From Univariate to Multivariate imaging Genetic Analyses
advertisement

From Univariate to
Multivariate imaging
Genetic Analyses
Thomas Nichols
Department of Statistics,
Warwick Manufacturing Group
University of Warwick
Joint with
Maria Vounou, Giovanni Montana
Statistics Section, Dept. of Mathematics
Imperial College
Imaging Genetics Menu
Imaging
Candidate ROI
Many ROI
Voxelwise
Genetics
Candidate SNP
Candidate Gene
Genome-wide SNP
[Filippini et al. 2009]
29,812 voxels
1 SNP
[Joyner et al. 2009]
4 ROIs, 11 SNPs
[Potkin et al. 2009]
1 BOLD ROI
317, 503 SNPs
[Stein et al. 2010]
31,622 voxels
448,293 SNPs
Genome-wide Gene
(Jason Stein/Andy Saykin/Bertrand Thirion)
• Voxel/Region QTL
– Whole genome
association
– Must have right ROI
100,000
voxels
• Candidate SNP
– Full image result
– Must have right SNP
500,000 SNPs
≈ 1010
tests!
500,000 SNPs
≈ 106
tests
500,000 SNPs
100,000
voxels
• Full cross analysis
– Massive multiple testing
problem!
100,000
voxels
Possible MassUnivariate Analyses
≈ 105
tests
Multivariate Regression
Genotypes
Images
Y
=
N × NV
Regression
Coefficients
X
+
N × NG
• Silly…
– If N > NG, fit equivalent
to NV univariate models
fit independently
– Much redundancy in C
• rank{C} ≤ min(NV, NG)
≪ NV ∙ NG
Error
E
N × NV
C
NG × NV
N # subjects
NV # voxels/ROIs
NG # genes/SNPs
Reduced Rank Regression
Images
Y
Genotypes
=
N × NV
• Fix rank r
• Approximate
Image
Coefficients
X
A
r × NV
N × NG
Error
+
E
N × NV
Genotype
Coefficients B
C≈BA
B & A each rank r
N×r
N # subjects
NV # voxels/ROIs
NG # genes/SNPs
Sparse Reduced Rank
Regression
Images
Y
Genotypes
=
N × NV
• Fix rank r
• Approximate
X
Sparse Image
Coefficients
+
A
N × NG
r × NV
Error
E
N × NV
Sparse
Genotype B
Coefficients
C≈BA
B & A each rank r
NG × r
• Enforce sparsity
Vounou, M., Nichols, T. E., & Montana, G. (2010). Discovering genetic associations with high-dimensional
neuroimaging phenotypes: A sparse reduced-rank regression approach. NeuroImage, 53(3), 1147-59.
N # subjects
NV # voxels/ROIs
NG # genes/SNPs
Sparse Reduced Rank
Regression - Estimation
• RRR
– Y = X A B + E
– For fixed rank r, find A & B that minimize
M = tr { (Y−XBA) Γ (Y−XBA)’ }
for some NV × NV matrix Γ, e.g. Γ = I
• SRRR
– For rank 1, find a & b that minimize
M = tr { (Y−Xba’) Γ (Y−Xba’)’ }
+ λa||a||1 + λb||b||1
– Then subtract Xba’ from the data, and repeat
– Need to specify final rank r, λa & λb
• Can set λa & λbin terms of #|a|>0 & #|b|>0
Simulation: Phenotype & SNPs
• Simulated MRI data
– ADNI T1 images through SPM5 VBM pipeline
– NV = 111 ROIs, placed on VBM data from 189 MCI
ADNI subjects
• GSK CIC Atlas, based on Harvard-Oxford atlas
– Estimate covariance Σ after adjusting for age &
gender
– Simulate ROI data (for arbitrary N) with covariance Σ
• Evaluate with realistic genetic population w/
FREGENE
– Simulates sequence-level data in large population
– Provides 10K individuals, 20Mb chromosome (~180K
SNPs)
• Chadeau-Hyam, et al. BMC Bioinformatics, 9:364, 2009
Simulation: Phenotype & SNPs
• FREGENE SNP simulation
–
–
–
–
Population of 10,000 evolved over 200,000 generations
20Mb simulated
37,748 SNPs with MAF>0.05
Select k=10 causative SNPs
• From all possible having MAF=0.2
• Used to induce phenotypic effect
– But then dropped from consideration
• Represents realistic setting, where causative SNP is not seen, but effect
captured through local LD
– From population of 10,000, repeatedly sample cohorts of size N
• Simulated association in MRI data
– Add genetic effect to Frontal and Temporal ROIs with causative
SNPs
• γ = 0.06, 0.08, or 0.1 reduction in mean GM in affected ROI
• Calibrated to Filipini et al. (2009)
– 10% reduction in GM ApoE ε4/ε4 subjects relative to subjects with no ε4 alleles
Out of Africa (OoA)
split & bottleneck
Founding population
in Africa
Expansion
Expansion
Expansion
Chadeau-Hyam, et al. BMC Bioinformatics, 9:364, 2009
Asian &
European split
FREGENE: Evolutionary
model of world population
• Linkage disequilibrium (LD)
– SNPs not independent
– Highly structured,
heterogeneous
dependence
• Population sub-structure
– Ethnic differences &
migration patterns
induce systematic
variation
• Multivariate analysis
– Want realistic multivariate
structure in our simulations
The Wellcome Trust Case Control Consortium, Nature 447, 661-678, 2007.
Why try so hard?
Why not rand{0,1,2}500,000 ?
Realistic Phenotype
• All pairwise GM
correlations
among NV = 111
ROIs
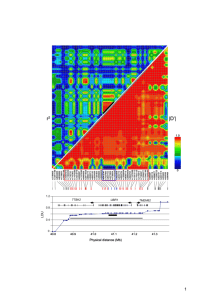
Realistic Genotypes
• Correlation of first
1000 simulated
SNPs
Simulation Setting:
Horse shoes & Imaging Genetics
• “True positive” with
missing causative
SNP
– Declare true positive
if LD coefficient close
enough
• LD-linked SNPs
– Of 1990 SNPs
– 51 linked (r>0.8) to
one or more the 10
causative SNPs
SRRR Simulation Results
• Power to detect 1 or more SNPs (NG=1990)
• For ranks r = 1,2,3 dominates Mass Uni.
– Better for higher r
SRRR Simulation Results
• Power to detect 1 or more SNPs (NG=1990)
• For ranks r = 1,2,3 dominates Mass Uni.
– Better for higher r; here r = 3
SRRR Simulation Results
• Power to detect 1 or more SNPs (NG=1990)
• For ranks r = 1,2,3 dominates Mass Uni.
– Better for higher r; here r = 3, high eff. size.
SRRR Simulation Results
• Power to detect 1 or more ROIs
• Less difference
– Power can be manipulated by varying λ by
rank
SRRR: Multivariate vs. MassUnivariate
• Does this NG=1990
result generalize?
• For up to 40k SNPs
– r = 3, med. effect
size, N=1000
– Power 2-5 greater
– Absolute power still
tiny
Sparse Reduced Rank Regression
for SNP – MRI Association
• Detailed simulation of imaging & genetic
correlations structure
– Suggests multivariate approach will outperform mass-univariate
– Power tiny, in any event
• Much work to do
– Haven’t addressed how to optimize phenotype
– Haven’t tried to estimate penalty parameters
λa, λb or r
• Currently investigating stability selection
– See #316 Le Floch et al
Conclusions
• Much work to do
– New sequence-level data increases NG
– But also numbers of rare SNPs