗ ( 1 + H13 ( u n c o n v e r g e d I n d i c e s ) /...
kH7H13 ) . ∗ H7( u n c o n v e r g e d I n d i c e s ) − kH7H7∗ H7Total ( ...
210
u n c o n v e r g e d I n d i c e s ) /2 >= r e s i d u a l T o l ) | ( H13 ( u n c o n v e r g e d I n d i c e s ) ...
. ˆ 2 + kH13H13 / 2 . ∗ ( 1 + H7( u n c o n v e r g e d I n d i c e s ) /kH7H13 ) . ∗ H13 ( ...
u n c o n v e r g e d I n d i c e s ) − kH13H13∗ H13Total ( u n c o n v e r g e d I n d i c e s ) /2 >= ...
residualTol ) ;
end
else
i f H13TotalSize (2) > 1
kH7H7= repmat ( kH7H7 , 1 , H 1 3 T o t a l S i z e ( 2 ) ) ;
kH13H13= repmat ( kH13H13 , 1 , H 1 3 T o t a l S i z e ( 2 ) ) ;
kH7H13= repmat ( kH7H13 , 1 , H 1 3 T o t a l S i z e ( 2 ) ) ;
end
k= 0 ; % I t e r a t i o n c o u n t e r
while any ( u n c o n v e r g e d I n d i c e s ( : ) )
k= k + 1 ;
i f promptFlag && ˜rem( k , 1 0 0 0 )
while t r u e % User i n p u t l o o p
r e s p o n s e= menu ( [ ’No convergence to root after ’ ...
num2str ( k ) ’ iterations . Iterate further ? ’ ] , ’Yes ’ , ’Yes , turn...
off prompting ’ , ’No , terminate execution ’ ) ;
switch response
case 1
break ;
case 2
promptFlag= f a l s e ;
break ;
case 3
211
disp ( ’Parameter Dump:’ ) ;
disp ( [ ’H13Total = ’ m a t 2 s t r ( H13Total ) ] ) ;
disp ( [ ’H7Total = ’ m a t 2 s t r ( H7Total ) ] ) ;
disp ( [ ’kH13H13 = ’ m a t 2 s t r ( kH13H13 ) ] ) ;
disp ( [ ’kH7H7 = ’ m a t 2 s t r ( kH7H7 ) ] ) ;
disp ( [ ’kH7H13 = ’ m a t 2 s t r ( kH7H13 ) ] ) ;
error ( ’Failed to converge to root.’ ) ;
otherwise
continue ;
end
end
end
% Only u p d a t e un c on v er ge d i n d i c e s
H7( u n c o n v e r g e d I n d i c e s )= ( sqrt ( ( kH7H7 ( u n c o n v e r g e d I n d i c e s ) ...
. ∗ ( 1 + H13 ( u n c o n v e r g e d I n d i c e s ) . / kH7H13 ( u n c o n v e r g e d I n d i c e s ) ) ) . ˆ 2 ...
+ 8∗kH7H7 ( u n c o n v e r g e d I n d i c e s ) . ∗ H7Total ( u n c o n v e r g e d I n d i c e s ) ) − ...
kH7H7 ( u n c o n v e r g e d I n d i c e s ) . ∗ ( 1 + H13 ( u n c o n v e r g e d I n d i c e s ) . / kH7H13 ( ...
unconvergedIndices ) ) ) /4;
H13 ( u n c o n v e r g e d I n d i c e s )= ( sqrt ( ( kH13H13 ( u n c o n v e r g e d I n d i c e s ...
) . ∗ ( 1 + H7( u n c o n v e r g e d I n d i c e s ) . / kH7H13 ( u n c o n v e r g e d I n d i c e s ) ) ) . ˆ 2 ...
+ 8∗kH13H13 ( u n c o n v e r g e d I n d i c e s ) . ∗ H13Total ( u n c o n v e r g e d I n d i c e s ) ) −...
kH13H13 ( u n c o n v e r g e d I n d i c e s ) . ∗ ( 1 + H7( u n c o n v e r g e d I n d i c e s ) . / ...
kH7H13 ( u n c o n v e r g e d I n d i c e s ) ) ) / 4 ;
% Use r e s i d u a l t o compute remainin g un c on v er ge d i n d i c e s
u n c o n v e r g e d I n d i c e s ( u n c o n v e r g e d I n d i c e s )= (H7( ...
u n c o n v e r g e d I n d i c e s ) . ˆ 2 + kH7H7 ( u n c o n v e r g e d I n d i c e s ) / 2 . ∗ ( 1 + H13 ( ...
u n c o n v e r g e d I n d i c e s ) . / kH7H13 ( u n c o n v e r g e d I n d i c e s ) ) . ∗ H7( ...
212
u n c o n v e r g e d I n d i c e s ) − kH7H7 ( u n c o n v e r g e d I n d i c e s ) . ∗ H7Total ( ...
u n c o n v e r g e d I n d i c e s ) /2 >= r e s i d u a l T o l ) | ( H13 ( u n c o n v e r g e d I n d i c e s ) ...
. ˆ 2 + kH13H13 ( u n c o n v e r g e d I n d i c e s ) / 2 . ∗ ( 1 + H7( u n c o n v e r g e d I n d i c e s ) ...
. / kH7H13 ( u n c o n v e r g e d I n d i c e s ) ) . ∗ H13 ( u n c o n v e r g e d I n d i c e s ) − kH13H13...
( u n c o n v e r g e d I n d i c e s ) . ∗ H13Total ( u n c o n v e r g e d I n d i c e s ) /2 >= ...
residualTol ) ;
end
end
% Compute h e t e r o d i m e r s
H7H13= H7 . ∗ H13 . / kH7H13 ;
H7H7= ( H7Total − H7 − H7H13 ) / 2 ;
H13H13= ( H13Total − H13 − H7H13 ) / 2 ;
end
calcPerMedAmp.m: This computes the component-wise period, median, and
amplitude of simply periodic or steady-state solutions computed with somitogenesisPCW.m.
function [ p e r i o d s , medians , a m p l i t u d e s ]= calcPerMedAmp ( s o l )
% Usage :
%
[ p e r i o d s , medians , a m p l i t u d e s ]= calcPerMedAmp ( s o l )
%
% C a l c u l a t e p e r i o d s , medians , and a m p l i t u d e s o f s i m p l y p e r i o d i c
%
( i n c l u d i n g e q u i l i b r a t e d ) s t a t e v a r i a b l e s in the
%
dde23 ( ) / ddsd ( ) s o l u t i o n s o l .
% This a l g o r i t h m assumes i t i s a n a l y z i n g ( i n each component o f
213
%
s o l . y ) a time s e r i e s r e a c h i n g an e q u i l i b r i u m or a s i m p l y
%
p e r i o d i c b e h a v i o r w i t h a t l e a s t t h r e e i n t e r n a l peak−to−peak
%
o s c i l l a t i o n s ( two i n t e r n a l max and two i n t e r n a l min , i n
%
e i t h e r a l t e r n a t i n g order ) .
% Simply p e r i o d i c time s e r i e s have i s o l a t e d extrema , w i t h a
%
s i n g l e maximum and a s i n g l e minimum i n each f u n d a m e n t a l
%
period .
% Assumes (& r e q u i r e s ) a t l e a s t t h r e e d a t a p o i n t s i n s o l . x .
% E q u i l i b r i a t e d s t a t e v a r i a b l e s are assigned zero period ,
%
a l t h o u g h i n c e r t a i n s i t u a t i o n s an e q u i l i b r i u m may be
%
r e p r e s e n t e d by a p e r i o d i c s o l u t i o n w i t h v e r y s m a l l a m p l i t u d e ...
%
i n which c a s e t h e median e s t i m a t e s t h e e q u i l i b r i u m .
,
% T o l e r a n c e s ( componentwise )
r e l T o l= 1 e −2; % R e l a t i v e t o l e r a n c e f o r d e t e c t i o n o f ...
equilibrium
absTol= 1 e −2; % A b s o l u t e t o l e r a n c e f o r d e t e c t i o n o f ...
equilibrium
t o l= 1 e −2; % R e l a t i v e t o l e r a n c e f o r d e t e c t i o n o f p e r i o d i c ...
signal
[M N]= s i z e ( s o l . y ) ; % Record d i m e n s i o n s o f time−s e r i e s
minMask= zeros ( 1 , N) ; % Pre−a l l o c a t e
maxMask= minMask ; % Pre−a l l o c a t e
% D e f a u l t v a l u e s (NaN)
p e r i o d s= nan (M, 1 ) ; % P r o t e i n ( s ) & mRNA( s ) f o r each c e l l
214
medians= p e r i o d s ;
a m p l i t u d e s= medians ;
t a i l T i m e= s o l . params . s e t t l i n g T i m e / 5 ; % Somewhat a r b i t r a r y
t a i l S t a r t I n d e x= find ( ( s o l . x ( end ) − t a i l T i m e ) <= s o l . x , 1 ) ;
i f isempty ( t a i l S t a r t I n d e x )
t a i l S t a r t I n d e x= 1 ; % Use w h o le time−s e r i e s
end
% TBD: V e c t o r i z e f o r l o o p ?
fo r k= 1 :M
% Check f i r s t f o r an e q u i l i b r i u m
t a i l M e a n= mean( s o l . y ( k , t a i l S t a r t I n d e x : end ) ) ;
i f ( t a i l M e a n ˜= 0 )
i f a l l ( abs ( ( s o l . y ( k , t a i l S t a r t I n d e x : end ) − t a i l M e a n ) /...
tailMean ) < r e l T o l )
p e r i o d s ( k )= 0 ;
medians ( k )= t a i l M e a n ;
a m p l i t u d e s ( k )= 0 ;
continue ;
end
e l s e i f a l l ( abs ( s o l . y ( k , t a i l S t a r t I n d e x : end ) ) < absTol )
p e r i o d s ( k )= 0 ;
medians ( k )= 0 ;
a m p l i t u d e s ( k )= 0 ;
continue ;
end
215
% Not an e q u i l i b r i u m , or e q u i l i b r i u m not s u f f i c i e n t l y
%
” r e a c h e d ” , so c h e c k f o r a s i m p l y p e r i o d i c component
% Note : Assumes t h a t n e i t h e r minMask nor maxMask m a t r i c e s a r e
%
empty , and t h e i s o l a t e d extrema a ssump ti on i s key t o t h e
%
following detection logic .
% Find l o c a l minima
minMask ( 2 : end−1)= ( s o l . y ( k , 1 : end−2) >= s o l . y ( k , 2 : end−1) ) & ( ...
s o l . y ( k , 2 : end−1) <= s o l . y ( k , 3 : end ) ) ;
minMask ( 1 )= s o l . y ( k , 1 ) <= s o l . y ( k , 2 ) ;
minMask ( end )= s o l . y ( k , end−1) >= s o l . y ( k , end ) ;
m i n I n d i c e s= find ( minMask ) ;
% Find l o c a l maxima
maxMask ( 2 : end−1)= s o l . y ( k , 1 : end−2) <= s o l . y ( k , 2 : end−1) & s o l ...
. y ( k , 2 : end−1) >= s o l . y ( k , 3 : end ) ;
maxMask ( 1 )= s o l . y ( k , 1 ) <= s o l . y ( k , 2 ) ;
maxMask ( end )= s o l . y ( k , end−1) <= s o l . y ( k , end ) ;
m a x I n d i c e s= find ( maxMask ) ;
% C l e a r any i n i t i a l c o n s t a n t a r t i f a c t from c o n s t a n t h i s t o r y
f o r i= 1 : min( length ( m i n I n d i c e s ) , length ( m a x I n d i c e s ) )
i f m i n I n d i c e s ( i ) ˜= m a x I n d i c e s ( i )
m i n I n d i c e s= m i n I n d i c e s ( i : end ) ;
m a x I n d i c e s= m ax I n d i c e s ( i : end ) ;
break ;
end
end
% Check f o r a t l e a s t 1 . 5 o s c i l l a t i o n s f o r a n a l y s i s
i f isempty ( m i n I n d i c e s ) | | isempty ( m a x I n d i c e s )
216
%
d i s p ( [ ’ Omega l i m i t s e t not found i n component ’ num2str (...
k) ’; equilibrium ? ’] ) ;
continue ;
else
numEdgeMinima= ( m i n I n d i c e s ( 1 ) < m a x I n d i c e s ( 1 ) ) + ( ...
m i n I n d i c e s ( end ) == N) ;
numEdgeMaxima= ( m a x I n d i c e s ( 1 ) < m i n I n d i c e s ( 1 ) ) + ( ...
m ax I n d i c e s ( end ) == N) ;
end
i f ( ( length ( m i n I n d i c e s ) − numEdgeMinima <= 1 ) && ( length ( ...
m ax I n d i c e s ) − numEdgeMaxima <= 1 ) ) | | ( m i n I n d i c e s ( end ) == ...
m ax I n d i c e s ( end ) )
%
d i s p ( [ ’ Omega l i m i t s e t not found i n component ’ num2str (...
k) ’; equilibrium ? ’] ) ;
else % O s c i l l a t o r y − analyze using l a s t 1.5 o s c i l l a t i o n s
i f m i n I n d i c e s ( end ) == N % I g n o r e any l a s t minimum a t end
l a s t M i n I n d e x= m i n I n d i c e s ( end−1) ;
lastMaxIndex= m a x I n d i c e s ( end ) ;
penultimateMaxIndex= m a x I n d i c e s ( end−1) ;
e l s e i f m a x I n d i c e s ( end ) == N % I g n o r e any l a s t maximum a t ...
end
lastMaxIndex= m a x I n d i c e s ( end−1) ;
l a s t M i n I n d e x= m i n I n d i c e s ( end ) ;
penultimat eMinInde x= m i n I n d i c e s ( end−1) ;
e l s e % TBD: This doesn ’ t happen ?
i f s o l . x ( lastMaxIndex ) < s o l . x ( l a s t M i n I n d e x ) % L a s t ...
extremum i s a minimum
217
l a s t M i n I n d e x= m i n I n d i c e s ( end ) ;
penultimat eMinInde x= m i n I n d i c e s ( end−1) ;
lastMaxIndex= m a x I n d i c e s ( end ) ;
e l s e % L a s t extremum i s a maximum
lastMaxIndex= m a x I n d i c e s ( end ) ;
penultimateMaxIndex= m a x I n d i c e s ( end−1) ;
l a s t M i n I n d e x= m i n I n d i c e s ( end ) ;
end
end
% Function h a n d l e s f o r fminbnd ( )
s o l F n= @( x ) d e v a l ( s o l , x , k ) ;
minusSolFn= @( x ) −d e v a l ( s o l , x , k ) ;
% Try t o f i n d and c h a r a c t e r i z e a l i m i t c y c l e o s c i l l a t i o n
i f s o l . x ( lastMaxIndex ) < s o l . x ( l a s t M i n I n d e x ) % L a s t ...
extremum i s a minimum
% Use l a s t two minima and i n t e r v e n i n g maximum
% Use l e f t or r i g h t a d j a c e n t p o i n t d e p e n d i n g on s l o p e
i f s o l . yp ( k , l a s t M i n I n d e x ) < 0
xLastMin= fminbnd ( solFn , s o l . x ( l a s t M i n I n d e x ) , s o l . x ( ...
l a s t M i n I n d e x +1) ) ;
e l s e i f s o l . yp ( k , l a s t M i n I n d e x ) > 0
xLastMin= fminbnd ( solFn , s o l . x ( l a s t M i n I n d e x −1) , s o l . x ( ...
lastMinIndex ) ) ;
else
xLastMin= s o l . x ( l a s t M i n I n d e x ) ;
end
218
i f s o l . yp ( k , penultimat eMinInde x ) < 0
xPenultimateMin= fminbnd ( solFn , s o l . x ( ...
penultimat eMinInde x ) , s o l . x ( penultimat eMinInde x +1) ) ;
e l s e i f s o l . yp ( k , penultimat eMinInde x ) > 0
xPenultimateMin= fminbnd ( solFn , s o l . x ( ...
penultimateMinIndex −1) , s o l . x ( penultimat eMinInde x ) ) ;
else
xPenultimateMin= s o l . x ( penultimat eMinInde x ) ;
end
% l a s t M a x I n d e x s h o u l d be i n b e t w e e n
% I n v e r t time s e r i e s so t h a t maxima become minima
i f s o l . yp ( k , lastMaxIndex ) < 0
xLastMax= fminbnd ( minusSolFn , s o l . x ( lastMaxIndex −1) , ...
s o l . x ( lastMaxIndex ) ) ;
e l s e i f s o l . yp ( k , lastMaxIndex ) > 0
xLastMax= fminbnd ( minusSolFn , s o l . x ( lastMaxIndex ) , s o l ...
. x ( lastMaxIndex +1) ) ;
else
xLastMax= s o l . x ( lastMaxIndex ) ;
end
l a s t M i n= d e v a l ( s o l , xLastMin , k ) ;
lastMax= d e v a l ( s o l , xLastMax , k ) ;
penultimateMin= d e v a l ( s o l , xPenultimateMin , k ) ;
l a s t A m p l i t u d e= lastMax − l a s t M i n ;
p e n u l t i m a t e A m p l i t u d e= lastMax − penultimateMin ;
219
i f ( l a s t A m p l i t u d e ˜= 0 ) && ( abs ( ( l a s t A m p l i t u d e − ...
penultimateAmplitude ) / lastAmplitude ) < t o l )
p e r i o d s ( k )= xLastMin − xPenultimateMin ;
medians ( k )= ( l a s t M i n + lastMax ) / 2 ;
a m p l i t u d e s ( k )= l a s t A m p l i t u d e ;
else
%
d i s p ( [ ’ Omega l i m i t s e t not found i n component ’ ...
num2str ( k ) ’ ; damped o s c i l l a t i o n s ? ’ ] ) ;
end
e l s e % L a s t extremum i s a maximum
% Use l a s t two maxima and i n t e r v e n i n g minimum
% Use l e f t or r i g h t a d j a c e n t p o i n t d e p e n d i n g on s l o p e
% Time s e r i e s i s i n v e r t e d t o t u r n maxima i n t o minima
i f s o l . yp ( k , lastMaxIndex ) < 0
xLastMax= fminbnd ( minusSolFn , s o l . x ( lastMaxIndex −1) , ...
s o l . x ( lastMaxIndex ) ) ;
e l s e i f s o l . yp ( k , lastMaxIndex ) > 0
xLastMax= fminbnd ( minusSolFn , s o l . x ( lastMaxIndex ) , s o l ...
. x ( lastMaxIndex +1) ) ;
else
xLastMax= s o l . x ( lastMaxIndex ) ;
end
i f s o l . yp ( k , penultimateMaxIndex ) < 0
xPenultimateMax= fminbnd ( minusSolFn , s o l . x ( ...
penultimateMaxIndex −1) , s o l . x ( penultimateMaxIndex ) ) ;
220
e l s e i f s o l . yp ( k , penultimateMaxIndex ) > 0
xPenultimateMax= fminbnd ( minusSolFn , s o l . x ( ...
penultimateMaxIndex ) , s o l . x ( penultimateMaxIndex +1) ) ;
else
xPenultimateMax= s o l . x ( penultimateMaxIndex ) ;
end
% l a s t M i n I n d e x s h o u l d be i n b e t w e e n
i f s o l . yp ( k , l a s t M i n I n d e x ) < 0
xLastMin= fminbnd ( solFn , s o l . x ( l a s t M i n I n d e x ) , s o l . x ( ...
l a s t M i n I n d e x +1) ) ;
e l s e i f s o l . yp ( k , l a s t M i n I n d e x ) > 0
xLastMin= fminbnd ( solFn , s o l . x ( l a s t M i n I n d e x −1) , s o l . x ( ...
lastMinIndex ) ) ;
else
xLastMin= s o l . x ( l a s t M i n I n d e x ) ;
end
lastMax= d e v a l ( s o l , xLastMax , k ) ;
l a s t M i n= d e v a l ( s o l , xLastMin , k ) ;
penultimateMax= d e v a l ( s o l , xPenultimateMax , k ) ;
l a s t A m p l i t u d e= lastMax − l a s t M i n ;
p e n u l t i m a t e A m p l i t u d e= penultimateMax − l a s t M i n ;
i f ( l a s t A m p l i t u d e ˜= 0 ) && ( abs ( ( l a s t A m p l i t u d e − ...
penultimateAmplitude ) / lastAmplitude ) < t o l )
p e r i o d s ( k )= xLastMax − xPenultimateMax ;
medians ( k )= ( l a s t M i n + lastMax ) / 2 ;
221
a m p l i t u d e s ( k )= l a s t A m p l i t u d e ;
else
%
d i s p ( [ ’ Omega l i m i t s e t not found i n component ’ ...
num2str ( k ) ’ ; damped o s c i l l a t i o n s ? ’ ] ) ;
end
end
end
end
end
sigmoidalGradient.m: This computes the total protein level using a sigmoidal spatiotemporal gradient.
function H13Total= s i g m o i d a l G r a d i e n t ( t , H13TotalMax , H13TotalMin ...
, H13TotalHalfLife )
% Usage :
%
H13Total= s i g m o i d a l G r a d i e n t ( t , H13TotalMax , H13TotalMin , ...
H13TotalHalfLife )
%
% C a l c u l a t e s t o t a l Her13 . 2 g r a d i e n t a t time t .
% 2D V e c t o r i z e d b a s e d on d i m e n s i o n s o f t :
%
1) The f i r s t ( row ) dimension r e p r e s e n t s c e l l number .
%
2) The second ( column ) dimension r e p r e s e n t s time .
%
% H13TotalMax , H13TotalMin , and H 1 3 T o t a l H a l f L i f e may be s c a l a r s
%
or column v e c t o r s , and a r e assumed t o have t h e same
222
%
d i m e n s i o n s . I f t h e y a r e column v e c t o r s , t h e n t h e y s h o u l d have
%
t h e same number o f rows as t .
%
% This f u n c t i o n c a l c u l a t e s :
%
%
%
%
I f t <= 0
H13Total= H13TotalMax
E l s e i f 0 < t <= H 1 3 T o t a l H a l f L i f e
H13Total= H13TotalMin + ( H13TotalMax − H13TotalMin ) /2 ∗ (2 ...
− exp ( 2 ∗ ( 1 − H 1 3 T o t a l H a l f L i f e / t ) ) )
%
%
E l s e i f H 1 3 T o t a l H a l f L i f e <= t < 2∗ H 1 3 T o t a l H a l f L i f e
H13Total= H13TotalMin + ( H13TotalMax − H13TotalMin ) /2 ∗ exp...
( 2 ∗ ( 1 + H 1 3 T o t a l H a l f L i f e /( t − 2∗ H 1 3 T o t a l H a l f L i f e ) ) )
%
%
%
Else
H13Total= H13TotalMin
End
i f i s s c a l a r ( H13TotalMax )
H13Total= H13TotalMax∗ o n e s ( s i z e ( t ) ) ; % Pre−a l l o c a t e
tMask= ( 0 < t ) & ( t <= H 1 3 T o t a l H a l f L i f e ) ;
H13Total ( tMask )= H13TotalMin + ( H13TotalMax − H13TotalMin ) ...
/ 2 ∗ ( 2 − exp ( 2 ∗ ( 1 − H 1 3 T o t a l H a l f L i f e . / t ( tMask ) ) ) ) ;
tMask= ( H 1 3 T o t a l H a l f L i f e < t ) & ( t < 2∗ H 1 3 T o t a l H a l f L i f e ) ;
H13Total ( tMask )= H13TotalMin + ( H13TotalMax − H13TotalMin ) ...
/2∗exp ( 2 ∗ ( 1 + H 1 3 T o t a l H a l f L i f e . / ( t ( tMask ) − 2∗ H 1 3 T o t a l H a l f L i f e ) ) ...
);
223
tMask= ( 2 ∗ H 1 3 T o t a l H a l f L i f e <= t ) ;
H13Total ( tMask )= H13TotalMin ;
else
t S i z e= s i z e ( t ) ;
i f t S i z e ( 2 ) > 1 % Match second dimension l e n g t h
H13TotalMax= repmat ( H13TotalMax , 1 , t S i z e ( 2 ) ) ;
H13TotalMin= repmat ( H13TotalMin , 1 , t S i z e ( 2 ) ) ;
H 1 3 T o t a l H a l f L i f e= repmat ( H 1 3 T o t a l H a l f L i f e , 1 , t S i z e ( 2 ) ) ;
end
H13Total= H13TotalMax ; % Pre−a l l o c a t e
tMask= ( 0 < t ) & ( t <= H 1 3 T o t a l H a l f L i f e ) ;
H13Total ( tMask )= H13TotalMin ( tMask ) + ( H13TotalMax ( tMask ) − ...
H13TotalMin ( tMask ) ) / 2 . ∗ ( 2 − exp ( 2 ∗ ( 1 − H 1 3 T o t a l H a l f L i f e ( tMask ) . / ...
t ( tMask ) ) ) ) ;
tMask= ( H 1 3 T o t a l H a l f L i f e < t ) & ( t < 2∗ H 1 3 T o t a l H a l f L i f e ) ;
H13Total ( tMask )= H13TotalMin ( tMask ) + ( H13TotalMax ( tMask ) − ...
H13TotalMin ( tMask ) ) / 2 . ∗ exp ( 2 ∗ ( 1 + H 1 3 T o t a l H a l f L i f e ( tMask ) . / ( t ( ...
tMask ) − 2∗ H 1 3 T o t a l H a l f L i f e ( tMask ) ) ) ) ;
tMask= ( 2 ∗ H 1 3 T o t a l H a l f L i f e <= t ) ;
H13Total ( tMask )= H13TotalMin ( tMask ) ;
end
end
224
exponentialGradient.m: This computes the total protein level using an exponential spatiotemporal gradient.
function H13Total= e x p o n e n t i a l G r a d i e n t ( t , H13TotalMax , ...
H13TotalMin , H 1 3 T o t a l H a l f L i f e )
% Usage :
%
H13Total= e x p o n e n t i a l G r a d i e n t ( t , H13TotalMax , H13TotalMin , ...
H13TotalHalfLife )
%
% C a l c u l a t e s t o t a l Her13 . 2 g r a d i e n t a t time t .
% 2D V e c t o r i z e d b a s e d on d i m e n s i o n s o f t :
%
1) The f i r s t ( row ) dimension r e p r e s e n t s c e l l number .
%
2) The second ( column ) dimension r e p r e s e n t s time .
%
% H13TotalMax , H13TotalMin , and H 1 3 T o t a l H a l f L i f e may be s c a l a r s
%
or column v e c t o r s , and a r e assumed t o have t h e same
%
d i m e n s i o n s . I f t h e y a r e column v e c t o r s , t h e n t h e y s h o u l d have
%
t h e same number o f rows as t .
%
% This f u n c t i o n c a l c u l a t e s :
%
%
%
%
I f t <= 0
H13Total= H13TotalMax
Else
H13Total= H13TotalMin + ( H13TotalMax − H13TotalMin ) ∗ exp(−...
ln (2) ∗ t / H13TotalHalfLife )
%
End
225
i f i s s c a l a r ( H13TotalMax )
H13Total= H13TotalMax∗ o n e s ( s i z e ( t ) ) ; % Pre−a l l o c a t e
tMask= ( 0 < t ) ;
H13Total ( tMask )= H13TotalMin + ( H13TotalMax − H13TotalMin ) ...
∗...
exp(−log ( 2 ) ∗ t ( tMask ) / H 1 3 T o t a l H a l f L i f e ) ;
else
t S i z e= s i z e ( t ) ;
i f t S i z e ( 2 ) > 1 % Match second dimension l e n g t h
H13TotalMax= repmat ( H13TotalMax , 1 , t S i z e ( 2 ) ) ;
H13TotalMin= repmat ( H13TotalMin , 1 , t S i z e ( 2 ) ) ;
H 1 3 T o t a l H a l f L i f e= repmat ( H 1 3 T o t a l H a l f L i f e , 1 , t S i z e ( 2 ) ) ;
end
H13Total= H13TotalMax ; % Pre−a l l o c a t e
tMask= ( 0 < t ) ;
H13Total ( tMask )= H13TotalMin ( tMask ) + . . .
( H13TotalMax ( tMask ) − H13TotalMin ( tMask ) ) / 2 . ∗ . . .
exp(−log ( 2 ) ∗ t ( tMask ) . / H 1 3 T o t a l H a l f L i f e ( tMask ) ) ;
end
end
zeroCrossingEvent.m: This short routine, called internally by ddesd or
dde23, ensures that computed solutions are non-negative.
226
function [ value , i s T e r m i n a l , d i r e c t i o n ]= z e r o C r o s s i n g E v e n t ( t , z , ...
Z)
% Stop i n t e g r a t i o n i f t h e r e i s a z e r o c r o s s i n g b e c a u s e ...
solutions
%
s h o u l d be non−n e g a t i v e . Going n e g a t i v e => s o m e t h i n g i s wrong...
.
v a l u e= z ; % D e t e c t z e r o c r o s s i n g i n any component
i s T e r m i n a l= o n e s ( s i z e ( z ) ) ; % Stop t h e i n t e g r a t i o n
d i r e c t i o n= −o n e s ( s i z e ( z ) ) ; % N e g a t i v e d i r e c t i o n
end
Parameter Estimation Code
parameterSamplerParallel.m: For parameter estimation and model selection purposes, this code runs multiple simulations using somitogenesisPCW.m at parameter combinations selected from a uniformly distributed parameter space. Matlab’s parfor loop structure is used to run simulations with different parameters in
parallel on multiple processors, if they are available.
function sample= p a r a m e t e r S a m p l e r P a r a l l e l ( argIn1 , v a r a r g i n )
%
% Usage :
%
%
sample= p a r a m e t e r S a m p l e r P a r a l l e l ( argIn1 , v a r a r g i n )
227
% Compares one and two b i n d i n g s i t e s c e n a r i o s f o r a random ...
sample
%
o f s e l e c t p a r a m e t e r s . L i n e a r t o t a l p r o t e i n decay v s .
%
d i f f e r e n t i a l decay i s chosen w i t h a b o o l e a n v a r i a b l e b e l o w . ...
An
%
range from an e x i s t i n g parameter sample may be used by ...
passing
%
t h e parameter sample s t r u c t i n a r g I n 1 and t h e f i r s t and l a s t
%
sample i n d i c e s t o use i n t h e second and t h i r d arguments ,
%
r e s p e c t i v e l y . Otherwise , p a s s i n g a s i n g l e p o s i t i v e i n t e g e r i n
%
a r g I n 1 g e n e r a t e s a new parameter sample o f s i z e a r g I n 1 .
%
S i m u l a t i o n s f o r d i f f e r e n t s a m p l e s can run i n p a r a l l e l by use
%
of the parfor loop .
parameterSamplerTic= t i c ;
% Initialize
timeStamp= d a t e s t r ( now , 3 0 ) ;
H13TotalMaxSweepStepSize= 1 0 ;
H13TotalMaxSweepLimit= 2 0 0 0 ;
l i n e a r D e c a y B o o l= f a l s e ; % l i n e a r OR d i f f e r e n t i a l decay
% Parameters o b j e c t t o b r o a d c a s t a c r o s s p a r f o r l o o p s
paramsBroadcast= Params ( f a l s e ) ; % f a l s e => no parameter n o i s e
paramsBroadcast . t i m e I n t e r v a l= [ 0 0 ] ;
paramsBroadcast . s e t t l i n g T i m e= 5 0 0 ;
paramsBroadcast . fastModeBool= t r u e ;
% Parameter sample r a n g e s ( params i s a s t r u c t )
228
params . rhoRange= [ 1 / 3 3 ] ;
params . kH13H13LogRange= [ 1 3 ] ;
params . kH7H7LogRange= [ 1 3 ] ;
params . kH7H13LogRange= [ 1 3 ] ;
params . tau h7Range= [ 2 . 3 8 . 1 ] ;
params . tau H7Range= paramsBroadcast . tau H7Mean ;
params . aH7Range= paramsBroadcast . aH7Mean ;
params . bH7Range= [ 0 . 2 0 0 . 5 0 ] ;
i f linearDecayBool
params . bH7H7Range= params . bH7Range ;
params . bH7H13Range= params . bH7Range ;
else
params . bH7H7Range= 0 ;
params . bH7H13Range= 0 ;
end
params . rhoMean= params . rhoRange ( 1 ) ;
params . gH7Range ( 2 )= paramsBroadcast . gH7Mean ;
params . rhoMean= params . rhoRange ( 2 ) ;
params . gH7Range ( 1 )= paramsBroadcast . gH7Mean ;
params . rH7H7LogRange= [−2 0 ] ;
params . wH7H7 H7H7LogRange= [ 0 2 ] ;
params . dH7Range= paramsBroadcast . dH7Mean ;
params . tau dRange= paramsBroadcast . tau dMean ;
params . tau DRange= paramsBroadcast . tau DMean ;
params . aDRange= paramsBroadcast . aDMean ;
params . bDRange= paramsBroadcast . bDMean ;
params . rhoMean= params . rhoRange ( 1 ) ;
229
params . gDRange ( 2 )= paramsBroadcast . gDMean ;
params . rhoMean= params . rhoRange ( 2 ) ;
params . gDRange ( 1 )= paramsBroadcast . gDMean ;
params . rNLogRange= [−1 1 ] ;
params . wNRange= paramsBroadcast . wNMean ;
params . wN H7H7Range= paramsBroadcast . wN H7H7Mean ;
params . wN H7H7 H7H7Range= paramsBroadcast . wN H7H7 H7H7Mean ;
params . dDRange= paramsBroadcast . dDMean ;
params . H13TotalMaxSweep= 0 : H13TotalMaxSweepStepSize : ...
H13TotalMaxSweepLimit ;
H13TotalMaxSweepLength= length ( params . H13TotalMaxSweep ) ;
i f ˜ i s s t r u c t ( a r g I n 1 ) % Generate a random sample o f s i z e a r g I n 1
sample . s a m p l e S i z e= a r g I n 1 ;
% Generate s a m p l i n g from uniform d i s t r i b u t i o n s
[ params . rhoStream , params . kH13H13Stream , params . kH7H7Stream , ...
params . kH7H13Stream , params . tau h7Stream , params . bH7Stream , ...
params . rH7H7Stream , params . wH7H7 H7H7Stream]= RandStream . c r e a t e ( ...
’mrg32k3a ’ , ’NumStreams ’ , 8 , ’Seed ’ ,sum( 1 0 0 ∗ clock ) ) ;
rhoSample= params . rhoRange ( 1 ) + ( params . rhoRange ( 2 ) − params...
. rhoRange ( 1 ) ) ∗rand ( params . rhoStream , sample . s a m p l e S i z e , 1 ) ;
kH13H13Sample= 1 0 . ˆ ( params . kH13H13LogRange ( 1 ) + ( params . ...
kH13H13LogRange ( 2 ) − params . kH13H13LogRange ( 1 ) ) ∗rand ( params . ...
kH13H13Stream , sample . s a m p l e S i z e , 1 ) ) ;
kH7H7Sample= 1 0 . ˆ ( params . kH7H7LogRange ( 1 ) + ( params . ...
kH7H7LogRange ( 2 ) − params . kH7H7LogRange ( 1 ) ) ∗rand ( params . ...
kH7H7Stream , sample . s a m p l e S i z e , 1 ) ) ;
230
kH7H13Sample= 1 0 . ˆ ( params . kH7H13LogRange ( 1 ) + ( params . ...
kH7H13LogRange ( 2 ) − params . kH7H13LogRange ( 1 ) ) ∗rand ( params . ...
kH13H13Stream , sample . s a m p l e S i z e , 1 ) ) ;
tau h7Sample= params . tau h7Range ( 1 ) + ( params . tau h7Range ( 2 ) ...
− params . tau h7Range ( 1 ) ) ∗rand ( params . tau h7Stream , sample . ...
sampleSize , 1) ;
bH7Sample= params . bH7Range ( 1 ) + ( params . bH7Range ( 2 ) − params...
. bH7Range ( 1 ) ) ∗rand ( params . bH7Stream , sample . s a m p l e S i z e , 1 ) ;
rH7H7Sample= 1 0 . ˆ ( params . rH7H7LogRange ( 1 ) + ( params . ...
rH7H7LogRange ( 2 ) − params . rH7H7LogRange ( 1 ) ) ∗rand ( params . ...
rH7H7Stream , sample . s a m p l e S i z e , 1 ) ) ;
wH7H7 H7H7Sample= 1 0 . ˆ ( params . wH7H7 H7H7LogRange ( 1 ) + ( ...
params . wH7H7 H7H7LogRange ( 2 ) − params . wH7H7 H7H7LogRange ( 1 ) ) ∗...
rand ( params . wH7H7 H7H7Stream , sample . s a m p l e S i z e , 1 ) ) ;
rNSample= params . rH7H7Sample / 1 0 ;
e l s e % Use g i v e n sample
paramsIn= a r g I n 1 ;
i f nargin == 3
s t a r t I n d e x= v a r a r g i n { 1 } ;
endIndex= v a r a r g i n { 2 } ;
sample . s a m p l e S i z e= length ( s t a r t I n d e x : endIndex ) ;
else
sample . s a m p l e S i z e= length ( paramsIn . rho ) ;
s t a r t I n d e x= 1 ;
endIndex= sample . s a m p l e S i z e ;
end
231
% Generate s a m p l i n g from e x i s t i n g sample , WIP: M i g r a t e t o ...
Streams
rhoSample= paramsIn . rho ( s t a r t I n d e x : endIndex ) ;
kH13H13Sample= paramsIn . kH13H13 ( s t a r t I n d e x : endIndex ) ;
kH7H7Sample= paramsIn . kH7H7 ( s t a r t I n d e x : endIndex ) ;
kH7H13Sample= paramsIn . kH7H13 ( s t a r t I n d e x : endIndex ) ;
tau h7Sample= paramsIn . t a u h 7 ( s t a r t I n d e x : endIndex ) ;
bH7Sample= paramsIn . bH7 ( s t a r t I n d e x : endIndex ) ;
rH7H7Sample= paramsIn . rH7H7 ( s t a r t I n d e x : endIndex ) ;
wH7H7 H7H7Sample= paramsIn . wH7H7 H7H7( s t a r t I n d e x : endIndex ) ;
rNSample= rH7H7Sample / 1 0 ;
end
% Initialize
o n e S i t e P r o b l e m P o i n t s= [ ] ;
oneSiteH13TotalMaxSweepLimitPoints= [ ] ;
t w o S i t e P r o b l e m P o i n t s= [ ] ;
twoSiteH13TotalMaxSweepLimitPoints= [ ] ;
o n e S i t e P e r i o d s= zeros ( [ paramsBroadcast . numEquations ...
H13TotalMaxSweepLength sample . s a m p l e S i z e ] ) ;
o n e S i t e M e d i a n s= o n e S i t e P e r i o d s ;
o n e S i t e A m p l i t u d e s= o n e S i t e P e r i o d s ;
t w o S i t e P e r i o d s= o n e S i t e P e r i o d s ;
twoSiteMedians= o n e S i t e P e r i o d s ;
t w o S i t e A m p l i t u d e s= o n e S i t e P e r i o d s ;
% Run s i m u l a t i o n f o r parameter sample , f o r b o t h one & two
232
%
b i n d i n g s i t e s ; p a r f o r l o o p s can e x e c u t e i n p a r a l l e l and i n
%
any o r d e r .
p a r f o r sampleCtr= 1 : sample . s a m p l e S i z e
paramsLocal= paramsBroadcast ;
% I n i t i a l i z e f o r 1 or 2 b i n d i n g s i t e s
paramsLocal . rhoMean= rhoSample ( sampleCtr ) ;
paramsLocal . kH13H13Mean= kH13H13Sample ( sampleCtr ) ;
paramsLocal . kH7H7Mean= kH7H7Sample ( sampleCtr ) ;
paramsLocal . kH7H13Mean= kH7H13Sample ( sampleCtr ) ;
paramsLocal . tau h7Mean= tau h7Sample ( sampleCtr ) ;
paramsLocal . bH7Mean= bH7Sample ( sampleCtr ) ;
i f linearDecayBool
paramsLocal . bH7H7Mean= paramsLocal . bH7Mean ;
paramsLocal . bH7H13Mean= paramsLocal . bH7Mean ;
else
paramsLocal . bH7H7Mean= 0 ;
paramsLocal . bH7H13Mean= 0 ;
end
paramsLocal . rH7H7Mean= rH7H7Sample ( sampleCtr ) ;
paramsLocal . wH7H7 H7H7Mean= wH7H7 H7H7Sample ( sampleCtr ) ;
paramsLocal . rNMean= rNSample ( sampleCtr ) ;
% One b i n d i n g −s i t e
paramsLocal . numBindingSites= 1 ;
[ o n e S i t e P e r i o d s ( : , : , sampleCtr ) , o n e S i t e M e d i a n s ( : , : , sampleCtr...
) , o n e S i t e A m p l i t u d e s ( : , : , sampleCtr ) , nanFlag , s w e e p L i m i t F l a g ]= ...
sweepH13TotalMax ( paramsLocal , params . H13TotalMaxSweep ) ;
233
i f nanFlag
o n e S i t e P r o b l e m P o i n t s= [ o n e S i t e P r o b l e m P o i n t s sampleCtr ] ;
e l s e i f s w e e p L i m i t F l ag
oneSiteH13TotalMaxSweepLimitPoints= [ ...
oneSiteH13TotalMaxSweepLimitPoints sampleCtr ] ;
end
% Two b i n d i n g −s i t e s
paramsLocal . numBindingSites= 2 ;
[ t w o S i t e P e r i o d s ( : , : , sampleCtr ) , twoSiteMedians ( : , : , sampleCtr...
) , t w o S i t e A m p l i t u d e s ( : , : , sampleCtr ) , nanFlag , s w e e p L i m i t Fl a g ]= ...
sweepH13TotalMax ( paramsLocal , params . H13TotalMaxSweep ) ;
i f nanFlag
t w o S i t e P r o b l e m P o i n t s= [ t w o S i t e P r o b l e m P o i n t s sampleCtr ] ;
e l s e i f s w e e p L i m i t F l ag
twoSiteH13TotalMaxSweepLimitPoints= [ ...
twoSiteH13TotalMaxSweepLimitPoints sampleCtr ] ;
end
end
params . rhoSample= rhoSample ;
params . kH13H13Sample= kH13H13Sample ;
params . kH7H7Sample= kH7H7Sample ;
params . kH7H13Sample= kH7H13Sample ;
params . tau h7Sample= tau h7Sample ;
params . bH7Sample= bH7Sample ;
params . rH7H7Sample= rH7H7Sample ;
234
params . wH7H7 H7H7Sample= wH7H7 H7H7Sample ;
params . rNSample= rNSample ;
sample . params= params ;
sample . o n e S i t e . p e r i o d s=o n e S i t e P e r i o d s ;
sample . o n e S i t e . medians= o n e S i t e M e d i a n s ;
sample . o n e S i t e . a m p l i t u d e s= o n e S i t e A m p l i t u d e s ;
sample . o n e S i t e . p r o b l e m P o i n t s= o n e S i t e P r o b l e m P o i n t s ;
sample . o n e S i t e . H13TotalMaxSweepLimitPoints= ...
oneSiteH13TotalMaxSweepLimitPoints ;
sample . t w o S i t e . p e r i o d s= t w o S i t e P e r i o d s ;
sample . t w o S i t e . medians= twoSiteMedians ;
sample . t w o S i t e . a m p l i t u d e s= t w o S i t e A m p l i t u d e s ;
sample . t w o S i t e . p r o b l e m P o i n t s= t w o S i t e P r o b l e m P o i n t s ;
sample . t w o S i t e . H13TotalMaxSweepLimitPoints= ...
twoSiteH13TotalMaxSweepLimitPoints ;
toc ( parameterSamplerTic )
% Save sample t o a f i l e w i t h a time−stamp
[ s u c c e s s , message , messageID ]= mkdir ( ’datasets ’ ) ;
if success
save ( [ ’datasets ’ f i l e s e p ’parameterSample_ ’ timeStamp ’_’ ...
num2str ( s t a r t I n d e x ) ’to ’ num2str ( endIndex ) ] , ’sample ’ ) ;
else
disp ( ’WARNING : Failed to save the dataset to disk.’ ) ;
end
235
% Warn a b o u t problem p o i n t s
i f ˜ ( isempty ( sample . o n e S i t e . p r o b l e m P o i n t s ) && isempty ( sample . ...
twoSite . problemPoints ) )
disp ( ’WARNING : There are problem points !’ ) ;
end
end
parameterStatistics.m: This code statistically analyzes parameter sampling results for modeling scenarios I–IV using outputs from parameterSamplerParallel.m.
The results of these analyses enable model selection.
function p a r a m e t e r S t a t i s t i c s ( n o n l i n e a r S a m p l e , l i n e a r S a m p l e )
% Usage :
%
p a r a m e t e r S t a t i s t i c s ( nonlinearSample , linearSample )
%
% This f u n c t i o n d o e s a c o m p a r a t i v e s t a t i s t i c a l a n a l y s i s on model
%
s c e n a r i o s I−IV . Each o f t h e two i n p u t d a t a s e t s ,
%
n o n l i n e a r S a m p l e and l i n e a r S a m p l e , i n c l u d e s one and two
%
b i n d i n g s i t e d a t a . n o n l i n e a r S a m p l e and l i n e a r S a m p l e a r e
%
r e q u i r e d t o have t h e same dimension d a t a s e t s .
disp ( ’ ’ ) ;
i f ˜isempty ( n o n l i n e a r S a m p l e . o n e S i t e . p r o b l e m P o i n t s ) | | ˜isempty...
( n o n l i n e a r S a m p l e . t w o S i t e . p r o b l e m P o i n t s ) | | ˜isempty ( l i n e a r S a m p l e ...
236
. o n e S i t e . p r o b l e m P o i n t s ) | | ˜isempty ( l i n e a r S a m p l e . t w o S i t e . ...
problemPoints )
disp ( ’WARNING : There are problem points !’ ) ;
disp ( ’ ’ ) ;
end
amplitudeThresh= 5 ; % R e q u i r e a t l e a s t t h i s peak−to−peak ...
a m p l i t u d e o f mRNA & p r o t e i n m o l e c u l e s
p e r i o d T o l= 0 . 0 1 ; % R e q u i r e t h a t mRNA & p r o t e i n o s c i l l a t i o n ...
periods agree to within t h i s
t a r g e t P e r i o d= 3 0 ; % D e s i r e d p e r i o d o f o s c i l l a t i o n
d e l t a P e r i o d= 3 ; % A l l o w a b l e +/− range i n o s c i l l a t i o n p e r i o d
t a r g e t D e l t a= 1 5 ; % F i l t e r o u t c h a n g e s i n p e r i o d l e s s than t h i s
[ numEquations numSweeps numSamples]= s i z e ( n o n l i n e a r S a m p l e . ...
oneSite . amplitudes ) ;
% One b i n d i n g s i t e , n o n l i n e a r decay
disp ( ’MODEL I, 1 binding site , nonlinear decay :’ ) ;
i f ˜isempty ( n o n l i n e a r S a m p l e . o n e S i t e . ...
H13TotalMaxSweepLimitPoints )
disp ( ’ Notice : H13TotalMax sweep limit reached .’ ) ;
end
% P e r i o d mask
periodMeans1n= mean( n o n l i n e a r S a m p l e . o n e S i t e . p e r i o d s , 1 ) ;
f u l l P e r i o d M e a n s 1 n = repmat ( periodMeans1n , [ numEquations , 1 , ...
1]) ;
237
periodMeans1n= s q u e e z e ( periodMeans1n ) ’ ;
periodMask1n= zeros ( s i z e ( f u l l P e r i o d M e a n s 1 n ) ) ;
periodMask1n ( f u l l P e r i o d M e a n s 1 n ˜= 0 )= abs ( ( n o n l i n e a r S a m p l e . ...
o n e S i t e . p e r i o d s ( f u l l P e r i o d M e a n s 1 n ˜= 0 ) − f u l l P e r i o d M e a n s 1 n ( ...
f u l l P e r i o d M e a n s 1 n ˜= 0 ) ) . / f u l l P e r i o d M e a n s 1 n ( f u l l P e r i o d M e a n s 1 n ˜=...
0) ) < periodTol ;
periodMask1n ( f u l l P e r i o d M e a n s 1 n == 0 )= abs ( n o n l i n e a r S a m p l e . ...
o n e S i t e . p e r i o d s ( f u l l P e r i o d M e a n s 1 n == 0 ) ) < p e r i o d T o l ;
periodMask1n= s q u e e z e ( a l l ( periodMask1n , 1 ) ) ’ ;
clear fullPeriodMeans1n
% Amplitude mask
amplitudeMask1n= s q u e e z e ( a l l ( amplitudeThresh <=n o n l i n e a r S a m p l e ...
. oneSite . amplitudes , 1) ) ’ ;
% Combine masks and f i n d o s c i l l a t i n g p r o p o r t i o n
fullOscMask1n= periodMask1n & amplitudeMask1n ;
oscMask1n= any ( fullOscMask1n , 2 ) ;
o s c I n d i c e s 1 n= find ( oscMask1n ) ;
oscProp1n= length ( o s c I n d i c e s 1 n ) /numSamples ;
disp ( [ ’Oscillating proportion : ’ num2str ( length ( o s c I n d i c e s 1 n ) ...
) ’/’ num2str ( numSamples ) ’= ’ num2str ( oscProp1n ) ] ) ;
[ maxArray1n , maxIndices1n ]= max( fullOscMask1n . ∗ periodMeans1n , ...
[ ] , 2) ;
globalMax1n= max( maxArray1n ) ;
minArray1n= zeros ( numSamples , 1 ) ;
m i n I n d i c e s 1 n= o n e s ( numSamples , 1 ) ;
fo r k= 1 : numSamples
238
% I g n o r e z e r o p e r i o d s ; p o s i t i v e I n d i c e s v e c t o r s h o u l d c o n s i s t ...
o f 0 or more ones f o l l o w e d by 0 or more z e r o s
p o s i t i v e I n d i c e s= fullOscMask1n ( k , : ) > 0 ;
i f any ( p o s i t i v e I n d i c e s )
[ minArray1n ( k ) , m i n I n d i c e s 1 n ( k ) ]= min( periodMeans1n ( k , ...
positiveIndices ) ) ;
end
end
globalMin1n= min( minArray1n ( 0 < minArray1n ) ) ;
i f isempty ( globalMin1n )
disp ( ’No period range .’ ) ;
else
disp ( [ ’Nontrivial period range : [’ num2str ( globalMin1n ) ’,...
’ num2str ( globalMax1n ) ’]’ ] ) ;
end
[ deltaMax1n , deltaMaxIndex1n ]= max( maxArray1n − minArray1n ) ;
i f ˜isempty ( o s c I n d i c e s 1 n )
disp ( [ ’Maximum Delta T: [’ num2str ( minArray1n ( ...
deltaMaxIndex1n ) ) ’, ’ num2str ( maxArray1n ( deltaMaxIndex1n ) ) ’],...
Delta T= ’ num2str ( deltaMax1n ) ’, k= ’ num2str ( deltaMaxIndex1n ) ...
’, j= [’ num2str ( m i n I n d i c e s 1 n ( deltaMaxIndex1n ) ) ’,’ num2str ( ...
maxIndices1n ( deltaMaxIndex1n ) ) ’]’ ] ) ;
else
disp ( ’Maximum Delta T: NaN ’ ) ;
end
239
% Analyze s u b s e t o f d a t a w i t h t a i l b u d p e r i o d i n d e s i r e d range
%
Uses IVT which assumes p e r i o d i s a c t s . f n . o f H13TotalMax
periodOscMask1n= ( minArray1n <= t a r g e t P e r i o d + d e l t a P e r i o d ) & ...
( t a r g e t P e r i o d − d e l t a P e r i o d <= maxArray1n ) ;
p e r i o d O s c I n d i c e s 1 n= find ( periodOscMask1n ) ;
periodOscProp1n= length ( p e r i o d O s c I n d i c e s 1 n ) /numSamples ;
disp ( [ num2str ( t a r g e t P e r i o d ) ’ +/- ’ num2str ( d e l t a P e r i o d ) ’ ...
min period oscillating proportion : ’ num2str ( length ( ...
p e r i o d O s c I n d i c e s 1 n ) ) ’/’ num2str ( numSamples ) ’= ’ num2str ( ...
periodOscProp1n ) ] ) ;
periodMaxArray1n= periodOscMask1n . ∗ maxArray1n ;
p e r i o d M a x I n d i c e s 1 n= maxIndices1n ;
p e r i o d M a x I n d i c e s 1 n ( periodOscMask1n == 0 )= 1 ;
periodMinArray1n= periodOscMask1n . ∗ minArray1n ;
p e r i o d M i n I n d i c e s 1 n= m i n I n d i c e s 1 n ;
p e r i o d M i n I n d i c e s 1 n ( periodOscMask1n == 0 )= 1 ;
p e r i o d D e l t a A r r a y 1 n= periodMaxArray1n − periodMinArray1n ;
[ periodDeltaMax1n , periodDeltaMaxIndex1n ]= max( ...
periodDeltaArray1n ) ;
i f ˜isempty ( p e r i o d O s c I n d i c e s 1 n )
disp ( [ ’Maximum Delta T containing ’ num2str ( t a r g e t P e r i o d ) ...
’ +/- ’ num2str ( d e l t a P e r i o d ) ’ : [’ num2str ( periodMinArray1n ( ...
periodDeltaMaxIndex1n ) ) ’, ’ num2str ( periodMaxArray1n ( ...
periodDeltaMaxIndex1n ) ) ’], Delta T= ’ num2str ( periodDeltaMax1n ) ...
’, k= ’ num2str ( periodDeltaMaxIndex1n ) ’, j= [’ num2str ( ...
240
p e r i o d M i n I n d i c e s 1 n ( periodDeltaMaxIndex1n ) ) ’,’ num2str ( ...
p e r i o d M a x I n d i c e s 1 n ( periodDeltaMaxIndex1n ) ) ’] @’ 10 ’ rho= ’ ...
num2str ( n o n l i n e a r S a m p l e . params . rhoSample ( periodDeltaMaxIndex1n ) ) ...
’, kH13H13 = ’ num2str ( n o n l i n e a r S a m p l e . params . kH13H13Sample ( ...
periodDeltaMaxIndex1n ) ) ’, kH7H7 = ’ num2str ( n o n l i n e a r S a m p l e . ...
params . kH7H7Sample ( periodDeltaMaxIndex1n ) ) ’, kH7H13 = ’ num2str ( ...
n o n l i n e a r S a m p l e . params . kH7H13Sample ( periodDeltaMaxIndex1n ) ) ’, ...
tau_h7 = ’ num2str ( n o n l i n e a r S a m p l e . params . tau h7Sample ( ...
periodDeltaMaxIndex1n ) ) ’, bH7= ’ num2str ( n o n l i n e a r S a m p l e . params...
. bH7Sample ( periodDeltaMaxIndex1n ) ) ’, rH7H7 = ’ num2str ( ...
n o n l i n e a r S a m p l e . params . rH7H7Sample ( periodDeltaMaxIndex1n ) ) ’, ...
wH7H7_H7H7 = ’ num2str ( n o n l i n e a r S a m p l e . params . wH7H7 H7H7Sample ( ...
periodDeltaMaxIndex1n ) ) ’, rN= ’ num2str ( n o n l i n e a r S a m p l e . params . ...
rNSample ( periodDeltaMaxIndex1n ) ) ] ) ;
else
disp ( [ ’Maximum Delta T containing ’ num2str ( t a r g e t P e r i o d ) ...
’ +/- ’ num2str ( d e l t a P e r i o d ) ’ : NaN ’ ] ) ;
end
periodDeltaCountFuncH1n= @( deltaTMin ) nnz ( deltaTMin <= ...
periodDeltaArray1n ( periodOscIndices1n ) ) ;
disp ( [ num2str ( t a r g e t P e r i o d ) ’ +/- ’ num2str ( d e l t a P e r i o d ) ’ ...
min period oscillating proportion with Delta T >= ’ num2str ( ...
t a r g e t D e l t a ) ’: ’ num2str ( periodDeltaCountFuncH1n ( t a r g e t D e l t a ) ) ...
’/’ num2str ( numSamples ) ’= ’ num2str ( periodDeltaCountFuncH1n ( ...
t a r g e t D e l t a ) /numSamples ) ] ) ;
241
disp ( ’ ’ ) ;
% One b i n d i n g s i t e , l i n e a r decay
disp ( ’MODEL II , 1 binding site , linear decay :’ ) ;
i f ˜isempty ( l i n e a r S a m p l e . o n e S i t e . H13TotalMaxSweepLimitPoints )
disp ( ’ Notice : H13TotalMax sweep limit reached .’ ) ;
end
% P e r i o d mask
p e r i o d M e a n s 1 l= mean( l i n e a r S a m p l e . o n e S i t e . p e r i o d s , 1 ) ;
f u l l P e r i o d M e a n s 1 l= repmat ( periodMe ans 1l , [ numEquations , 1 , 1 ] ) ...
;
p e r i o d M e a n s 1 l= s q u e e z e ( p e r i o d M e a n s 1 l ) ’ ;
p e r i o d M a s k 1 l= zeros ( s i z e ( f u l l P e r i o d M e a n s 1 l ) ) ;
p e r i o d M a s k 1 l ( f u l l P e r i o d M e a n s 1 l ˜= 0 )= abs ( ( l i n e a r S a m p l e . ...
o n e S i t e . p e r i o d s ( f u l l P e r i o d M e a n s 1 l ˜= 0 ) − f u l l P e r i o d M e a n s 1 l ( ...
f u l l P e r i o d M e a n s 1 l ˜= 0 ) ) . / f u l l P e r i o d M e a n s 1 l ( f u l l P e r i o d M e a n s 1 l ˜=...
0) ) < periodTol ;
p e r i o d M a s k 1 l ( f u l l P e r i o d M e a n s 1 l == 0 )= abs ( l i n e a r S a m p l e . o n e S i t e ...
. p e r i o d s ( f u l l P e r i o d M e a n s 1 l == 0 ) ) < p e r i o d T o l ;
p e r i o d M a s k 1 l= s q u e e z e ( a l l ( periodMask1l , 1 ) ) ’ ;
clear fullPeriodMeans1l
% Amplitude mask
amplitudeMask1l= s q u e e z e ( a l l ( amplitudeThresh <= l i n e a r S a m p l e . ...
oneSite . amplitudes , 1) ) ’ ;
% Combine masks and f i n d o s c i l l a t i n g p r o p o r t i o n
f u l l O s c M a s k 1 l= p e r i o d M a s k 1 l & amplitudeMask1l ;
242
oscMask1l= any ( f u l l O s c M a s k 1 l , 2 ) ;
o s c I n d i c e s 1 l= find ( oscMask1l ) ;
o s c P r o p 1 l= length ( o s c I n d i c e s 1 l ) /numSamples ;
disp ( [ ’Oscillating proportion : ’
num2str ( length ( o s c I n d i c e s 1 l ...
) ) ’/’ num2str ( numSamples ) ’= ’ num2str ( o s c P r o p 1 l ) ] ) ;
[ maxArray1l , m a x I n d i c e s 1 l ]= max( f u l l O s c M a s k 1 l . ∗ periodMe ans 1l , ...
[ ] , 2) ;
g l o b a l M a x 1 l= max( maxArray1l ) ;
minArray1l= zeros ( numSamples , 1 ) ;
m i n I n d i c e s 1 l= o n e s ( numSamples , 1 ) ;
fo r k= 1 : numSamples
p o s i t i v e I n d i c e s= f u l l O s c M a s k 1 l ( k , : ) > 0 ;
i f any ( p o s i t i v e I n d i c e s )
[ minArray1l ( k ) , m i n I n d i c e s 1 l ( k ) ]= min( p e r i o d M e a n s 1 l ( k , ...
positiveIndices ) ) ;
end
end
g l o b a l M i n 1 l= min( minArray1l ( 0 < minArray1l ) ) ;
i f isempty ( g l o b a l M i n 1 l )
disp ( ’No period range .’ ) ;
else
disp ( [ ’Nontrivial period range : [’ num2str ( g l o b a l M i n 1 l ) ’,...
’ num2str ( g l o b a l M a x 1 l ) ’]’ ] ) ;
end
[ deltaMax1l , d e l t a M a x I n d e x 1 l ]= max( maxArray1l − minArray1l ) ;
243
i f ˜isempty ( o s c I n d i c e s 1 l )
disp ( [ ’Maximum Delta T: [’ num2str ( minArray1l ( ...
d e l t a M a x I n d e x 1 l ) ) ’, ’ num2str ( maxArray1l ( d e l t a M a x I n d e x 1 l ) ) ’],...
Delta T= ’ num2str ( d e l t a M a x 1 l ) ’, k= ’ num2str ( d e l t a M a x I n d e x 1 l ) ...
’, j= [’ num2str ( m i n I n d i c e s 1 l ( d e l t a M a x I n d e x 1 l ) ) ’,’ num2str ( ...
m a x I n d i c e s 1 l ( d e l t a M a x I n d e x 1 l ) ) ’]’ ] ) ;
else
disp ( ’Maximum Delta T: NaN ’ ) ;
end
periodOscMask1l= ( minArray1l <= t a r g e t P e r i o d + d e l t a P e r i o d ) & ...
( t a r g e t P e r i o d − d e l t a P e r i o d <= maxArray1l ) ;
p e r i o d O s c I n d i c e s 1 l= find ( periodOscMask1l ) ;
p e r i o d O s c P r o p 1 l= length ( p e r i o d O s c I n d i c e s 1 l ) /numSamples ;
disp ( [ num2str ( t a r g e t P e r i o d ) ’ +/- ’ num2str ( d e l t a P e r i o d ) ’ ...
min period oscillating proportion : ’ num2str ( length ( ...
p e r i o d O s c I n d i c e s 1 l ) ) ’/’ num2str ( numSamples ) ’= ’ num2str ( ...
periodOscProp1l ) ] ) ;
periodMaxArray1l= periodOscMask1l . ∗ maxArray1l ;
p e r i o d M a x I n d i c e s 1 l= m a x I n d i c e s 1 l ;
p e r i o d M a x I n d i c e s 1 l ( periodOscMask1l == 0 )= 1 ;
p e r i o d M i n A r r a y 1 l= periodOscMask1l . ∗ minArray1l ;
p e r i o d M i n I n d i c e s 1 l= m i n I n d i c e s 1 l ;
p e r i o d M i n I n d i c e s 1 l ( periodOscMask1l == 0 )= 1 ;
p e r i o d D e l t a A r r a y 1 l= periodMaxArray1l − p e r i o d M i n A r r a y 1 l ;
244
[ pe riodDe ltaMax1l , p e r i o d D e l t a M a x I n d e x 1 l ]= max( ...
periodDeltaArray1l ) ;
i f ˜isempty ( p e r i o d O s c I n d i c e s 1 l )
disp ( [ ’Maximum Delta T containing ’ num2str ( t a r g e t P e r i o d ) ...
’ +/- ’ num2str ( d e l t a P e r i o d ) ’ : [’ num2str ( p e r i o d M i n A r r a y 1 l ( ...
p e r i o d D e l t a M a x I n d e x 1 l ) ) ’, ’ num2str ( periodMaxArray1l ( ...
p e r i o d D e l t a M a x I n d e x 1 l ) ) ’], Delta T= ’ num2str ( p e r i o d D e l t a M a x 1 l ) ...
’, k= ’ num2str ( p e r i o d D e l t a M a x I n d e x 1 l ) ’, j= [’ num2str ( ...
p e r i o d M i n I n d i c e s 1 l ( p e r i o d D e l t a M a x I n d e x 1 l ) ) ’,’ num2str ( ...
p e r i o d M a x I n d i c e s 1 l ( p e r i o d D e l t a M a x I n d e x 1 l ) ) ’] @’ 10 ’ rho= ’ ...
num2str ( l i n e a r S a m p l e . params . rhoSample ( p e r i o d D e l t a M a x I n d e x 1 l ) ) ’...
, kH13H13 = ’ num2str ( l i n e a r S a m p l e . params . kH13H13Sample ( ...
p e r i o d D e l t a M a x I n d e x 1 l ) ) ’, kH7H7 = ’ num2str ( l i n e a r S a m p l e . params . ...
kH7H7Sample ( p e r i o d D e l t a M a x I n d e x 1 l ) ) ’, kH7H13 = ’ num2str ( ...
l i n e a r S a m p l e . params . kH7H13Sample ( p e r i o d D e l t a M a x I n d e x 1 l ) ) ’, ...
tau_h7 = ’ num2str ( l i n e a r S a m p l e . params . tau h7Sample ( ...
p e r i o d D e l t a M a x I n d e x 1 l ) ) ’, bH7= ’ num2str ( l i n e a r S a m p l e . params . ...
bH7Sample ( p e r i o d D e l t a M a x I n d e x 1 l ) ) ’, rH7H7 = ’ num2str ( ...
l i n e a r S a m p l e . params . rH7H7Sample ( p e r i o d D e l t a M a x I n d e x 1 l ) ) ’, ...
wH7H7_H7H7 = ’ num2str ( l i n e a r S a m p l e . params . wH7H7 H7H7Sample ( ...
p e r i o d D e l t a M a x I n d e x 1 l ) ) ’, rN= ’ num2str ( l i n e a r S a m p l e . params . ...
rNSample ( p e r i o d D e l t a M a x I n d e x 1 l ) ) ] ) ;
else
disp ( [ ’Maximum Delta T containing ’ num2str ( t a r g e t P e r i o d ) ...
’ +/- ’ num2str ( d e l t a P e r i o d ) ’ : NaN ’ ] ) ;
end
245
periodDeltaCountFuncH1l= @( deltaTMin ) nnz ( deltaTMin <= ...
periodDeltaArray1l ( periodOscIndices1l ) ) ;
disp ( [ num2str ( t a r g e t P e r i o d ) ’ +/- ’ num2str ( d e l t a P e r i o d ) ’ ...
min period oscillating proportion with Delta T >= ’ num2str ( ...
t a r g e t D e l t a ) ’: ’ num2str ( periodDeltaCountFuncH1l ( t a r g e t D e l t a ) ) ...
’/’ num2str ( numSamples ) ’= ’ num2str ( periodDeltaCountFuncH1l ( ...
t a r g e t D e l t a ) /numSamples ) ] ) ;
disp ( ’ ’ ) ;
% Two b i n d i n g s i t e s , n o n l i n e a r decay
disp ( ’MODEL III , 2 binding sites , nonlinear decay :’ ) ;
i f ˜isempty ( n o n l i n e a r S a m p l e . t w o S i t e . ...
H13TotalMaxSweepLimitPoints )
disp ( ’ Notice : H13TotalMax sweep limit reached .’ ) ;
end
% P e r i o d mask
periodMeans2n= mean( n o n l i n e a r S a m p l e . t w o S i t e . p e r i o d s , 1 ) ;
f u l l P e r i o d M e a n s 2 n = repmat ( periodMeans2n , [ numEquations , 1 , ...
1]) ;
periodMeans2n= s q u e e z e ( periodMeans2n ) ’ ;
periodMask2n= zeros ( s i z e ( f u l l P e r i o d M e a n s 2 n ) ) ;
periodMask2n ( f u l l P e r i o d M e a n s 2 n ˜= 0 )= abs ( ( n o n l i n e a r S a m p l e . ...
t w o S i t e . p e r i o d s ( f u l l P e r i o d M e a n s 2 n ˜= 0 ) − f u l l P e r i o d M e a n s 2 n ( ...
246
f u l l P e r i o d M e a n s 2 n ˜= 0 ) ) . / f u l l P e r i o d M e a n s 2 n ( f u l l P e r i o d M e a n s 2 n ˜=...
0) ) < periodTol ;
periodMask2n ( f u l l P e r i o d M e a n s 2 n == 0 )= abs ( n o n l i n e a r S a m p l e . ...
t w o S i t e . p e r i o d s ( f u l l P e r i o d M e a n s 2 n == 0 ) ) < p e r i o d T o l ;
periodMask2n= s q u e e z e ( a l l ( periodMask2n , 1 ) ) ’ ;
clear fullPeriodMeans2n ;
% Amplitude mask
amplitudeMask2n= s q u e e z e ( a l l ( amplitudeThresh <= ...
nonlinearSample . twoSite . amplitudes , 1) ) ’ ;
% Combine masks and f i n d o s c i l l a t i n g p r o p o r t i o n
fullOscMask2n= periodMask2n & amplitudeMask2n ;
oscMask2n= any ( fullOscMask2n , 2 ) ;
o s c I n d i c e s 2 n= find ( oscMask2n ) ;
oscProp2n= length ( o s c I n d i c e s 2 n ) /numSamples ;
disp ( [ ’Oscillating proportion : ’
num2str ( length ( o s c I n d i c e s 2 n ...
) ) ’/’ num2str ( numSamples ) ’= ’ num2str ( oscProp2n ) ] ) ;
[ maxArray2n , maxIndices2n ]= max( fullOscMask2n . ∗ periodMeans2n , ...
[ ] , 2) ;
globalMax2n= max( maxArray2n ) ;
minArray2n= zeros ( numSamples , 1 ) ;
m i n I n d i c e s 2 n= o n e s ( numSamples , 1 ) ;
fo r k= 1 : numSamples
p o s i t i v e I n d i c e s= fullOscMask2n ( k , : ) > 0 ;
i f any ( p o s i t i v e I n d i c e s )
[ minArray2n ( k ) , m i n I n d i c e s 2 n ( k ) ]= min( periodMeans2n ( k , ...
positiveIndices ) ) ;
247
end
end
globalMin2n= min( minArray2n ( 0 < minArray2n ) ) ;
i f isempty ( globalMin2n )
disp ( ’No period range .’ ) ;
else
disp ( [ ’Nontrivial period range : [’ num2str ( globalMin2n ) ’,...
’ num2str ( globalMax2n ) ’]’ ] ) ;
end
[ deltaMax2n , deltaMaxIndex2n ]= max( maxArray2n − minArray2n ) ;
i f ˜isempty ( o s c I n d i c e s 2 n )
disp ( [ ’Maximum Delta T: [’ num2str ( minArray2n ( ...
deltaMaxIndex2n ) ) ’, ’ num2str ( maxArray2n ( deltaMaxIndex2n ) ) ’],...
Delta T= ’ num2str ( deltaMax2n ) ’, k= ’ num2str ( deltaMaxIndex2n ) ...
’, j= [’ num2str ( m i n I n d i c e s 2 n ( deltaMaxIndex2n ) ) ’, ’ num2str ( ...
maxIndices2n ( deltaMaxIndex2n ) ) ’]’ ] ) ;
else
disp ( ’Maximum Delta T: NaN ’ ) ;
end
periodOscMask2n= ( minArray2n <= t a r g e t P e r i o d + d e l t a P e r i o d ) & ...
( t a r g e t P e r i o d − d e l t a P e r i o d <= maxArray2n ) ;
p e r i o d O s c I n d i c e s 2 n= find ( periodOscMask2n ) ;
periodOscProp2n= length ( p e r i o d O s c I n d i c e s 2 n ) /numSamples ;
disp ( [ num2str ( t a r g e t P e r i o d ) ’ +/- ’ num2str ( d e l t a P e r i o d ) ’ ...
min period oscillating proportion : ’ num2str ( length ( ...
248
p e r i o d O s c I n d i c e s 2 n ) ) ’/’ num2str ( numSamples ) ’= ’ num2str ( ...
periodOscProp2n ) ] ) ;
periodMaxArray2n= periodOscMask2n . ∗ maxArray2n ;
p e r i o d M a x I n d i c e s 2 n= maxIndices2n ;
p e r i o d M a x I n d i c e s 2 n ( periodOscMask2n == 0 )= 1 ;
periodMinArray2n= periodOscMask2n . ∗ minArray2n ;
p e r i o d M i n I n d i c e s 2 n= m i n I n d i c e s 2 n ;
p e r i o d M i n I n d i c e s 2 n ( periodOscMask2n == 0 )= 1 ;
p e r i o d D e l t a A r r a y 2 n= periodMaxArray2n − periodMinArray2n ;
[ periodDeltaMax2n , periodDeltaMaxIndex2n ]= max( ...
periodDeltaArray2n ) ;
i f ˜isempty ( p e r i o d O s c I n d i c e s 2 n )
disp ( [ ’Maximum Delta T containing ’ num2str ( t a r g e t P e r i o d ) ...
’ +/- ’ num2str ( d e l t a P e r i o d ) ’ : [’ num2str ( periodMinArray2n ( ...
periodDeltaMaxIndex2n ) ) ’, ’ num2str ( periodMaxArray2n ( ...
periodDeltaMaxIndex2n ) ) ’], Delta T= ’ num2str ( periodDeltaMax2n ) ...
’, k= ’ num2str ( periodDeltaMaxIndex2n ) ’, j= [’ num2str ( ...
p e r i o d M i n I n d i c e s 2 n ( periodDeltaMaxIndex2n ) ) ’,’ num2str ( ...
p e r i o d M a x I n d i c e s 2 n ( periodDeltaMaxIndex2n ) ) ’] @’ 10 ’ rho= ’ ...
num2str ( n o n l i n e a r S a m p l e . params . rhoSample ( periodDeltaMaxIndex2n ) ) ...
’, kH13H13 = ’ num2str ( n o n l i n e a r S a m p l e . params . kH13H13Sample ( ...
periodDeltaMaxIndex2n ) ) ’, kH7H7 = ’ num2str ( n o n l i n e a r S a m p l e . ...
params . kH7H7Sample ( periodDeltaMaxIndex2n ) ) ’, kH7H13 = ’ num2str ( ...
n o n l i n e a r S a m p l e . params . kH7H13Sample ( periodDeltaMaxIndex2n ) ) ’, ...
tau_h7 = ’ num2str ( n o n l i n e a r S a m p l e . params . tau h7Sample ( ...
249
periodDeltaMaxIndex2n ) ) ’, bH7= ’ num2str ( n o n l i n e a r S a m p l e . params...
. bH7Sample ( periodDeltaMaxIndex2n ) ) ’, rH7H7 = ’ num2str ( ...
n o n l i n e a r S a m p l e . params . rH7H7Sample ( periodDeltaMaxIndex2n ) ) ’, ...
wH7H7_H7H7 = ’ num2str ( n o n l i n e a r S a m p l e . params . wH7H7 H7H7Sample ( ...
periodDeltaMaxIndex2n ) ) ’, rN= ’ num2str ( n o n l i n e a r S a m p l e . params . ...
rNSample ( periodDeltaMaxIndex2n ) ) ] ) ;
else
disp ( [ ’Maximum Delta T containing ’ num2str ( t a r g e t P e r i o d ) ...
’ +/- ’ num2str ( d e l t a P e r i o d ) ’ : NaN ’ ] ) ;
end
periodDeltaCountFuncH2n= @( deltaTMin ) nnz ( deltaTMin <= ...
periodDeltaArray2n ( periodOscIndices2n ) ) ;
disp ( [ num2str ( t a r g e t P e r i o d ) ’ +/- ’ num2str ( d e l t a P e r i o d ) ’ ...
min period oscillating proportion with Delta T >= ’ num2str ( ...
t a r g e t D e l t a ) ’: ’ num2str ( periodDeltaCountFuncH2n ( t a r g e t D e l t a ) ) ...
’/’ num2str ( numSamples ) ’= ’ num2str ( periodDeltaCountFuncH2n ( ...
t a r g e t D e l t a ) /numSamples ) ] ) ;
disp ( ’ ’ ) ;
% Two b i n d i n g s i t e s , l i n e a r decay
disp ( ’MODEL IV , 2 binding sites , linear decay :’ ) ;
i f ˜isempty ( l i n e a r S a m p l e . t w o S i t e . H13TotalMaxSweepLimitPoints )
disp ( ’ Notice : H13TotalMax sweep limit reached .’ ) ;
end
250
% P e r i o d mask
p e r i o d M e a n s 2 l= mean( l i n e a r S a m p l e . t w o S i t e . p e r i o d s , 1 ) ;
f u l l P e r i o d M e a n s 2 l = repmat ( periodMe ans 2l , [ numEquations , 1 , ...
1]) ;
p e r i o d M e a n s 2 l= s q u e e z e ( p e r i o d M e a n s 2 l ) ’ ;
p e r i o d M a s k 2 l= zeros ( s i z e ( f u l l P e r i o d M e a n s 2 l ) ) ;
p e r i o d M a s k 2 l ( f u l l P e r i o d M e a n s 2 l ˜= 0 )= abs ( ( l i n e a r S a m p l e . ...
t w o S i t e . p e r i o d s ( f u l l P e r i o d M e a n s 2 l ˜= 0 ) − f u l l P e r i o d M e a n s 2 l ( ...
f u l l P e r i o d M e a n s 2 l ˜= 0 ) ) . / f u l l P e r i o d M e a n s 2 l ( f u l l P e r i o d M e a n s 2 l ˜=...
0) ) < periodTol ;
p e r i o d M a s k 2 l ( f u l l P e r i o d M e a n s 2 l == 0 )= abs ( l i n e a r S a m p l e . t w o S i t e ...
. p e r i o d s ( f u l l P e r i o d M e a n s 2 l == 0 ) ) < p e r i o d T o l ;
p e r i o d M a s k 2 l= s q u e e z e ( a l l ( periodMask2l , 1 ) ) ’ ;
clear fullPeriodMeans2l ;
% Amplitude mask
amplitudeMask2l= s q u e e z e ( a l l ( amplitudeThresh <= l i n e a r S a m p l e . ...
twoSite . amplitudes , 1) ) ’ ;
% Combine masks and f i n d o s c i l l a t i n g p r o p o r t i o n
f u l l O s c M a s k 2 l= p e r i o d M a s k 2 l & amplitudeMask2l ;
oscMask2l= any ( f u l l O s c M a s k 2 l , 2 ) ;
o s c I n d i c e s 2 l= find ( oscMask2l ) ;
o s c P r o p 2 l= length ( o s c I n d i c e s 2 l ) /numSamples ;
disp ( [ ’Oscillating proportion : ’ num2str ( length ( o s c I n d i c e s 2 l ) ...
) ’/’ num2str ( numSamples ) ’= ’ num2str ( o s c P r o p 2 l ) ] ) ;
[ maxArray2l , m a x I n d i c e s 2 l ]= max( f u l l O s c M a s k 2 l . ∗ periodMe ans 2l , ...
[ ] , 2) ;
251
g l o b a l M a x 2 l= max( maxArray2l ) ;
minArray2l= zeros ( numSamples , 1 ) ;
m i n I n d i c e s 2 l= o n e s ( numSamples , 1 ) ;
fo r k= 1 : numSamples
p o s i t i v e I n d i c e s= f u l l O s c M a s k 2 l ( k , : ) > 0 ;
i f any ( p o s i t i v e I n d i c e s )
[ minArray2l ( k ) , m i n I n d i c e s 2 l ( k ) ]= min( p e r i o d M e a n s 2 l ( k , ...
positiveIndices ) ) ;
end
end
g l o b a l M i n 2 l= min( minArray2l ( 0 < minArray2l ) ) ;
i f isempty ( g l o b a l M i n 2 l )
disp ( ’No period range .’ ) ;
else
disp ( [ ’Nontrivial period range : [’ num2str ( g l o b a l M i n 2 l ) ’,...
’ num2str ( g l o b a l M a x 2 l ) ’]’ ] ) ;
end
[ deltaMax2l , d e l t a M a x I n d e x 2 l ]= max( maxArray2l − minArray2l ) ;
i f ˜isempty ( o s c I n d i c e s 2 l )
disp ( [ ’Maximum Delta T: [’ num2str ( minArray2l ( ...
d e l t a M a x I n d e x 2 l ) ) ’, ’ num2str ( maxArray2l ( d e l t a M a x I n d e x 2 l ) ) ’],...
Delta T= ’ num2str ( d e l t a M a x 2 l ) ’, k= ’ num2str ( d e l t a M a x I n d e x 2 l ) ...
’, j= [’ num2str ( m i n I n d i c e s 2 l ( d e l t a M a x I n d e x 2 l ) ) ’, ’ num2str ( ...
m a x I n d i c e s 2 l ( d e l t a M a x I n d e x 2 l ) ) ’]’ ] ) ;
else
disp ( ’Maximum Delta T: NaN ’ ) ;
252
end
periodOscMask2l= ( minArray2l <= t a r g e t P e r i o d + d e l t a P e r i o d ) & ...
( t a r g e t P e r i o d − d e l t a P e r i o d <= maxArray2l ) ;
p e r i o d O s c I n d i c e s 2 l= find ( periodOscMask2l ) ;
p e r i o d O s c P r o p 2 l= length ( p e r i o d O s c I n d i c e s 2 l ) /numSamples ;
disp ( [ num2str ( t a r g e t P e r i o d ) ’ +/- ’ num2str ( d e l t a P e r i o d ) ’ ...
min period oscillating proportion : ’ num2str ( length ( ...
p e r i o d O s c I n d i c e s 2 l ) ) ’/’ num2str ( numSamples ) ’= ’ num2str ( ...
periodOscProp2l ) ] ) ;
periodMaxArray2l= periodOscMask2l . ∗ maxArray2l ;
p e r i o d M a x I n d i c e s 2 l= m a x I n d i c e s 2 l ;
p e r i o d M a x I n d i c e s 2 l ( periodOscMask2l == 0 )= 1 ;
p e r i o d M i n A r r a y 2 l= periodOscMask2l . ∗ minArray2l ;
p e r i o d M i n I n d i c e s 2 l= m i n I n d i c e s 2 l ;
p e r i o d M i n I n d i c e s 2 l ( periodOscMask2l == 0 )= 1 ;
p e r i o d D e l t a A r r a y 2 l= periodMaxArray2l − p e r i o d M i n A r r a y 2 l ;
[ pe riodDe ltaMax2l , p e r i o d D e l t a M a x I n d e x 2 l ]= max( ...
periodDeltaArray2l ) ;
i f ˜isempty ( p e r i o d O s c I n d i c e s 2 l )
disp ( [ ’Maximum Delta T containing ’ num2str ( t a r g e t P e r i o d ) ...
’ +/- ’ num2str ( d e l t a P e r i o d ) ’ : [’ num2str ( p e r i o d M i n A r r a y 2 l ( ...
p e r i o d D e l t a M a x I n d e x 2 l ) ) ’, ’ num2str ( periodMaxArray2l ( ...
p e r i o d D e l t a M a x I n d e x 2 l ) ) ’], Delta T= ’ num2str ( p e r i o d D e l t a M a x 2 l ) ...
’, k= ’ num2str ( p e r i o d D e l t a M a x I n d e x 2 l ) ’, j= [’ num2str ( ...
253
p e r i o d M i n I n d i c e s 2 l ( p e r i o d D e l t a M a x I n d e x 2 l ) ) ’,’ num2str ( ...
p e r i o d M a x I n d i c e s 2 l ( p e r i o d D e l t a M a x I n d e x 2 l ) ) ’] @’ 10 ’ rho= ’ ...
num2str ( l i n e a r S a m p l e . params . rhoSample ( p e r i o d D e l t a M a x I n d e x 2 l ) ) ’...
, kH13H13 = ’ num2str ( l i n e a r S a m p l e . params . kH13H13Sample ( ...
p e r i o d D e l t a M a x I n d e x 2 l ) ) ’, kH7H7 = ’ num2str ( l i n e a r S a m p l e . params . ...
kH7H7Sample ( p e r i o d D e l t a M a x I n d e x 2 l ) ) ’, kH7H13 = ’ num2str ( ...
l i n e a r S a m p l e . params . kH7H13Sample ( p e r i o d D e l t a M a x I n d e x 2 l ) ) ’, ...
tau_h7 = ’ num2str ( l i n e a r S a m p l e . params . tau h7Sample ( ...
p e r i o d D e l t a M a x I n d e x 2 l ) ) ’, bH7= ’ num2str ( l i n e a r S a m p l e . params . ...
bH7Sample ( p e r i o d D e l t a M a x I n d e x 2 l ) ) ’, rH7H7 = ’ num2str ( ...
l i n e a r S a m p l e . params . rH7H7Sample ( p e r i o d D e l t a M a x I n d e x 2 l ) ) ’, ...
wH7H7_H7H7 = ’ num2str ( l i n e a r S a m p l e . params . wH7H7 H7H7Sample ( ...
p e r i o d D e l t a M a x I n d e x 2 l ) ) ’, rN= ’ num2str ( l i n e a r S a m p l e . params . ...
rNSample ( p e r i o d D e l t a M a x I n d e x 2 l ) ) ] ) ;
else
disp ( [ ’Maximum Delta T containing ’ num2str ( t a r g e t P e r i o d ) ...
’ +/- ’ num2str ( d e l t a P e r i o d ) ’ : NaN ’ ] ) ;
end
periodDeltaCountFuncH2l= @( deltaTMin ) nnz ( deltaTMin <= ...
periodDeltaArray2l ( periodOscIndices2l ) ) ;
disp ( [ num2str ( t a r g e t P e r i o d ) ’ +/- ’ num2str ( d e l t a P e r i o d ) ’ ...
min period oscillating proportion with Delta T >= ’ num2str ( ...
t a r g e t D e l t a ) ’: ’ num2str ( periodDeltaCountFuncH2l ( t a r g e t D e l t a ) ) ...
’/’ num2str ( numSamples ) ’= ’ num2str ( periodDeltaCountFuncH2l ( ...
t a r g e t D e l t a ) /numSamples ) ] ) ;
254
% F i g u r e s f o r main paper
f i g u r e H= f i g u r e ( 1 ) ; c l f ( f i g u r e H ) ; set ( f i g u r e H , ’Position ’ , [ 4 5 ...
305 775 2 3 1 ] ) ;
colormap ( [ 0 . 2 5 0 . 2 5 0 . 2 5 ; 0 . 7 5 0 . 7 5 0 . 7 5 ] ) ;
axisH= subplot ( 1 , 3 , 1 ) ;
bar ( axisH , 1 0 0 ∗ [ oscProp1n periodOscProp1n ; o s c P r o p 1 l ...
p e r i o d O s c P r o p 1 l ; oscProp2n periodOscProp2n ; o s c P r o p 2 l ...
periodOscProp2l ] ) ;
ylim ( axisH , [ 0 7 5 ] ) ;
set ( axisH , ’YTick ’ , 0 : 1 0 : 7 0 ) ;
ylabel ( axisH , ’% of Simulations ’ , ’FontWeight ’ , ’demi ’ , ’...
FontSize ’ , 1 2 ) ;
xlim ( axisH , [ 0 . 5 4 . 5 ] ) ;
xlabel ( axisH , ’Model Number ’ , ’FontWeight ’ , ’demi ’ , ’...
FontSize ’ , 1 2 ) ;
set ( axisH , ’XTickLabel ’ , { ’I’ ’II ’ ’III ’ ’IV ’ } ) ;
legend ( axisH , ’Periodic ’ , [ num2str ( t a r g e t P e r i o d ) ’\pm ’ ...
num2str ( d e l t a P e r i o d ) ’ min.’ ] , ’Location ’ , ’NorthWest ’ ) ;
t i t l e ( axisH , ’Model Comparison ’ , ’FontWeight ’ , ’demi ’ , ’...
FontSize ’ , 1 2 ) ;
set ( axisH , ’FontWeight ’ , ’demi ’ ) ;
axisH= subplot ( 1 , 3 , 2 ) ;
xEdges= 0 : 2 . 5 : 3 0 ;
xMidpoints= xEdges ( 1 : end−1) + 1 . 2 5 ;
count2n= zeros ( length ( xMidpoints ) , 1 ) ;
255
c o u n t 2 l= count2n ;
f o r k= 1 : length ( xMidpoints )
count2n ( k )= periodDeltaCountFuncH2n ( xEdges ( k ) ) − ...
periodDeltaCountFuncH2n ( xEdges ( k+1) ) ;
c o u n t 2 l ( k )= periodDeltaCountFuncH2l ( xEdges ( k ) ) − ...
periodDeltaCountFuncH2l ( xEdges ( k+1) ) ;
end
bar ( axisH , xMidpoints , 100/ numSamples ∗ [ count2n c o u n t 2 l ] , ...
’BarLayout ’ , ’stacked ’ , ’BarWidth ’ , 1 ) ;
ylabel ( axisH , ’% of Simulations ’ , ’FontWeight ’ , ’demi ’ , ’...
FontSize ’ , 1 2 ) ;
xlim ( axisH , [ 0 3 0 ] ) ;
set ( axisH , ’XTick ’ , 0 : 2 . 5 : 3 0 ) ;
set ( axisH , ’XTickLabel ’ , { ’0’ ’’ ’5’ ’’ ’10 ’ ’’ ’15 ’ ’’ ’...
20’ ’’ ’25 ’ ’’ ’30 ’ } ) ;
xlabel ( axisH , ’{\ Delta }T (min .) ’ , ’FontWeight ’ , ’demi ’ , ’...
FontSize ’ , 1 2 ) ;
legend ( axisH , ’Model III ’ , ’Model IV ’ ) ;
t i t l e ( axisH , ’{\ Delta }T Distributions ’ , ’FontWeight ’ , ’...
demi ’ , ’FontSize ’ , 1 2 ) ;
set ( axisH , ’FontWeight ’ , ’demi ’ ) ;
% Tabulations
disp ( ’ ’ ) ;
disp ( [ ’Model III Delta T tabulation (out of ’ num2str ( ...
numSamples ) ’ samples ):’ ] ) ;
disp ( [ ’Bin edges : ’ , m a t 2 s t r ( xEdges ) ] ) ;
disp ( [ ’Counts : ’ , m a t 2 s t r ( count2n ’ ) ] ) ;
256
disp ( ’ ’ ) ;
disp ( [ ’Model IV Delta T tabulation (out of ’ num2str ( ...
numSamples ) ’ samples ):’ ] ) ;
disp ( [ ’Bin edges : ’ , m a t 2 s t r ( xEdges ) ] ) ;
disp ( [ ’Counts : ’ , m a t 2 s t r ( c o u n t 2 l ’ ) ] ) ;
axisH= subplot ( 1 , 3 , 3 ) ;
n o n l i n e a r M e a n P e r i o d s= periodMeans2n ( periodDeltaMaxIndex2n ...
,:) ;
l i n e a r M e a n P e r i o d s= p e r i o d M e a n s 2 l ( p e r i o d D e l t a M a x I n d e x 2 l , : ) ;
s e r i e s H= plot ( axisH , n o n l i n e a r S a m p l e . params . ...
H13TotalMaxSweep ( 1 : p e r i o d M i n I n d i c e s 2 n ( periodDeltaMaxIndex2n ) ) , ...
n o n l i n e a r M e a n P e r i o d s ( 1 : p e r i o d M i n I n d i c e s 2 n ( periodDeltaMaxIndex2n ) ...
) , ’k-’ , l i n e a r S a m p l e . params . H13TotalMaxSweep ( 1 : ...
p e r i o d M i n I n d i c e s 2 l ( p e r i o d D e l t a M a x I n d e x 2 l ) ) , l i n e a r M e a n P e r i o d s ( 1 : ...
p e r i o d M i n I n d i c e s 2 l ( p e r i o d D e l t a M a x I n d e x 2 l ) ) , ’k--’ ) ;
set ( s e r i e s H ( 1 ) , ’LineWidth ’ , 2 ) ;
set ( s e r i e s H ( 2 ) , ’LineWidth ’ , 1 ) ;
ylabel ( axisH , ’Period (min .) ’ , ’FontWeight ’ , ’demi ’ , ’...
FontSize ’ , 1 2 ) ;
xlabel ( axisH , ’Total Control Protein (copy #) ’ , ’...
FontWeight ’ , ’demi ’ , ’FontSize ’ , 1 2 ) ;
t i t l e ( axisH , ’Solutions for {\ Delta }T_{III }^{ max} & {\...
Delta }T_{IV }^{ max} ’ , ’FontWeight ’ , ’demi ’ , ’FontSize ’ , 1 2 ) ;
legend ( axisH , ’Model III ’ , ’Model IV ’ ) ;
set ( axisH , ’FontWeight ’ , ’demi ’ ) ;
257
f i g u r e H= f i g u r e ( 2 ) ; c l f ( f i g u r e H ) ; set ( f i g u r e H , ’Position ’ , [ 3 0 ...
325 512 2 4 0 ] ) ;
colormap ( [ 0 . 5 0 . 5 0 . 5 ] ) ;
axisH= subplot ( 1 , 2 , 1 ) ;
e d g e s= 2 : 0 . 5 : 9 ;
bar ( axisH , edges , h i s t c ( n o n l i n e a r S a m p l e . params . ...
tau h7Sample ( p e r i o d O s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
%
s e t ( f i n d a l l ( a l l c h i l d ( gca ) , ’ Marker ’ , ’ ∗ ’ ) , ’ Marker ’ , ’ none ...
’) ;
ylabel ( axisH , ’Simulation Count ’ , ’FontWeight ’ , ’demi ’ , ’...
FontSize ’ , 1 2 ) ;
xlim ( axisH , [ 2 . 3 8 . 1 ] ) ;
set ( axisH , ’XTick ’ , [ 2 . 3 3 : 1 : 7 8 . 1 ] ) ;
xlabel ( axisH , ’\ tau_c ’ , ’FontSize ’ , 1 2 ) ;
t i t l e ( axisH , ’Sensitive , Model III ’ , ’FontWeight ’ , ’demi ’ , ...
’FontSize ’ , 1 2 ) ;
set ( axisH , ’FontWeight ’ , ’demi ’ ) ;
axisH= subplot ( 1 , 2 , 2 ) ;
e d g e s= 1 / 3 : 1 / 3 : 3 ;
bar ( axisH , edges , h i s t c ( n o n l i n e a r S a m p l e . params . rhoSample ( ...
p e r i o d O s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
ylabel ( axisH , ’Simulation Count ’ , ’FontWeight ’ , ’demi ’ , ’...
FontSize ’ , 1 2 ) ;
xlim ( axisH , [ 0 . 9 9 / 3 3 ] ) ;
set ( axisH , ’XTick ’ , [ 0 . 3 3 1 2 3 ] ) ;
xlabel ( axisH , ’\rho ’ , ’FontWeight ’ , ’demi ’ , ’FontSize ’ , 1 2 ) ;
258
t i t l e ( axisH , ’Insensitive , Model III ’ , ’FontWeight ’ , ’demi...
’ , ’FontSize ’ , 1 2 ) ;
set ( axisH , ’FontWeight ’ , ’demi ’ ) ;
% F i g u r e s f o r Supplement
t o p G r a p h T i t l e= [ ’Periodic ’ ] ;
midGraphTitle= [ num2str ( t a r g e t P e r i o d ) ’\pm ’ num2str ( ...
d e l t a P e r i o d ) ’ min ’ ] ;
b o t G r a p h T i t l e= [ ’\ DeltaT \geq ’ num2str ( t a r g e t D e l t a ) ’ min ’ ] ;
f i g u r e ( 5 ) ; c l f ; colormap ( [ 0 . 5 0 . 5 0 . 5 ] ) ;
axisH= subplot ( 3 , 4 , 1 ) ;
e d g e s= 1 / 3 : 1 / 3 : 3 ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . rhoSample ( ...
o s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 0 . 9 9 / 3 3 ] ) ;
set ( axisH , ’XTick ’ , [ 0 . 3 3 1 2 3 ] ) ;
xlabel ( ’\rho ’ ) ;
t i t l e ( topGraphTitle ) ;
axisH= subplot ( 3 , 4 , 5 ) ;
e d g e s= 1 / 3 : 1 / 3 : 3 ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . rhoSample ( ...
p e r i o d O s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 0 . 9 9 / 3 3 ] ) ;
set ( axisH , ’XTick ’ , [ 0 . 3 3 1 2 3 ] ) ;
xlabel ( ’\rho ’ ) ;
t i t l e ( midGraphTitle ) ;
259
axisH= subplot ( 3 , 4 , 9 ) ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . rhoSample ( ...
p e r i o d D e l t a A r r a y 2 n >= t a r g e t D e l t a ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 0 . 9 9 / 3 3 ] ) ;
set ( axisH , ’XTick ’ , [ 0 . 3 3 1 2 3 ] ) ;
xlabel ( ’\rho ’ ) ;
t i t l e ( botGraphTitle ) ;
axisH= subplot ( 3 , 4 , 2 ) ;
e d g e s= 2 : 0 . 5 : 9 ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . tau h7Sample ( ...
o s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 2 . 3 8 . 1 ] ) ;
set ( axisH , ’XTick ’ , [ 2 . 3 4 : 1 : 6 8 . 1 ] ) ;
xlabel ( ’\ tau_c ’ ) ;
t i t l e ( topGraphTitle ) ;
axisH= subplot ( 3 , 4 , 6 ) ;
e d g e s= 2 : 0 . 5 : 9 ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . tau h7Sample ( ...
p e r i o d O s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 2 . 3 8 . 1 ] ) ;
set ( axisH , ’XTick ’ , [ 2 . 3 4 : 1 : 6 8 . 1 ] ) ;
xlabel ( ’\ tau_c ’ ) ;
t i t l e ( midGraphTitle ) ;
axisH= subplot ( 3 , 4 , 1 0 ) ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . tau h7Sample ( ...
p e r i o d D e l t a A r r a y 2 n >= t a r g e t D e l t a ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 2 . 3 8 . 1 ] ) ;
260
set ( axisH , ’XTick ’ , [ 2 . 3 4 : 1 : 6 8 . 1 ] ) ;
xlabel ( ’\ tau_c ’ ) ;
t i t l e ( botGraphTitle ) ;
axisH= subplot ( 3 , 4 , 3 ) ;
e d g e s= 0 : 0 . 1 : 1 ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . rH7H7Sample ( ...
o s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 0 . 0 1 1 ] ) ;
set ( axisH , ’XTick ’ , [ 0 . 0 1 0 . 5 1 ] ) ;
xlabel ( ’\rho_{C:C}’ ) ;
t i t l e ( topGraphTitle ) ;
axisH= subplot ( 3 , 4 , 7 ) ;
e d g e s= 0 : 0 . 1 : 1 ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . rH7H7Sample ( ...
p e r i o d O s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 0 . 0 1 1 ] ) ;
set ( axisH , ’XTick ’ , [ 0 . 0 1 0 . 5 1 ] ) ;
xlabel ( ’\rho_{C:C}’ ) ;
t i t l e ( midGraphTitle ) ;
axisH= subplot ( 3 , 4 , 1 1 ) ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . rH7H7Sample ( ...
p e r i o d D e l t a A r r a y 2 n >= t a r g e t D e l t a ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 0 . 0 1 1 ] ) ;
set ( axisH , ’XTick ’ , [ 0 . 0 1 0 . 5 1 ] ) ;
xlabel ( ’\rho_{C:C}’ ) ;
t i t l e ( botGraphTitle ) ;
axisH= subplot ( 3 , 4 , 4 ) ;
261
e d g e s= 0 : 1 0 : 1 0 0 ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . wH7H7 H7H7Sample ( ...
o s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 1 1 0 0 ] ) ;
set ( axisH , ’XTick ’ , [ 1 50 1 0 0 ] ) ;
xlabel ( ’\ omega_ {C:C}’ ) ;
t i t l e ( topGraphTitle ) ;
axisH= subplot ( 3 , 4 , 8 ) ;
e d g e s= 0 : 1 0 : 1 0 0 ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . wH7H7 H7H7Sample ( ...
p e r i o d O s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 1 1 0 0 ] ) ;
set ( axisH , ’XTick ’ , [ 1 50 1 0 0 ] ) ;
xlabel ( ’\ omega_ {C:C}’ ) ;
t i t l e ( midGraphTitle ) ;
axisH= subplot ( 3 , 4 , 1 2 ) ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . wH7H7 H7H7Sample ( ...
p e r i o d D e l t a A r r a y 2 n >= t a r g e t D e l t a ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 1 1 0 0 ] ) ;
set ( axisH , ’XTick ’ , [ 1 50 1 0 0 ] ) ;
xlabel ( ’\ omega_ {C:C}’ ) ;
t i t l e ( botGraphTitle ) ;
f i g u r e ( 6 ) ; c l f ; colormap ( [ 0 . 5 0 . 5 0 . 5 ] ) ;
axisH= subplot ( 3 , 4 , 1 ) ;
e d g e s= 0 . 2 : 0 . 0 5 : 0 . 5 ;
262
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . bH7Sample ( ...
o s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 0 . 2 0 . 5 ] ) ;
set ( axisH , ’XTick ’ , [ 0 . 2 0 . 3 5 0 . 5 ] ) ;
xlabel ( ’\ beta_C ’ ) ;
t i t l e ( topGraphTitle ) ;
axisH= subplot ( 3 , 4 , 5 ) ;
e d g e s= 0 . 2 : 0 . 0 5 : 0 . 5 ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . bH7Sample ( ...
p e r i o d O s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 0 . 2 0 . 5 ] ) ;
set ( axisH , ’XTick ’ , [ 0 . 2 0 . 3 5 0 . 5 ] ) ;
xlabel ( ’\ beta_C ’ ) ;
t i t l e ( midGraphTitle ) ;
axisH= subplot ( 3 , 4 , 9 ) ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . bH7Sample ( ...
p e r i o d D e l t a A r r a y 2 n >= t a r g e t D e l t a ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 0 . 2 0 . 5 ] ) ;
set ( axisH , ’XTick ’ , [ 0 . 2 0 . 3 5 0 . 5 ] ) ;
xlabel ( ’\ beta_C ’ ) ;
t i t l e ( botGraphTitle ) ;
axisH= subplot ( 3 , 4 , 2 ) ;
e d g e s= 0 : 5 0 : 1 0 0 0 ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . kH13H13Sample ( ...
o s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 1 0 1 0 0 0 ] ) ;
set ( axisH , ’XTick ’ , [ 1 0 500 1 0 0 0 ] ) ;
263
xlabel ( ’\ kappa_ {G:G}’ ) ;
t i t l e ( topGraphTitle ) ;
axisH= subplot ( 3 , 4 , 6 ) ;
e d g e s= 0 : 5 0 : 1 0 0 0 ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . kH13H13Sample ( ...
p e r i o d O s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 1 0 1 0 0 0 ] ) ;
set ( axisH , ’XTick ’ , [ 1 0 500 1 0 0 0 ] ) ;
xlabel ( ’\ kappa_ {G:G}’ ) ;
t i t l e ( midGraphTitle ) ;
axisH= subplot ( 3 , 4 , 1 0 ) ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . kH13H13Sample ( ...
p e r i o d D e l t a A r r a y 2 n >= t a r g e t D e l t a ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 1 0 1 0 0 0 ] ) ;
set ( axisH , ’XTick ’ , [ 1 0 500 1 0 0 0 ] ) ;
xlabel ( ’\ kappa_ {G:G}’ ) ;
t i t l e ( botGraphTitle ) ;
axisH= subplot ( 3 , 4 , 3 ) ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . kH7H7Sample ( ...
o s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 1 0 1 0 0 0 ] ) ;
set ( axisH , ’XTick ’ , [ 1 0 500 1 0 0 0 ] ) ;
xlabel ( ’\ kappa_ {C:C}’ ) ;
t i t l e ( topGraphTitle ) ;
axisH= subplot ( 3 , 4 , 7 ) ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . kH7H7Sample ( ...
p e r i o d O s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
264
xlim ( [ 1 0 1 0 0 0 ] ) ;
set ( axisH , ’XTick ’ , [ 1 0 500 1 0 0 0 ] ) ;
xlabel ( ’\ kappa_ {C:C}’ ) ;
t i t l e ( midGraphTitle ) ;
axisH= subplot ( 3 , 4 , 1 1 ) ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . kH7H7Sample ( ...
p e r i o d D e l t a A r r a y 2 n >= t a r g e t D e l t a ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 1 0 1 0 0 0 ] ) ;
set ( axisH , ’XTick ’ , [ 1 0 500 1 0 0 0 ] ) ;
xlabel ( ’\ kappa_ {C:C}’ ) ;
t i t l e ( botGraphTitle ) ;
axisH= subplot ( 3 , 4 , 4 ) ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . kH7H13Sample ( ...
o s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 1 0 1 0 0 0 ] ) ;
set ( axisH , ’XTick ’ , [ 1 0 500 1 0 0 0 ] ) ;
xlabel ( ’\ kappa_ {C:G}’ ) ;
t i t l e ( topGraphTitle ) ;
axisH= subplot ( 3 , 4 , 8 ) ;
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . kH7H13Sample ( ...
p e r i o d O s c I n d i c e s 2 n ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 1 0 1 0 0 0 ] ) ;
set ( axisH , ’XTick ’ , [ 1 0 500 1 0 0 0 ] ) ;
xlabel ( ’\ kappa_ {C:G}’ ) ;
t i t l e ( midGraphTitle ) ;
axisH= subplot ( 3 , 4 , 1 2 ) ;
265
bar ( edges , h i s t c ( n o n l i n e a r S a m p l e . params . kH7H13Sample ( ...
p e r i o d D e l t a A r r a y 2 n >= t a r g e t D e l t a ) , e d g e s ) , ’histc ’ ) ;
xlim ( [ 1 0 1 0 0 0 ] ) ;
set ( axisH , ’XTick ’ , [ 1 0 500 1 0 0 0 ] ) ;
xlabel ( ’\ kappa_ {C:G}’ ) ;
t i t l e ( botGraphTitle ) ;
i f t r u e % f i s e m p t y ( v e r ( ’ s t a t s ’ ) ) % S t a t i s t i c s Toolbox not ...
available
disp ( [ 10 ’Statistics Toolbox is not available for making ...
box plots .’ ] ) ;
e l s e % S t a t i s t i c s Toolbox a v a i l a b l e
modelNames= c e l l ( length ( find ( periodOscMask1 ) ) + length ( find ( ...
periodOscMask2 ) ) , 1 ) ;
modelNames ( 1 : length ( find ( periodOscMask1 ) ) )= { ’1 Site ’ } ;
modelNames ( length ( find ( periodOscMask1 ) ) +1:end )= { ’2 Site ’ } ;
figure (5) ; c l f ;
subplot ( 2 , 2 , 1 ) ;
b o x p l o t ( [ n o n l i n e a r S a m p l e . params . t a u h 7 ( p e r i o d O s c I n d i c e s 1 ...
) ; n o n l i n e a r S a m p l e . params . t a u h 7 ( p e r i o d O s c I n d i c e s 2 ) ] , modelNames...
);
t i t l e ( ’\tau_{h7} delay ’ ) ;
ylabel ( ’\tau_{h7} delay (min .) ’ ) ;
subplot ( 2 , 2 , 3 ) ;
266
b o x p l o t ( [ n o n l i n e a r S a m p l e . params . rho ( p e r i o d O s c I n d i c e s 1 ) ; ...
n o n l i n e a r S a m p l e . params . rho ( p e r i o d O s c I n d i c e s 2 ) ] , modelNames ) ;
t i t l e ( ’\rho ’ ) ;
ylabel ( ’\rho ’ ) ;
subplot ( 2 , 2 , 2 )
b o x p l o t ( [ n o n l i n e a r S a m p l e . params . kH13H13 ( ...
p e r i o d O s c I n d i c e s 1 ) ; n o n l i n e a r S a m p l e . params . kH13H13 ( ...
p e r i o d O s c I n d i c e s 2 ) ] , modelNames ) ;
t i t l e ( ’\ kappa_ {13:13} ’ ) ;
ylabel ( ’\ kappa_ {13:13} ’ ) ;
subplot ( 2 , 2 , 4 )
b o x p l o t ( [ n o n l i n e a r S a m p l e . params . rH7H7 ( p e r i o d O s c I n d i c e s 1 ) ...
; n o n l i n e a r S a m p l e . params . rH7H7 ( p e r i o d O s c I n d i c e s 2 ) ] , modelNames ) ;
t i t l e ( ’\rho_ {7:7} ’ ) ;
ylabel ( ’\rho_ {7:7} ’ ) ;
% Show range o f D e l t a s
figure (6) ; c l f ;
b o x p l o t ( [ p e r i o d D e l t a A r r a y 1 ( p e r i o d O s c I n d i c e s 1 ) ; ...
p e r i o d D e l t a A r r a y 2 ( p e r i o d O s c I n d i c e s 2 ) ] , modelNames ) ;
t i t l e ( ’\ Delta Period across viable H_ {13.2} range ’ ) ;
ylabel ( ’\ Delta Period (min .) ’ ) ;
end
disp ( ’ ’ ) ;
267
end
Plotting Code
plotSolution.m: A simple plotting function called by somitogenesisPCW.m.
function p l o t S o l u t i o n ( s o l )
% Usage :
%
plotSolution ( sol )
%
% P l o t s v a r i o u s time s e r i e s f o r t h e f i r s t c e l l o n l y
i n d i c e s= [ s o l . params . H 7 T o t a l I n d i c e s ( 1 ) ; s o l . params . h 7 I n d i c e s ...
(1) ] ;
i f s o l . params . numGenes == 2
i n d i c e s= [ i n d i c e s ; s o l . params . D I n d i c e s ( 1 ) ; s o l . params . ...
dIndices (1) ] ;
end
s t a r t T i m e I n d e x= find ( s o l . x >= s o l . params . t i m e I n t e r v a l ( 1 ) , 1 ) ;
i f isempty ( s t a r t T i m e I n d e x )
s t a r t T i m e I n d e x= 1 ;
end
maxY= max(max( s o l . y ( i n d i c e s , s t a r t T i m e I n d e x : end ) ) ) ;
figure (1) ;
a x i s 1= subplot ( 3 , 1 , 1 ) ;
s e r i e s 1= plot ( a x i s 1 , s o l . x , s o l . y ( i n d i c e s , : ) , ’LineWidth ’...
,2) ;
268
ylim ( a x i s 1 , [ 0 , 1 . 1 ∗maxY ] ) ;
set ( a x i s 1 , ’FontWeight ’ , ’demi ’ ) ;
xlabel ( ’time (min)’ , ’FontSize ’ , 1 2 ) ;
ylabel ( ’Copy #’ , ’FontSize ’ , 1 2 ) ;
i f s o l . params . c o u p l i n g B o o l
t i t l e ( [ num2str ( s o l . params . numCells , ’%d’ ) , ’ Coupled ...
Cell(s) -- Cell 1’ ] , ’FontSize ’ , 1 4 ) ;
else
t i t l e ( [ num2str ( s o l . params . numCells , ’%d’ ) , ’ Uncoupled ...
Cell(s) -- Cell 1’ ] , ’FontSize ’ , 1 4 ) ;
end
i f s o l . params . numGenes == 1
legend ( s e r i e s 1 ( end−1:end ) , [ ’$H_ {\ widehat {7}}= ’ num2str...
( s o l . s t a t i s t i c s . medians ( 1 ) , ’%.1f’ ) ’ \pm ’ num2str ( s o l . ...
s t a t i s t i c s . a m p l i t u d e s ( 1 ) / 2 , ’%.1f’ ) ’; ’ num2str ( s o l . s t a t i s t i c s . ...
p e r i o d s ( 1 ) , ’%.1f’ ) ’$ min ’ ] , [ ’$h_7= ’ num2str ( s o l . s t a t i s t i c s . ...
medians ( 2 ) , ’%.1f’ ) ’ \pm ’ num2str ( s o l . s t a t i s t i c s . a m p l i t u d e s ( 2 ) ...
/ 2 , ’%.1f’ ) ’; ’ num2str ( s o l . s t a t i s t i c s . p e r i o d s ( 2 ) , ’%.1f’ ) ’$ ...
min ’ ] ) ;
set ( legend ( a x i s 1 ) , ’Location ’ , ’West ’ , ’Interpreter ’ , ’...
latex ’ ) ;
e l s e % s o l . params . numGenes == 2
legend ( s e r i e s 1 ( end−3:end ) , [ ’$H_ {\ widehat {7}}= ’ num2str...
( s o l . s t a t i s t i c s . medians ( 1 ) , ’%.1f’ ) ’ \pm ’ num2str ( s o l . ...
s t a t i s t i c s . a m p l i t u d e s ( 1 ) / 2 , ’%.1f’ ) ’; ’ num2str ( s o l . s t a t i s t i c s . ...
p e r i o d s ( 1 ) , ’%.1f’ ) ’$ min ’ ] , [ ’$h_7= ’ num2str ( s o l . s t a t i s t i c s . ...
medians ( 2 ) , ’%.1f’ ) ’ \pm ’ num2str ( s o l . s t a t i s t i c s . a m p l i t u d e s ( 2 ) ...
269
/ 2 , ’%.1f’ ) ’; ’ num2str ( s o l . s t a t i s t i c s . p e r i o d s ( 2 ) , ’%.1f’ ) ’$ ...
min ’ ] , [ ’$D= ’ num2str ( s o l . s t a t i s t i c s . medians ( 3 ) , ’%.1f’ ) ’ \pm...
’ num2str ( s o l . s t a t i s t i c s . a m p l i t u d e s ( 3 ) / 2 , ’%.1f’ ) ’; ’ num2str ( ...
s o l . s t a t i s t i c s . p e r i o d s ( 3 ) , ’%.1f’ ) ’$ min ’ ] , [ ’$d= ’ num2str ( s o l ...
. s t a t i s t i c s . medians ( 4 ) , ’%.1f’ ) ’ \pm ’ num2str ( s o l . s t a t i s t i c s . ...
a m p l i t u d e s ( 4 ) / 2 , ’%.1f’ ) ’; ’ num2str ( s o l . s t a t i s t i c s . p e r i o d s ( 4 ) , ...
’%.1f’ ) ’$ min ’ ] ) ;
set ( legend ( a x i s 1 ) , ’Location ’ , ’West ’ , ’Orientation ’ , ’...
vertical ’ , ’Interpreter ’ , ’latex ’ ) ;
end
x L i m i t s= xlim ; % Used i n p l o t s b e l o w
a x i s 2= subplot ( 3 , 1 , 2 ) ;
a r e a ( a x i s 2 , s o l . x , [ s o l . output . H7H13 ( 1 , : ) ’ , s o l . output . ...
H7H7 ( 1 , : ) ’ , s o l . output . H7 ( 1 , : ) ’ ] ) ;
xlim ( a x i s 2 , x L i m i t s ) ;
ylim ( a x i s 2 , [ 0 , 1 . 1 ∗maxY ] ) ;
set ( a x i s 2 , ’FontWeight ’ , ’demi ’ ) ;
xlabel ( ’time (min)’ , ’FontSize ’ , 1 2 ) ;
ylabel ( ’Copy #’ , ’FontSize ’ , 1 2 ) ;
t i t l e ( ’Her7 Distribution -- Cell 1’ , ’FontSize ’ , 1 4 ) ;
legend ( ’$H_ {7:13} $’ , ’$H_ {7:7} $’ , ’$H_7$ ’ ) ;
set ( legend ( a x i s 2 ) , ’Location ’ , ’West ’ , ’Interpreter ’ , ’...
latex ’ ) ;
a x i s 3= subplot ( 3 , 1 , 3 ) ;
270
a r e a ( a x i s 3 , s o l . x , [ s o l . output . H7H13 ( 1 , : ) ’ , s o l . output . ...
H13H13 ( 1 , : ) ’ , s o l . output . H13 ( 1 , : ) ’ ] ) ;
xlim ( a x i s 3 , x L i m i t s ) ;
set ( a x i s 3 , ’FontWeight ’ , ’demi ’ ) ;
xlabel ( ’time (min)’ , ’FontSize ’ , 1 2 ) ;
ylabel ( ’Copy #’ , ’FontSize ’ , 1 2 ) ;
t i t l e ( ’Her13 .2 Distribution -- Cell 1’ , ’FontSize ’ , 1 4 ) ;
legend ( ’$H_ {7:13} $’ , ’$H_ {13:13} $’ , ’$H_ {13}$’ ) ;
set ( legend ( a x i s 3 ) , ’Location ’ , ’NorthEast ’ , ’Interpreter ’ , ...
’latex ’ ) ;
end