Accurate and Computationally Efficient Analysis of Longitudinal fMRI data
advertisement

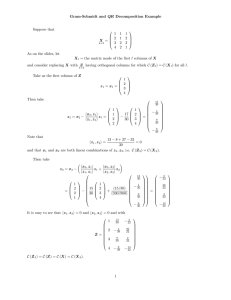
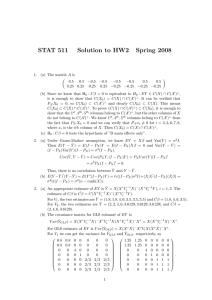
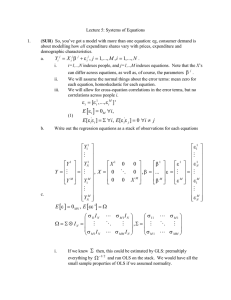
Accurate and Computationally Efficient Analysis of Longitudinal fMRI data Thomas Nichols and Lourens Waldorp University of Warwick, Coventry, England and University of Amsterdam, The Netherlands Introduction There are a growing number of longitudinal fMRI studies collecting rich datasets. Standard software, SPM and FSL in particular, cannot accurately model these data when there are k > 2 visits (repeated measurements). Specifically, FSL can only accomodate a single contrast (COPE) at the second level (e.g. for visit 2 - visit 1) and SPM, though it models repeated measures correlation, unrealistically assumes that the correlation is equal over the whole brain. While proper longitudinal modelling is available in standard statistics software (e.g. SAS’s proc mix, or R’s lme), the iterative estimation proceedures can be prohibiatively slow. 10 k=5 5 110 5 0 -5 120 110 100 90 80 12 50 100 200 12 50 # Subjects 100 200 # Subjects Methods It is common practice to fit longitudinal data via OLS by including dummy variables for each subject to account for subject-specific differences in the overall mean. While this ”poor man’s MFX” approach is not correct in general, it happens to be valid when the data form a balanced 1-way ANOVA and intra-visit correlation is the same for all pairs of visit (the condition of ”compound symmetry”). When compound symmetry doesn’t hold, the estimates will be suboptimal (inefficient) and standard errors biased. We compare three univariate models for MFX inference on longitudinal data: OLS, GLS, and SW via Monte Carlo simulation. For a range of subject sample sizes (n = 12, 25, 50, 100, 200) and number of visits (k = 3, 5, 8), and compound symmetric and non-compound symmetric (autoregressive structure, AR(1)) intra-visit correlation 25 20 25 50 100 # Subjects 200 OLS GLS SW 15 % SE Stdev 5 0 5 12 k=8 10 20 k=5 0 # Subjects % SE Stdev 25 20 200 15 100 10 50 k=3 5 12 Figure 1: With compound symmetry, in the rows are relative bias in the standard error, FPR, and effeiciency of OLS (red), GLS (green) and SW (blue) for k = 3, 5, 8 time points. http://home.medewerker.uva.nl/l.j.waldorp 200 0 # Subjects 200 % SE Stdev # Subjects 100 110 110 25 15 5 0 50 200 10 % SE Stdev 15 12 100 # Subjects OLS GLS SW 5 200 50 k=8 0 10 % SE Stdev 12 20 25 20 25 20 15 10 100 % relative FPR 200 5 50 100 k=8 100 100 0 % SE Stdev 12 50 # Subjects 90 50 # Subjects k=5 12 120 120 12 # Subjects k=3 200 80 200 80 # Subjects 100 100 k=5 % relative FPR 50 50 100 80 12 12 # Subjects 100 200 200 90 100 100 k=3 % relative FPR 50 50 # Subjects 90 % relative FPR 110 100 80 90 % relative FPR 110 100 90 80 % relative FPR 12 12 k=8 5 5 120 120 120 k=5 -10 # Subjects % relative Bias 200 0 100 -5 50 -10 12 # Subjects k=3 10 10 10 0 200 -10 # Subjects 100 15 50 k=8 10 12 % relative Bias 200 0 100 % relative Bias 50 k=5 k=3 -10 -10 12 Results Fig. 1 shows the situation for compound symmetry. The Relative Bias in standard errors, FPR of the three estimators are approximately the same, indicating that the SW does not perform poorly when compound symmetry holds. When compound symmetry does not hold (Fig. 2), OLS is least accurate. The FPRs in Fig. 2 show that SW is reasonably robust against number of visits. All methods show stronger errors for few number of subjects. Note that the variation of SW is larger than both GLS and OLS. -5 0 % relative Bias 5 k=8 -5 % relative Bias 5 0 -5 -10 % relative Bias k=3 We use the R software to perform these evaluations in order to have access to state of the art GLS MFX modelling tools and sandwich estimators. Note that OLS is offered in all fMRI software, and SW does not require iteration and is easily implemented. -5 10 10 In this work we propose the use of Ordinary Least Squares (OLS) combined with the sandwich estimator (White, 1982), which should provide fast and valid P-values (with possibly sub-optimal sensitivity). We compare this approach to the commonly used “poor man’s” OLS longitudinal model for longitudinal data (which is fast, but possibly both inaccruate P-values and poor sensitivity), and Generalized Least Square (GLS) to fit a proper longintiudonal model that accounts for intrasubject correlation (which is the gold standard, but may be slow). structure, we evaluate the properties of the MFX estimates of the linear change in BOLD over time: (1) SE (Standard Error) Bias, the average error in the estimated variance of BOLD change, (2) FPR relative accuracy, the relative false positive rate (FPR) compared to the nominal level, and (3) SE Stdev, the standard deviation of the standard error estimates divided by the Monte Carlo mean of the standard error. We use 1000 simulations for each setting. 12 50 100 200 # Subjects 12 50 100 200 # Subjects Figure 2: Without compound symmetry (AR(1)), in the rows are relative bias in the standard error, FPR, and effeiciency of OLS (red), GLS (green) and SW (blue) for k = 3, 5, 8 visits. Discussion We have shown that the widely used OLS approach to modelling longitudinal data can be inaccurate in a variety of settings, due to bias in the standard errors of the effect. Although SW is not always significantly better than OLS, SW can be more robust than OLS to more extreme covariance structures than shown here. While GLS-based MFX provides optimal inferences, the use of the Sandwich Estimator Standard Errors with OLS provides a computationally efficient, robust, flexible and accurate approach to longitudinal fMRI. References [1] Laird and Ware (1982). Biometrics, 38(4), 963-974. [2] Verbeke and Molenberghs (2000). Linear mixed models. Springer. [3] Beckmann (2004). IEEE Trans. Med. Im. 23, 137-152. [4] White (1981). JASA, 76(374), 419-433. waldorp@uva.nl


![Job Evaluation [Opens in New Window]](http://s2.studylib.net/store/data/009982944_1-4058a11a055fef377b4f45492644a05d-300x300.png)