CARMEN: Code Analysis, Repository and Modelling for e-Neuroscience
advertisement

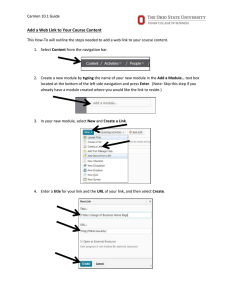
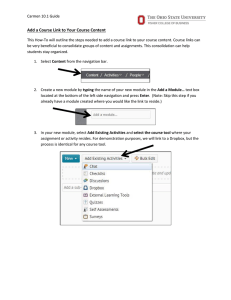
CARMEN: Code Analysis, Repository and Modelling for e-Neuroscience Research Challenge Understanding the brain may be the greatest informatics challenge of the 21st century Worldwide >100,000 neuroscientists (~ 5,000 in UK) are generating vast amounts of data Principal experimental data formats: molecular (genomic/proteomic) neurophysiological (time-series electrical measures of activity) anatomical (spatial) behavioural Neuroinformatics concerns how these data are handled and integrated, including the application of computational modelling Neuroinformatics In recent years new technological opportunities for data sharing have emerged with faster networks, improved database technologies, and affordable massive data storage capabilities Neuroinformatics is increasingly exploiting these opportunities to enable data sharing, re-use of data and novel analysis based on new combinations of data that can be performed via database systems Need for Cooperation Understanding the brain may be the greatest informatics challenge of the 21st century OECD identified a need to work cooperatively in order to achieve major advances and have established the International Neuroinformatics Coordinating Facility Cooperation will permit: development of common processes best value from data – long term curation ‘mega-analysis’ of large data sets integration of data sets across different scales and different approaches interdisciplinary research Potential Barriers to Cooperation Technical Multiple proprietary data formats Need for detailed, standardised and evolvable metadata Volume of the data to be analysed Cultural Multiple communities each acting independently Concerns about the consequences of sharing data Difficulty in appreciating how the science could be moved forwards by e-Science CARMEN – Focus on Neural Activity Understanding the brain may be the greatest informatics challenge of the 21st century raw voltage signal data is collected using single or multi-electrode array recording novel optical recording, particularly the activity dynamics of large networks resolving the ‘neural code’ from the timing of action potential activity neurone 1 neurone 2 neurone 3 Electrophysiological Data Much current knowledge about brain function is based on analysis of firing patterns of individual neurones. New computer-based data acquisition systems and techniques for recording simultaneously from many neurones means data are amassing rapidly. Neural modelling generates massive simulated data sets that need to be processed, analysed and compared with experimental data. Neuronal recordings can be intra- or extra-cellular recordings of single spikes, ensembles of neurones, or field potentials. All of these data are types of time-series data which require a specialised information handling system. CARMEN Objectives To demonstrate and sustain advances in neuroscience enabled by e-Science technology To create a grid-enabled, real time ‘virtual laboratory’ environment for neurophysiological data To develop an extensible, client-defined ‘toolkit’ for data extraction, analysis and modelling To provide a repository for archiving, sharing, integration and discovery of data To achieve wide community and commercial engagement in developing and using CARMEN Project Exemplar Recording from brain tissue removed from epileptic patients (scarce tissue and data rates up to 20 GB/h) On line analysis by distributed collaborators will enable experiment to be defined during data collection Repository will enable integration of rare case types from different laboratories New knowledge will lead to advances in treatment CARMEN Consortium Newcastle: Colin Ingram Paul Watson Stuart Baker Marcus Kaiser Phil Lord Evelyne Sernagor Tom Smulders Miles Whittington Cambridge: Stephen Eglen York: Leicester: Rodrigio Quian Quiroga Imperial: Simon Schultz Stirling: Jim Austin Tom Jackson Warwick: Jianfeng Feng Sheffield: Kevin Gurney Paul Overton Manchester: Stefano Panzeri Leslie Smith St. Andrews: Anne Smith Plymouth: Roman Borisyuk CARMEN Consortium Commercial Partners - applications in the pharmaceutical sector - interfacing of data acquisition software - application of database infrastructure - commercialisation of analysis tools Work Packages WP1 Spike Detection & Sorting WP 3 Data-Driven Parameter Determination in ConductanceBased Models WP2 Information Theoretic Analysis of Derived Signals Data Storage & Analysis WP5 Measurement and Visualisation of Spike Synchronisation WP4 Intelligent Database Querying WP6 Multilevel Analysis and Modelling in Networks CARMEN Structure Hub and Spoke Project Hub: A “CAIRN” repository for the storage and analysis of neuroscience data Spokes: A set of neuroscience projects that will produce data and analysis services for the hub, and use it to address key neuroscience questions e-Science Challenges Managing vast amounts of data > 50TB primary data Extracting value from the data discovery & interpretation analysis – harnessing compute resources curation of services as well as data Controlling access to the data & services CARMEN Active Information Repository Node OMII/ myGrid: Taverna/ BPEL OGSA-DAI & SRB Web Portal DAME: Signal Data Explorer Rich Clients ....... Web Portal Gold: Role & Task based Security OMII: Grimoire Security ....... Workflow Enactment Engine Compute Cluster on which Services are Dynamically Deployed Data myGrid Metadata Registry Service Repository & Gold: Feta, Provenance Dynasoar White Rose Grid Newcastle Grid A Typical Scenario we want to Support • Data Collection from Electrode Array • Spike Detection • with User Defined Threshold • Spike Sorting • Analysis • Visualisation Currently, this is a semi-manual process We have an initial prototype for automating this…. Signal Data Explorer Example Workflow Example Workflow Enactment External Client Workflow Engine BPEL / TAVERNA Repository INPUT Data Spike Sorting Service Security Available Services SRB FileSystem Registry Reporting Query RDBMS Dynamically Deployed Services in Dynasoar OUTPUT Metadata Example Graph Output Example Movie Output Some Remaining Challenges Extensible, standardised metadata for neuroscience data formats (timing, data channels, etc.) experimental design (e.g. stimuli or drug treatments) concurrent data (e.g. behaviour, physiological measures) experimental idiosyncrasies (e.g. artifacts) experimental conditions (animals, temperature, treatments etc.) Some Remaining Challenges (cont.) Locating patterns in time-series data across multiple levels of abstraction Reproducible e-Science curating services as well as data public repositories of deployable services dynamic service deployment Real-time expert collaboration CARMEN CARMEN is delivering an e-Science infrastructure that can be applied across a range of diverse and challenging applications (not only neuroscience) CARMEN enables cooperation and interdisciplinary working in ways currently not possible CARMEN will deliver new results in neuroscience, computer science and medicine