This Document Contains:
advertisement

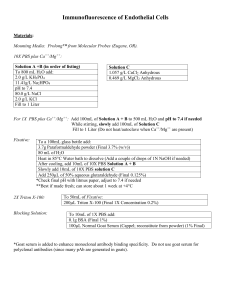
This Document Contains: 1. In-Cell Western Protocol II. Cell Seeding and Stimulation Supplemental Protocol III. Complete Assay Example: Detailing the Seeding, Stimulation and Detection of the A431 Cellular Response to Epidermal Growth Factor PI-140 0103 Doc #988-07083 ® I. In-Cell Western Protocol Materials needed: Odyssey® reagents: IR-labeled secondary antibodies Odyssey® Blocking Buffer Additional reagents needed: Primary antibodies 1X PBS wash buffer Tween-20 Triton X-100 37% formaldehyde 1. Grow cells in 96- or 384-well plates until confluent by standard procedures. Serum-starve cells if necessary and proceed to stimulate or monitor cells. • This step varies dramatically, depending on the experimental goal. Conditions such as cell type, dose of ligand(s) and inhibitor(s), stimulation time, and temperature should be decided according to the experimental system targeted. • See Supplemental Protocol II, page 5, for specification of 96-well plates and example condi tions of cell seeding, starvation, and stimulation. • Continue experimentation with step 2 immediately after completion of the desired cell stimulation. 2. Remove activation or stimulation media manually or by aspiration. Immediately fix cells with 4% formaldehyde in PBS for 20 minutes at room temperature. • Prepare fresh Fixing Solution as follows: 45 ml 5 ml Final 50 ml 1X PBS 37% Formaldehyde 3.7% Formaldehyde • Using multi-channel pipettor, add 150 µl of fresh Fixing Solution (room temperature solution, RT). Make sure to add solution to side of well to avoid disruption of cell attachment at well bottom. • Allow incubation on bench top for 20 minutes at RT with no shaking. 3. Wash four times with 1X PBS containing 0.1% Triton X-100 (cell permeablization) for 5 minutes per wash. • Prepare Triton Washing Solution as follows: 495 ml 5 ml Final 500 ml 1X PBS 10% Triton X-100 1X PBS + 0.1% Triton X-100 • Remove Fixing Solution into appropriate waste container (contains formaldehyde). • Using multi-channel pipettor, add 200 µl of Triton Washing Solution (RT). Make sure to add solution to side of well to avoid disruption of cell attachment at well bottom. • Allow wash to shake on a rotator for 5 minutes at RT. Continued Page 2 • Repeat washing steps 4 more times after removing wash manually. • Do not allow cells/wells to become dry during washing. Immediately add the next wash after disposal. 4. Add Odyssey Blocking Buffer and incubate for 1 hour at RT with moderate shaking. • Using multi-channel pipettor, block cells/wells with LI-COR® Odyssey Blocking Buffer (#927-40000) using 150 µl for each well. Make sure to add solution to side of well to avoid disruption of cell attachment. • Allow blocking for 60 minutes at room temperature with moderate shaking on a rotator. • Plates can be blocked overnight at 4°C if desired. • Do not add Tween-20 during blocking. • Odyssey blocking reagent will yield higher sensitivity, consistency, and performance than other blocking agents. Nonfat dry milk, casein solutions, or BSA are not recommended. 5. For simultaneous detection of two antigens, the primary and secondary antibodies must be carefully selected to meet the following criteria: • Design the experiment carefully if the analysis is targeting detection of two antigenic epitopes within the same protein (for example, both total ERK and phosphorylation specific ERK). It is important to choose two antibodies against two epitopes, so that the antigenic regions are separated enough that the antibodies do not compete or interfere with the bind ing of each other. This can be validated by one- and two-color Western blot experiments. • After antibodies have been chosen, test the primary antibodies by Western blotting with activated and resting lysates using the Odyssey system. This analysis brings confidence to the specificity of the primary antibodies within your experimental design and the detection system. • The two primary antibodies must be derived from different host species so they can be discriminated by secondary antibodies of different specificities (for example, primary antibod ies from rabbit and mouse will be discriminated by anti-rabbit and anti-mouse secondary antibodies). • For best results, avoid using primary antibodies from mouse and rat together for a two-color experiment. Because the species are so closely related, it is not possible to completely adsorb away cross-reactivity. Substantial cross-reactivity between bands may occur. Continued Page 3 6. Add the two primary antibodies into a tube containing Odyssey Blocking Buffer. The optimum dilution will depend on your antibody, and should be determined empirically. A suggested starting range is 1:20 – 1:500 with at least a 10-fold greater amount of antibody than that used in Western blotting. • Mix the primary antibody solution thoroughly before adding to wells. • Remove blocking buffer from the blocking step and add 50 µl of the desired primary antibody or antibodies in Odyssey Blocking Buffer to cover the bottom of each well (add 20 ul for 384-well format). 7. Incubate with primary antibody for 2 hours or longer with gentle shaking at RT. The optimum incubation times will vary for different primary antibodies. 8. Wash the plate 4 times with 1X PBS + 0.1% Tween-20 for 5 minutes at RT with gentle shaking, using a generous amount of buffer. • Prepare Tween Washing Solution as follows: 995 ml 5 ml 1000 ml 1X PBS 20% Tween-20 1X PBS with 0.1% Tween-20 • Using a multi-channel pipettor, add 200 µl of Tween Washing Solution (RT). Make sure to add solution to the side of wells to avoid disruption of cell attachment at well bottom. • Allow wash to shake on a rotator for 5 minutes at RT. Repeat washing steps 4 more times. 9. The two secondary antibodies used must be derived from the same host species (usually goat) so they will not react against each other. One secondary antibody must be labeled with IRDye™ 800CW, and the other with Alexa Fluor 680. We recommend that you use IRDye 800CW second ary to detect the more abundant antigen or stronger signal, and Alexa Fluor 680 secondary to detect the less abundant antigen, or weaker signal (such as protein phosphorylation). 10. Dilute the fluorescently labeled secondary antibody in Odyssey Blocking Buffer. Avoid prolonged exposure of the antibody vials to light. Suggested dilution range is 1:100 – 1:800 (usually 1:200) for Alexa Fluor 680 conjugates and 1:800 for IRDye 800CW conjugates). Add Tween-20 to the diluted antibody at a final concentration of 0.1 – 0.5%, to lower background. 11. Add 50 µl of the secondary antibody solution to each well and incubate for 60 minutes with gentle shaking at RT. Protect plate from light during incubation. 12. Wash the plate 4 times with 1X PBS containing 0.1% Tween-20 for 5 minutes at RT with gentle shaking, using a generous amount of buffer. • Using a multi-channel pipettor, add 200 µl of Tween Washing Solution (RT). Make sure to add solution to the side of wells to avoid disruption of cell attachment at well bottom. • Allow wash to shake on a rotator for 5 minutes at RT. Repeat washing steps 4 more times. • Protect plate from light during washing. 13. Scan in the plate using the appropriate channels for detection with the Odyssey Imager (700 nm detection for Alexa Fluor 680 antibody and 800 nm detection for IRDye 800CW antibody). • Before scanning, clean the bottom plate surface and also the glass surface of the Odyssey Imager with moist, lint-free paper to avoid obstructions during scanning or final image. Page 4 Continued • Image settings suggested for cell based assays include a 3.0 mm focus offset for optimal detec tion within plates (see plate recommendation in Supplemental Protocol). Plates other than LI-COR recommendations may require lower or higher focus offsets for optimal resolution and detection. • Intensity settings for both 700 and 800 nm channels should be set to 5 for initial image scan ning. If the image signal is saturated or too high, scan using a lower intensity setting (i.e., 2.5). If the image signal is too low, scan using a higher intensity setting (i.e., 7.5). • Scan settings of medium to lowest quality with a 169 mm resolution provide satisfactory results with the least necessary scan time. • Plates stored for future use should remain protected from light. The fluorescent signal will remain stable for several weeks or longer if protected from light. Plates may be stored dry at 4°C or -20°C. II. Supplemental Protocol: Example Protocol for Cell Seeding and Stimulation Cell Seeding: Proper selection of microwell plates is necessary as each plate holds its own characteristics including well depth, plate autofluorescence, and well-to-well signal crossover. For cell based assays requiring sterile plates for tissue culture growth, experimentation, and optimal imaging, use the following plates recommended by LI-COR Biosciences (plates must have clear or black wells with a clear detection surface): Nunc-Nalgene (Part No. 167008) for 96-well format Falcon (BD Biosciences Part No. 353961) for 384-well format. Through experience, we have found that a confluent T75 flask (~1.5x107 cells) can produce 2 to 4 96-well plates for use in the cell based assay. The range in number of plates generated will depend on the cell type and the desired time for expansion to well confluency. This example protocol generates a seeding density of approximately 1x104 to 2.5x104 cells per well. Cell growth should be monitored daily with expansion in complete media usually yielding well confluency with ~5x104 cells per well in 2 to 3 days. The following protocol can be used as an example for cell seeding, growth, and starvation prior to cell stimulation and analysis. Optimization may be required due to variation in cell line properties, scaling, and seeding density. 1. Allow cell growth in a T75 flask using standard tissue culture procedures until cells reach confluency (~1.5x107 cells). 2. Remove growth media, wash cells with 1X PBS, and trypsinize cells for displacement. 3. Neutralize trypsinized cells with culture media and clarify by centrifugation. Continued Page 5 4. Remove supernatant and disrupt the cell pellet manually by hand tapping the collection tube. To maintain cell integrity, avoid using pipet or vortex during pellet disruption. 5. Reconstitute cells in 40 ml of complete media (2 plates x 96 wells x 200 µl/well = ~40 ml). 6. 7. Thoroughly mix the cell suspension manually. Under sterile conditions, dispense 200 µl of the cell suspension per well (50 µl for 384-well format). 8. Incubate cells and monitor cell density until confluency is achieved with well-to-well consistency. Starvation and Activation: Both the starvation and activation of cells will greatly depend on the desired experimental conditions. In some situations, starvation would not be required if the stimulant is absent from the complete media. Variance in the time, manner, and source of activation will depend on the desired cell-based assay. 1. Warm serum-free media to 37ºC. 2. Remove complete media from plate wells by aspiration or manual displacement. 3. Replace media with 200 µl of pre-warmed, serum-free media per well. 4. Allow cells to incubate an additional 16 to 24 hours to ensure metabolism of complete media constituents. 5. Remove starvation media from plate wells by aspiration or manual displacement. 6. Add either serum-free media plus 1% BSA for resting cells (mock) or serum-free media plus 1% BSA with stimulation ligand such as EGF for activated cells. Use 100 µl of resting/activation media per well. 7. Allow incubation at 37ºC for desired stimulation time. 8. Remove resting or activation media and proceed immediately to the fixation step in Cell Based Assay Experimental Protocol. Page 6 III. Complete Assay Example: Detailing the Seeding, Stimulation and Detection of the A431 Cellular Response to Epidermal Growth Factor Materials Needed: Odyssey reagents: Additional reagents needed: IR-labeled secondary antibodies Odyssey Blocking Buffer Primary antibodies 1X PBS wash buffer Tween-20 Triton X-100 Epidermal Growth Factor Nunc-Nalgene 96-well Microplate (Part #167008) 1. Allow A431 (ATCC; CRL-1555) cell growth in a T75 flask using standard tissue culture procedures until cells reach confluency (~1.5x107 cells; DMEM, 16.5% FBS; Gibco). 2. Remove growth media, wash cells with 1X PBS, and trypsinize cells for displacement. 3. Neutralize trypsinized cells with culture media and clarify by centrifugation. 4. Remove supernatant and disrupt the cell pellet manually by hand tapping the collection tube. To maintain cell integrity, avoid using pipet or vortex during pellet disruption. 5. Reconstitute cells in 40 ml of complete media (2 plates x 96 wells x 200 µl/well = ~40 ml). 6. Thoroughly mix the cell suspension manually. 7. Under sterile conditions, dispense 200 µl of the cell suspension per well in Nunc-Nalgene 96-well microplate. 8. Incubate cells and monitor cell density until confluency is achieved with well-to-well consistency. 9. Warm serum-free media (D-MEM; Gibco) to 37ºC. 10. Remove complete media from plate wells by aspiration or manual displacement. 11. Replace media with 200 µl of pre-warmed, serum-free media per well. 12. Allow cells to incubate an additional 16 to 24 hours to ensure metabolism of complete media constituents. 13. Remove starvation media from plate wells by aspiration or manual displacement. 14. Add either serum-free media plus 1% BSA for resting cells (mock) or serum-free media plus 1% BSA with 100 ng/ml EGF for activated cells. Use 100 µl of resting/activation media per well. 15. Allow incubation at 37ºC for 7.5 minutes. Continued Page 7 16. Remove activation or stimulation media manually or by aspiration. Immediately fix cells with 4% formaldehyde in 1X PBS for 20 minutes at room temperature. • Prepare fresh Fixing Solution as follows: 45 ml 5 ml 50 ml 1X PBS 37% Formaldehyde 3.7% Formaldehyde • Using multi-channel pipettor, add 150 µl of fresh Fixing Solution (room temperature solution, RT). Make sure to add solution to side of well to avoid disruption of cell attachment at well bottom. • Allow incubation on bench top for 20 minutes at RT with no shaking. 17. Wash four times with 1X PBS containing 0.1% Triton X-100 (cell permeablization) for 5 minutes per wash. • Prepare Triton Washing Solution as follows: 495 ml 1X PBS 10% Triton X-100 5 ml 500 ml 1X PBS + 0.1% Triton X-100 • Remove Fixing Solution in appropriate waste container (contains formaldehyde). • Using multi-channel pipettor, add 200 µl of Triton Washing Solution (RT). Make sure to add solution to side of well to avoid disruption of cell attachment at well bottom. • Allow wash to shake on a rotator for 5 minutes at RT. • Repeat washing steps 4 more times after manually removing wash. • Do not allow cells/wells to become dry during washing. Immediately add the next wash after disposal. 18. Add Odyssey Blocking Buffer and incubate for 1 hour at RT with moderate shaking. • Using multi-channel pipettor, block cells/wells with LI-COR Blocking Buffer (#927-40000), using 150 µl for each well. Make sure to add solution to side of well to avoid disruption of cell attachment. • Allow blocking for 60 minutes at room temperature, with moderate shaking on a rotator. 19. Add the two primary antibodies into a tube containing Odyssey Blocking Buffer. Combine solutions as defined below for each target analysis: A. Phospho-EGFR Tyr1045 (Rabbit; 1:100 dilution; Cell Signaling Technology 2237) Total EGFR (Mouse; 1:500 dilution; Biosource International AHR5062) B. Phospho-EGFR Tyr1045 (Rabbit; 1:100 dilution; Cell Signaling Technology 2237) Total ERK (Mouse; 1:50 dilution; Santa Cruz Biotechnology SC-1647) C. Phospho-ERK (Mouse; 1:100 dilution; Santa Cruz Biotechnology SC-7383) Total ERK (Rabbit; 1:100 dilution; Santa Cruz Biotechnology SC-94) Continued Page 8 • Mix the primary antibody solution thoroughly before adding to wells. • Remove blocking buffer from the blocking step and add 50 µl of the desired primary antibody or antibodies in Odyssey Blocking Buffer to cover the bottom of each well. 20. Incubate with primary antibody for 2 hours with gentle shaking at RT. 21. Wash the plate 4 times with 1X PBS + 0.1% Tween-20 for 5 minutes at RT with gentle shaking, using a generous amount of buffer. • Prepare Tween Washing Solution as follows: 995 ml 1X PBS 20% Tween-20 5 ml 1000 ml 1X PBS with 0.1% Tween-20 • Using a multi-channel pipettor, add 200 µl of Tween Washing Solution (RT). Make sure to add solution to the side of wells to avoid disruption of cell attachment at well bottom. • Allow wash to shake on a rotator for 5 minutes at RT. Repeat washing steps 4 more times. 22. Dilute the fluorescently labeled secondary antibody in Odyssey Blocking Buffer and add 0.5% Tween-20 to the diluted antibody to lower background as specified below: • A. Goat anti-rabbit Alexa Fluor 680 (1:200 dilution; Molecular Probes) Goat anti-mouse IRDye 800CW (1:800 dilution; Rockland Immunochemicals) B. Goat anti-rabbit Alexa Fluor 680 (1:200 dilution; Molecular Probes) Goat anti-mouse IRDye 800CW (1:800 dilution; Rockland Immunochemicals) C. Goat anti-mouse Alexa Fluor 680 (1:200 dilution; Molecular Probes) Goat anti-rabbit IRDye 800CW (1:800 dilution; Rockland Immunochemicals) Avoid prolonged exposure of the antibody vials to light. 23. Mix the antibody solutions thoroughly, add 50 µl of the secondary antibody solution to each well and incubate for 60 minutes with gentle shaking at RT. Protect plate from light during incubation. 24. Wash the plate 4 times with 1X PBS + 0.1% Tween-20 for 5 minutes at RT with gentle shaking, using a generous amount of buffer. • Using a multi-channel pipettor, add 200 µl of Tween Washing Solution (RT). Make sure to add solution to the side of wells to avoid disruption of cell attachment at well bottom. • Allow wash to shake on a rotator for 5 minutes at RT. Repeat washing steps 4 more times. • Protect plate from light during washing. 25. Scan the plate with detection in both the 700 and 800 channels using the Odyssey Imager (700 nm detection for Alexa Fluor 680 antibody and 800 nm detection for IRDye 800CW antibody). • Before scanning, clean the bottom plate surface and the Odyssey Imager scanning bed with moist, lint-free paper to avoid obstructions during scanning. • Complete plate imaging with a scan setting of medium quality, 169 µm resolution, and intensity setting of 5 for both 700 and 800 nm channels. Page 9 Results Typical assay results after completing the above protocol for the quantitative and simultaneous measurements of total ERK and phosphorylation of ERK in response to EGF stimulation: Replicate1/2 1 2 3 4 5 6 7 8 9 10 11 12 Background 100% Sample Sample Sample Sample Sample Sample Sample Sample Sample Sample Rep 1 800 3.23 76.59 74.34 69.96 71.72 71.82 71.3 72.59 72.21 70.67 70.11 68.35 Rep 2 800 3.67 76.7 77.4 73.25 71.33 72.96 72.48 72.82 73.19 70.41 69.54 69.47 Rep 1 700 21.54 53.7 60.26 54.93 58.17 59.38 60.87 65.54 75.4 Rep 2 700 25.61 52.57 59.77 58.76 60.76 60.57 61.76 66.25 75.15 101.3 144.44 225.61 96.41 150.21 231.78 The previous chart details resultant raw data after completing the Complete Assay Example. Within the example, all wells contained confluent A431 cells. Columns 3 through 12 were treated with different doses of EGF as indicated, while columns 1 and 2 were left as resting cells. After stimulation, permeabilization, and blocking of cells, primary antibodies were added to wells 2 through 12. Secondary antibody was then added to all wells, resulting in the first well to serve for assay background subtraction. Final data would typically be interpreted after background subtraction and normalization against the total protein detection measurements. Page 10 Experimental Considerations • Proper selection of microplates can significantly affect the results of your analysis, as each plate has its own characteristics, including well depth, plate autofluorescence, and well-to-well signal crossover. Try to use the general considerations for microplate selection provided below. • In-Cell Western analyses use detection at the well surface with no liquid present. This results in minimal well-to-well signal spread, allowing the use of both clear and black well plates. However, do not use plates with white wells, since the autofluorescence from the white surface will create significant noise. • For In-Cell Western assays requiring sterile plates for tissue culture growth, LI-COR Biosciences recommends using the following plates: 96 well format 96 well format Nunc-Nalgene (Part Number 161093, 165305) Falcon (Part Number 353075, 353948) 384 well format 384 well format Nunc-Nalgene (Part Number 164688, 164730) Falcon (Part Number 353961, 353962) • Plate dimensions necessary for use with the Odyssey Imager include a maximum 4.0 mm distance from the Odyssey Imager glass bed to the target detection area of the plate. When using the plates specified above for In-Cell Western assays, the recommended focus offset is 3.0 mm. • Plates deviating from LI-COR recommendations may require lower or higher focus offsets for optimal resolution and detection. If alternative plates are used, an initial optimization scan will be necessary. Scan a plate containing experimental and control samples at 0.5, 1.0, 2.0, 3.0, and 4.0 mm focus offsets. Use the same intensity settings for each scan. After reviewing the collected scans, use the focus offset with the highest signal-to-noise as your focus offset for experiments with alternate plates. • Protect plates from light before imaging to ensure highest sensitivity. When storing plates after imag ing, they should be protected from light at room temperature or 4ºC. • Intensity for both 700 and 800 nm channels should be set to 5 for initial scanning. If your image sig nal is saturated or too high, re-scan using a lower intensity setting (i.e., 2.5). If your image signal is too low, re-scan using a higher intensity setting (i.e., 7.5). • Scan settings of medium to lowest quality, with 169 mm resolution, provide satisfactory results with minimal scan time. Higher scan quality and/or resolution may be used, but will increase scan time. • Establish the specificity of your primary antibody by screening plate-like lysates through Western blotting and detection on the Odyssey imager. If significant non-specific banding is present, choose alternative primary antibodies to avoid results with non-specific signal detection. Page 11