Data provenance in biomedical discovery Donald Dunbar Queen’s Medical Research Institute
advertisement
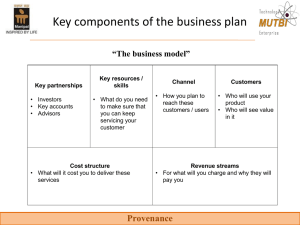
Data provenance in biomedical discovery Donald Dunbar Queen’s Medical Research Institute University of Edinburgh Workshop on Principles of Provenance in Databases May 21st 2008 Background biomedical research basic & clinical science animal, cell models, patients genes, proteins, pathways data analysis & mining publication Biomedical discovery • Looking for contribution to – human health and disease • In house experiments – data workflows – knowledge capture • Use public databases – many data types – integration is a problem Databases we use sequence structure function expression domain specific Data workflows database experiment 1 experiment 2 raw data spreadsheet publication processed data database calculations Data workflows IN copy and paste open from file ‘algorithm’ copy and paste BUT: web services automated tools & databases bioinformatics workflows save to file OUT Bioinformatics workflows Is our field changing? databases experiments knowledge knowledgebase Knowledge capture Knowledge capture What provenance to we need? Example: Gene expression in a transgenic animal gene expression measurements gene annotation where, when public databases which identifiers integration when, what, how output from machine how processing what and how did we select genes data mining … What provenance to we need? Example: Curated protein database database links expert data contributor, date curator input source, identifiers, dates development verify, add, delete, modify archive versions, dates Curated database schema & interface changes What do we do now (for provenance)? • We trust the main data providers a lot! – a pragmatic approach • We use tools and note the settings – rarely fully • We put extra fields in our databases – source, modify date • We deposit our data in public repositories – but only when we need to What might we do next? • Use workflow tools like Taverna – capture workflow provenance • Build provenance tool & database – widely applicable • Make provenance more visible to biologists – so they value and use it Conclusions • • • • • In biology we don’t do provenance well (yet) We use databases and manual workflows We implement rudimentary provenance We should build useful provenance tools We need to make provenance visible