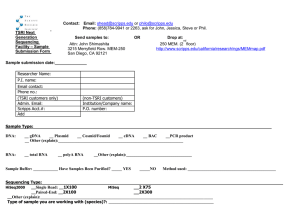
Scripps Florida 2008
advertisement

Scripps Florida 2008 • Cancer Biology • Chemistry • Infectology • Molecular Therapeutics • Molecular and Intregrative Neurosciences • Translational Research Institute Cancer Biology Aberrant cell division in a precancerous cell: Shown is a differential interference contrast image of an early-passage p53-null mouse embryo fibroblast. Note that the chromosomes in the cell are being pulled in 3 directions. Daughter cells that arise are aneuploid and/or polyploid. Work done by Frank C. Dorsey, Ph.D., research associate, in the laboratory of John L. Cleveland, Ph.D., professor. Kendall Nettles, Ph.D., Assistant Professor CANCER BIOLOGY 2008 THE SCRIPPS RESEARCH INSTITUTE 19 DEPAR TMENT OF CANCER BIOLOGY S TA F F John L. Cleveland, Ph.D. Professor and Chairman Jun-Li Luo, Ph.D., MD Assistant Professor Kendall Nettles, Ph.D. Assistant Professor HaJeung Park, Ph.D. Ann Griffith, Ph.D. Meredith A. Steeves, Ph.D. Mark A. Hall, Ph.D. Weilin Wu, Ph.D. Jun Hyuck Lee, Ph.D. Howard Petrie, Ph.D. Professor Tina Izard, Ph.D. Associate Professor Nagi G. Ayad, Ph.D. Assistant Professor Philippe R.J. Bois, Ph.D. Assistant Professor Michael Conkright, Ph.D. Assistant Professor Woonghee Lee, Ph.D. S C I E N T I F I C A S S O C I AT E R E S E A R C H A S S O C I AT E S Hiroshi Nakase, Ph.D. Chunying Yang, M.D. Antonio Amelio, Ph.D. Robert J. Rounbehler, Ph.D. Mi Ra Chang, Ph.D. Jianjun Shi, Ph.D. Frank C. Dorsey, Ph.D. Zhen Wu, Ph.D. Joanne R. Doherty, Ph.D. Rangarajan Erumbi, Ph.D. Ihn Kyung Jang, Ph.D. Irina Getun, Ph.D. Bhargavakrishna Yekkala, Ph.D. S TA F F SCIENTISTS German Gil, Ph.D. Sollepura Yogesha, Ph.D. Min Zhao, Ph.D. Chairman’s Overview he Department of Cancer Biology on the Florida campus was established in November 2006. The department has rapidly grown to now include 8 faculty members. The broad goals of the research programs of the department are to fully define the molecular events that underlie human cancer and then apply this knowledge to the development of novel therapeutic strategies and new agents for cancer prevention and John L. Cleveland, Ph.D. therapeutics. The programs include those that examine the roles of signal transduction pathways, oncogenes, and tumor suppressors that are altered in cancer and how these alterations control cell division, growth, survival, differentiation, cell migration and metastasis, tumor angiogenesis, transcriptional circuits, and genomic stability and how they modify the response to therapeutic agents. In addition, the interplay between tumors and the immune system in cancer is a new major thrust of research. Faculty members in the department use a T battery of state-of-the-art technologies for target discovery and validation, ranging from biochemistry and cell biology to preclinical models to x-ray crystallography. In addition, unique models have been developed to evaluate the efficacy of new leads in cancer prevention and therapeutics. Investigators in the department have interests in understanding the molecular underpinnings of all of the major human malignant neoplasms, including lung, breast, prostate, colon, and brain cancer, and several hematologic malignant neoplasms. Other interests include pediatric oncology, the interplay between malignant neoplasms and metabolism, and the relationships between aging and cancer. One of the many strengths at the Florida campus is high-throughput technologies that enable investigators to rapidly move forward potential leads by using both genetic and small-molecule screens. Strong collaborations with the major cancer centers in the State of Florida and with cancer researchers at the California campus of Scripps Research will allow leads that are identified to rapidly advance to translational and clinical studies. 20 CANCER BIOLOGY 2008 INVESTIGATORS’ R EPORTS Myc-Mediated Pathways in Cancer and Development J.L. Cleveland, M.A. Hall, F.C. Dorsey, R. Rounbehler, K. Yekkala, J. Doherty, M. Steeves, C. Yang, T. Bratton, S. Prater, W. Li yc oncoproteins function as master regulators of transcription and regulate up to 10%–15% of the genome. Three Myc oncogenes (c-Myc, N-Myc, L-Myc) are activated in about 70% of human cancers. Their activation can occur directly via gene amplification, chromosomal translocations, or somatic missense mutations or indirectly via alterations in signal transduction pathways or the loss of tumor suppressors that normally regulate and/or harness Myc expression. The pervasive selection for Myc activation in cancer in part reflects the essential roles of Myc as a regulator of cell growth and division, but overexpression of Myc also triggers accelerated rates of cell proliferation, tumor angiogenesis, and metastasis. Further, Myc regulates stem cell fate and supercompetition, a scenario in which cells that overexpress Myc kill their neighboring, normal cells. We have used mouse models to dissect the contribution of key targets downstream of Myc that control tumorigenesis. In normal cells, Myc triggers apoptosis through the Arf-p53 tumor suppressor pathway that is inactivated in most malignant tumors and by selectively affecting the expression of members of the Bcl-2 family of proteins that directly control the intrinsic apoptotic pathway. We have shown that these pathways hold Myc-induced tumorigenesis in check and that mutations in these apoptotic regulators are a hallmark of most malignant tumors. Although apoptotic regulators clearly serve as guardians against Myc-induced cancer, we have found that the ability of Myc to provoke accelerated cell growth is also critical for tumorigenesis. First, Myc coordinately regulates the expression of cytokines that direct cell growth and tumor angiogenesis. Second, Myc suppresses expression of the universal cyclin-dependent kinase (Cdk) inhibitor p27Kip1 that normally inhibits the activity of cyclin E–Cdk2 and cyclin A–Cdk2 complexes that are necessary for entry and progression through the DNA synthesis (S) phase of the cell cycle. Notably, we found that Myc suppresses p27Kip1 protein levels by inducing transcription of the Cks1 component of the M THE SCRIPPS RESEARCH INSTITUTE SCFSkp2 E3 ubiquitin ligase complex that targets p27Kip1 for destruction by the 26S proteasome. Accordingly, loss of Cks1 disables the ability of Myc to suppress p27Kip1 and markedly impairs Myc-induced proliferation and tumorigenesis, whereas loss of p27Kip1 accelerates Mycinduced tumorigenesis. Remarkably, Cks1 overexpression is a hallmark of all lymphomas with Myc involvement, suggesting this pathway is a general route by which Myc coordinates cell growth and division and that the pathway can be targeted by directed therapeutic agents. Because Myc regulates such a large number of genes and is essential for cell growth and division, the adverse effects of agents that directly target the transcription functions of Myc might be greater than the agents’ beneficial effects. We therefore have focused our efforts on key transcription targets of Myc that might be suitable therapeutic targets. We found that inhibiting ornithine decarboxylase, a direct transcription target of Myc and the rate-limiting enzyme of polyamine biosynthesis, impairs Myc-induced proliferation and tumorigenesis. These results were underscored by our findings that heterozygosity in the gene that encodes ornithine decarboxylase, a condition that only reduces the enzyme activity of ornithine decarboxylase and the generation of its product by half, triples the life span of tumor-prone mice. Thus, agents that target the polyamine pathway have promise in both the prevention and the treatment of cancer. Currently, we are defining the mechanism by which targeting ornithine decarboxylase disables the proliferative response of Myc. Our results indicate, quite remarkably, that targeting ornithine decarboxylase disables the ability of Myc to suppress p27Kip1 by shortcircuiting of the Myc-to-Cks1 pathway. Finally, we recently discovered that additional Myc transcription targets that can be exploited in cancer therapy include components of the autophagy pathway, an ancient survival pathway that directs the digestion of bulk cytoplasmic material and organelles when cells are faced with nutrient- or oxygen-deprived conditions, a scenario manifests in the tumor microenvironment. We have shown that agents that disable autophagy have tremendous potential in cancer prevention and treatment. Currently, we are defining the mechanisms by which Myc regulates the expression of genes that control the autophagy pathway and their potential as targets for agents to prevent and treat cancer. PUBLICATIONS Carew, J.S., Nawrocki, S.T., Reddy, V.K., Bush, D., Rehg, J.E., Goodwin, A., Houghton, J.A., Casero, R.A., Jr., Marton, L.J., Cleveland, J.L. The novel polyamine analogue CGC-11093 enhances the antimyeloma activity of bortezomib. Cancer Res. 68:4783, 2008. CANCER BIOLOGY 2008 Garrison, S.P., Jeffers, J.R., Yang, C., Nilsson, J.A., Hall, M.A., Rehg, J.E., Yue, W., Yu, J., Zhang, L., Onciu, M., Sample, J.T., Cleveland, J.L., Zambetti, G.P. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol. Cell. Biol. 28:5391, 2008. Klionsky, D.J., Abeliovich, H., Agostinis, P., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4:151, 2008. Maclean, K.H., Dorsey, F.C., Cleveland, J.L., Kastan, M.B. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J. Clin. Invest. 118:79, 2008. Nawrocki, S.T., Carew, J.S., Douglas, L., Cleveland, J.L., Humphreys, R., Houghton, J.A. Histone deacetylase inhibitors enhance lexatumumab-induced apoptosis via a p21Cip1-dependent decrease in survivin levels. Cancer Res. 67:6987, 2007. Nawrocki, S.T., Carew, J.S., Maclean, K.H., Courage, J.F., Huang, P., Houghton, J.A., Cleveland, J.L., Giles, F.J., McConkey, D.J. Myc regulates aggresome formation, the induction of Noxa, and apoptosis in response to the combination of bortezomib and SAHA. Blood 112:2917, 2008. Rodrigues, C.O., Nerlick, S.T.,White, E.L., Cleveland, J.L., King, M.L. A Myc-Slug (Snail2)/Twist regulatory circuit directs vascular development. Development 135:1903, 2008. Sanjuan, M.A., Dillon, C.P., Tait, S.W., Moshiach, S., Dorsey, F., Connell, S., Komatsu, M., Tanaka, K., Cleveland, J.L., Withoff, S., Green, D.R. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450:1253, 2007. THE SCRIPPS RESEARCH INSTITUTE 21 V I N C U L I N S T R U C T U R E A N D R E G U L AT I O N Our crystal structures, biochemical studies, and biological experiments have redefined vinculin structure and regulation. First, in its resting, inactive conformation, vinculin is held in a closed-clamp conformation through interactions of a 7-helical bundle domain present in its head domain (Vh1) with a 5-helical bundle in the tail domain (Vt); 3 additional helical bundle domains that were identified likely also serve as docking sites for interactions with partners. Second, contrary to dogma, we found that, talin itself is a direct activator of vinculin; α-helical vinculin-binding sites (VBSs) in the central rod domain of talin trigger vinculin activation by displacing Vt from a distance. More importantly, our structures revealed that this activation of vinculin and displacement of Vt occurred via a heretofore unknown change in protein structure, by a process we termed helical bundle conversion (Fig. 1). Third, our studies Schweers, R.L., Zhang, J., Randall, M.S., Loyd, M.R., Li, W., Dorsey, F.C., Kundu, M., Opferman, J.T., Cleveland, J.L., Miller, J.L., Ney, P.A. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U. S. A. 104:19500, 2007. Structural Dynamics in Adhesion Complexes T. Izard, P.R. Bois, J.H. Lee, G.T.V. Nhieu,* H. Park, E.S. Rangarajan, S.D. Yogesha * Pasteur Institute, Paris, France ell migration and morphogenesis are essential for the development, growth, and survival of metazoans, and these processes are also involved in pathophysiologic conditions such as cancer metastasis and myopathies. Migration and morphogenesis rely on the ability of a cell to dynamically form and break specific contacts, called adhesion junctions, with neighboring cells (adherens junctions) or the extracellular matrix (focal adhesions). Vinculin is an essential regulator of both cell-cell (cadherin-catenin mediated) and cell-matrix (integrin-talin mediated) junctions, where it provides links to the actin cytoskeleton by binding to talin in integrin complexes or to α-catenin and α-actinin in cadherin junctions. Previously, little was known about the structure and activation of vinculin, although the accepted belief was that activation required severing intramolecular interactions of the vinculin head and tail domains. C F i g . 1 . Vinculin activation by talin through helical bundle con- version. Ribbon drawing of the vinculin head (Vh1; cyan) and tail (Vt; yellow) domains activated by talin’s VBS (red). A, The crystal structure of the Vh1-Vt complex revealed that vinculin is held in a closed conformation through many hydrophobic interactions between the Vh1 and Vt domains. B, The crystal structure of talin-VBS3 bound to Vh1 shows that talin binds to vinculin at an accessible site distal from the Vh1-Vt interface and that talin-VBS3 binding displaces Vt from a distance. Wholesale structural changes occur upon talin binding, whereby the 4-helical bundle of Vh1 incorporates the amphipathic VBS helix of talin to form an entirely new 5-helical bundle via helical bundle conversion. 22 CANCER BIOLOGY 2008 of the complex composed of α-actinin and vinculin revealed that α-actinin activates vinculin and alters its structure in unique ways. These findings supported our model in which the vinculin Vh1 domain functions as a “molecular switch” that undergoes rapid and unique changes in its structure after binding to different activators, which then endow vinculin with the ability to bind to unique partners in adherens junctions vs focal adhesions. Finally, our studies indicated that adhesion signaling involves a chain reaction of structural alterations in which, after their activation, the VBSs of talin or α-actinin first unravel from their buried locations and then bind to and activate vinculin, which then undergoes wholesale changes in its structure (Fig. 2). THE SCRIPPS RESEARCH INSTITUTE studies showed that IpaA acts as a talin mimic that disrupts vinculin’s contacts with talin and α-actinin, and our results suggest that this mechanism is a general one that is exploited by other pathogens. Importantly, our biological studies have shown that this interaction is necessary for efficient entry of Shigella into host cells. Our recent studies have revealed additional layers of functional complexity of IpaA. First, we found that the second, somewhat lower affinity, VBS of IpaA can bind to a second motif of the vinculin Vh1 domain, a situation that would stabilize IpaA-vinculin interactions (Fig. 3). Second, our biochemical and genetic screens F i g . 2 . Relays in adherens junctions. Ribbon drawing of α-actinin (gray and black) and vinculin. Top, In their resting state, both α-actinin and vinculin are in a closed conformation. The VBS is shown in red. Bottom, When activated, α-actinin unfurls to expose its VBS, which then binds to and activates vinculin, resulting in helical bundle conversion of the vinculin Vh1 domain and severing of vinculin’s headtail interaction. The α-actinin antiparallel homodimer has 2 VBS sites. For clarity, only 1 vinculin molecule is shown bound to the α-actinin homodimer. TA R G E T I N G V I N C U L I N I N PAT H O G E N - H O S T INTERACTIONS We have also made significant inroads in understanding how vinculin is co-opted by pathogens. Initially, we have focused on Shigella flexneri, the principal pathogen of bacillary dysentery. Shigella organisms inject invasin proteins (IpaA-IpaD) that create pores in intestinal epithelial cells and that trigger the formation of filopodial and lamellopodial extensions that surround the bacteria. IpaA, a protein of approximately 70 kD essential for the pathogenesis of Shigella in vivo, facilitates entry of the bacteria into host cells by binding to vinculin. We established that IpaA has 2 high-affinity VBSs that bind to the Vh1 domain of vinculin and induce unique alterations in the domain’s structure. Strikingly, our F i g . 3 . A novel second binding site on the vinculin Vh1 domain allows IpaA to bind vinculin in its closed conformation. A, Ribbon drawing of the vinculin head domain (Vh1; yellow) bound by the 2 VBSs of S flexneri IpaA. The first IpaA-VBS (blue) binds to Vh1 with femtomolar affinity by molecular mimicry of the Vh1-talin interaction, via helical bundle conversion. In contrast, the second IpaA-VBS (red) binds vinculin by a helix addition mechanism, a scenario that allows the S flexneri invasin to also recruit pools of inactive vinculin and that would also facilitate the bridging of 2 molecules of vinculin by IpaA. B, Surface drawing of full-length human vinculin (yellow, orange, magenta, blue, gray, and cyan) bound to IpaA-VBS (ribbon drawing, red). The weaker binding IpaA-VBS binds vinculin by a helix addition mechanism, which has no allosteric effects on vinculin, allowing IpaA to bind to vinculin in its closed conformation. Reprinted from Nhieu, G.T., Izard, T. Vinculin binding in its closed conformation by a helix addition mechanism. EMBO J. 26:4588, 2007. CANCER BIOLOGY 2008 have revealed new cytoskeletal binding partners for IpaA. The physiologic roles of these interactions will be tested, and along with our structural analyses, these studies may point to new therapeutic avenues. PUBLICATIONS Nhieu, G.T., Izard, T. Vinculin binding in its closed conformation by a helix addition mechanism [published correction appears in EMBO J. 27:922, 2008]. EMBO J. 26:4588, 2007. Regulation of Mitotic Entry and Exit by Ubiquitin-Mediated Proteolysis N. Ayad, S. Simanski, N. Nagarsheth biquitin-mediated proteolysis is one of the main ways cells eliminate intracellular proteins. This elimination is important for cell homeostasis, development, and growth. Recent studies have also indicated that one or more components of this system are overexpressed in cancer cells, making the components attractive targets for pharmacologic inhibition. Although a fair amount is known about the pathways leading to ubiquitin-mediated degradation, many essential components have not been identified. We devised a means of identifying regulators of the anaphase-promoting complex (APC), an essential ubiquitin ligase required for the metaphase-to-anaphase transition and exit from mitosis. We fused cyclin B1, a known APC substrate, to luciferase and cotransfected this fusion construct with 14,000 cDNAs. This genomewide screen led to the identification of multiple regulators of the APC, including some that are overexpressed in breast cancer. Currently, we are identifying the mechanism by which these proteins regulate the APC during exit from mitosis and are developing high-throughput screens. Our eventual goal is to eliminate the activity of the proteins selectively in cancer cells. APC activity is inhibited before mitosis because its premature activation would lead to genomic instability. One way the APC is inhibited is by inhibiting the cdk1/cyclin B complex required for entry into mitosis. This complex is kept inactive before mitosis because it is phosphorylated by the tyrosine kinase Wee1. Wee1 is degraded during the G2 phase and mitosis to tip the balance to active cdk1 and allow mitotic entry to proceed. We analyzed Wee1 degradation in somatic cells and found that 2 separate ubiquitin ligases containing U THE SCRIPPS RESEARCH INSTITUTE 23 β-transducin repeat–containing protein and trigger of mitotic entry 1 are required for Wee1 destruction and mitotic entry. These findings indicated that eukaryotic cells have multiple means of regulating Wee1 degradation, because inactivation of a single critical component would lead to premature mitosis and disastrous consequences for the organism. We are determining the respective roles of these ligases in cancer progression. PUBLICATIONS Smith, A., Simanski, S., Fallahi, M., Ayad, N.G. Redundant ubiquitin ligase activities regulate Wee1 degradation and mitotic entry. Cell Cycle 6:2795, 2007. Anatomy and Regulation of Mouse Recombination Hot Spots P.R. Bois, M. Fallahi-Sichani, I. Getun, Z.K. Wu rossover events are necessary for meiosis progression and for genome reshuffling and diversity before the generation of gametes. A peculiarity of meiosis is the programmed nature of double-strand breaks, which are induced by the conserved endonuclease Spo11. Studies in a variety of model systems have shown that recombination occurs in distinct regions termed hot spots. An estimated 5%–10% of the genomes of higher eukaryotes are recombinogenic; the remainder reside in the “cold.” However, little is known about the nature and mechanisms that control recombination hot spots in mammalian genomes. We have focused on identifying novel hot spots in the mouse chromosome 19 by directly detecting crossover events. Characterization of a highly polymorphic hot spot (HS23.7) revealed quite unexpectedly that repair does not have to be complete for meiosis to proceed. The persistence of these unrepaired heteroduplex regions at crossover sites in mature spermatozoa promotes genome instability that was revealed by the high diversity and rearrangements observed in laboratory strains and wild mouse populations. We are identifying and characterizing more of these recombinogenic regions where chromosomes are reshuffled between generations and provide the driving mechanism at the origin of diversity. C PUBLICATIONS Bois, P.R. A highly polymorphic meiotic recombination mouse hot spot exhibits incomplete repair. Mol. Cell. Biol. 27:7053, 2007. 24 CANCER BIOLOGY 2008 Molecular Mechanisms of cAMP-Mediated Transcription THE SCRIPPS RESEARCH INSTITUTE novel compounds are also being used to explore the biological effects of inhibiting inflammatory programs in cancer cell proliferation and invasion and the role of estrogen signaling in immune function. M.D. Conkright, A.L. Amelio, N.E. Bruno lucose homeostasis is maintained by coordinating glucose metabolism in skeletal muscle, lipid storage in adipose tissue, and glucose production in the liver. Insulin and glucagon are central hormone regulators of glucose homeostasis. Glucagon initiates the gluconeogenic program in hepatocytes by activating the cAMP signaling pathway, whereas insulin inhibits hepatic glucose output. Our recent detection of a new component of the cAMP pathway, transducers of regulated CREB (TORCs), established that cAMP signaling is more sophisticated than previously recognized and provided new insights into glucose homeostasis. Collectively, recent studies have indicated that insulin, glucagon, and energy signals converge on TORC2 phosphorylation to modulate glucose output via CREBmediated expression of hepatic genes. However, the specific nuclear actions of TORC2 are unknown. Thus, the mechanisms involved in differentiating TORC2transmitted signals are of biological and clinical interest. G PUBLICATIONS Altarejos, J.Y., Goebel, N., Conkright, M.D., Inoue, H., Xie, J., Arias, C.M., Sawchenko, P.E., Montminy, M. The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nat. Med. 14:1112, 2008. Amelio, A.L., Miraglia, L.J., Conkright, J.J., Mercer, B.A., Batalov, S., Cavett, V., Orth, A.P., Busby, J., Hogenesch, J.B., Conkright M.D. A coactivator trap identifies NONO (p54nrb) as a new component of the cAMP signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 104:20314, 2007. Structural and Molecular Mechanisms of Nuclear Receptor Signaling K.W. Nettles, G. Gil, J. Nowak, M. Zhou ur overall goal is to understand how small-molecule ligands control specific physiologic outcomes through the chemical-structural interface of the ligand with specific nuclear receptors. We have developed new chemical probes for estrogen receptors that selectively suppress inflammatory gene expression programs. We characterized a series of these probes by using x-ray crystallography, revealing the structural features of signaling specificity. The O PUBLICATIONS Bruning, J.B., Chalmers, M.J., Prasad, S., Busby, S.A., Kamenecka, T.M., He, Y., Nettles, K.W., Griffin, P.R. Partial agonists activate PPARγ using a helix 12 independent mechanism. Structure 15:1258, 2007. Nettles, K.W., Bruning, J.B., Gil, G., Nowak, J., Sharma, S.K., Hahm, J.B., Kulp, K., Hochberg, R.B., Zhou, H., Katzenellenbogen, J.A., Katzenellenbogen, B.S., Kim, Y., Joachmiak, A., Greene, G.L. NF-κB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses [published correction appears in Nat. Chem. Biol. 4:379, 2008]. Nat. Chem. Biol. 4:241, 2008. Nettles, K.W., Gil, G., Nowak, J., Métivier, R., Sharma, V.B., Greene, G.L. CBP is a dosage-dependent regulator of nuclear factor-κB suppression by the estrogen receptor. Mol. Endocrinol. 22:263, 2008. Chemistry A chiral catalyst for enantioselective carbon-hydrogen activation. Work done in the laboratory of Jin-Quan Yu, Ph.D., associate professor. Jin-Quan Yu, Ph.D., Associate Professor CHEMISTRY DEPAR TMENT OF CHEMISTRY S TA F F K.C. Nicolaou, Ph.D.* Chairman Aline W. and L.S. Skaggs Professor of Chemical Biology Darlene Shiley Chair in Chemistry Dariush Ajami, Ph.D. Assistant Professor of Molecular Assembly Phil S. Baran, Ph.D. Professor Dale L. Boger, Ph.D.* Richard and Alice Cramer Professor of Chemistry Tobin J. Dickerson, Ph.D. Assistant Professor Albert Eschenmoser, Ph.D.* Professor Sheng Ding, Ph.D. Associate Professor M.G. Finn, Ph.D.* Professor Valery Fokin, Ph.D. Associate Professor M. Reza Ghadiri, Ph.D.* Professor William A. Greenberg, Ph.D. Assistant Professor of Chemistry Inkyu Hwang, Ph.D. Assistant Professor Kim D. Janda, Ph.D.** Professor Ely R. Callaway, Jr., Chair in Chemistry Director, Worm Institute of Research and Medicine Gunnar Kaufmann, Ph.D. Assistant Professor of Chemistry 2008 Jeffery W. Kelly, Ph.D.* Lita Annenberg Hazen Professor of Chemistry Ramanarayanan Krishnamurthy, Ph.D. Associate Professor Lucas J. Leman, Ph.D. Assistant Professor of Chemistry Richard A. Lerner, M.D.*** President, The Scripps Research Institute Lita Annenberg Hazen Professor of Immunochemistry Cecil H. and Ida M. Green Chair in Chemistry Roy Periana, Ph.D.***** Professor Evan T. Powers, Ph.D. Associate Professor of Chemistry Julius Rebek, Jr., Ph.D.* Professor Director, The Skaggs Institute for Chemical Biology Edward Roberts, Ph.D. Professor THE SCRIPPS RESEARCH INSTITUTE (Andrew) Bin Zhou, Ph.D. Assistant Professor of Immunochemistry 79 Deboshri Banerjee, Ph.D. Elizabeth Barrett, Ph.D.**** Roland Barth, Ph.D.***** SENIOR SCIENTIST Clay Bennett, Ph.D. Luis Martinez, Ph.D.***** Moritz Biskup, Ph.D. † Universität Karlsruhe Karlsruhe, Germany S TA F F S C I E N T I S T S Lisa Eubanks, Ph.D. Rajesh Grover, Ph.D. Sarah Hanson, Ph.D. Lubica Supekova, Ph.D. Wen Xiong, Ph.D. Anthony Boitano, Ph.D. † Genomics Institute of the Novartis Foundation San Diego, California Laure Bouchez, Ph.D. Kristopher Boyle, Ph.D. Christopher Burke, Ph.D SERVICE FACILITIES Antonio Burtoloso, Ph.D. † University of Sao Paulo Sao Paulo, Brazil Raj K. Chadha, Ph.D. Director, X-ray Crystallography Facility Mark Bushey, Ph.D. † Exxon, Inc. Union City, New Jersey Dee H. Huang, Ph.D. Director, Nuclear Magnetic Resonance Facility Darren Bykowski, Ph.D.***** Gary E. Siuzdak, Ph.D. Senior Director, Mass Spectrometry Facility Katerina Capkova, Ph.D. I N S T R U M E N TAT I O N / Floyd E. Romesberg, Ph.D. Associate Professor Petr Capek, Ph.D. Arani Chanda, Ph.D. Ke Chen, Ph.D. Peng Chen, Ph.D. William Roush, Ph.D.***** Professor SENIOR RESEARCH Peter G. Schultz, Ph.D.* Professor Scripps Family Chair Suresh Pitram, Ph.D. Govardhan Cherukupalli, Ph.D. † Epix Pharmaceuticals Lexington, Massachusetts R E S E A R C H A S S O C I AT E S Jodie Chin, Ph.D. Ramzey Abujarour, Ph.D. Srinivas Reddy Chirapu, Ph.D. Rajesh Ambasudhan, Ph.D. Chandramouli Chiruta, Ph.D. Manuel Amorin Lopez, Ph.D. Dong-Gyu Cho, Ph.D. Mark Ams, Ph.D. So-Hye Cho, Ph.D. Yoshio Ando, Ph.D. Sungwook Choi, Ph.D. Deepshikha Angrish, Ph.D. Joyanta Choudhury, Ph.D. Shinji Ashida, Ph.D. Sarwat Chowdhury, Ph.D. Micahel Baksh, Ph.D. Stepan Chuprakov, Ph.D. K. Barry Sharpless, Ph.D.* W.M. Keck Professor of Chemistry Anita D. Wentworth, Ph.D. Assistant Professor Paul Wentworth, Jr., Ph.D. Professor Chi-Huey Wong, Ph.D.* Professor of Chemistry Jin-Quan Yu, Ph.D. Associate Professor A S S O C I AT E 80 CHEMISTRY 2008 THE SCRIPPS RESEARCH INSTITUTE Petr Cigler, Ph.D. David Edmonds, Ph.D. Neil Grimster, Ph.D. Giltae Hwang, Ph.D. T. Ryan Cirz † Achaogen South San Francisco, California Jem Efe, Ph.D. Rajesh K. Grover, Ph.D. Jan Elsner, Ph.D. † Celgene Pharmaceuticals San Diego, California Jan Grunewald, Ph.D. Michael Jahnz, Ph.D. † NOXXON Pharma AG Berlin, Germany Daniel Ess, Ph.D. Richard Guy, Ph.D.**** Ph.D. † Scott Cockroft, University of Edinburgh Edinburgh, Scotland Ph.D. † David Colby, Purdue University West Lafayette, Indiana Kevin Cole, Ph.D. † Eli Lilly and Company Indianapolis, Indiana Christine Crane, Ph.D. Matthew Cremeens, Ph.D. † Gonzaga University Redmond, Washington Fernando Rodrigo Pinacho Crisostomo, Ph.D. † Burnham Institute for Medical Research La Jolla, California Cyrine Ezzili, Ph.D. Xingang Fang, Ph.D. Simon Ficht, Ph.D. † Sanofi-Aventis Deutschland GmbH Frankfurt, Germany Joseph Rodolph Fotsing, Ph.D. † Senomyx, Inc. San Diego, California Bozena Frackowiak, Ph.D. † Politechnika Opolska Opole, Poland Etzer Darout, Pfizer Inc. Groton, Connecticut Ph.D. † Amy DeBaillie, Eli Lilly and Company Indianapolis, Indiana Graeme Freestone, Metabasis Therapeutics, Inc. San Diego, California Amelia Fuller, Ph.D. † Santa Clara University Santa Clara, California Jianmin Gao, Ph.D. † Boston College Chestnut Hill, Massachusetts Haibo Ge, Ph.D. Judith Denery, Ph.D. Ross Denton, Ph.D. † University of Cambridge Cambridge, England Savvas Georgiades, Ph.D.**** Ola Ghoneim, Ph.D. † Qatar University Doha, Qatar Caroline Desponts, Ph.D. Antonia Di Mola, Ph.D. Deguo Du, Ph.D. Anna Dubrovska, Ph.D. Viktoriya Dubrovskaya, Ph.D. Joshua Dunetz, Ph.D. † Pfizer Inc. Groton, Connecticut Kyle Eastman, Ph.D. Yuanjun He, Ph.D. Jason Hein, Ph.D. Dube Henry, Ph.D. Nathan Gianneschi, Ph.D. † University of California San Diego, California Cristina Gil-Lamaignere, Ph.D. † University Hospital Nuestra Señora de la Candelaria Santa Cruz de Tenerife, Spain Rodolfo Gonzalez, Ph.D. Scott Grecian, Ph.D. Rong Jiang, Ph.D.***** Guo Jiantoa, Ph.D. Hiroyuki Kakei, Ph.D. † Takeda Pharmaceutical Company Limited Osaka, Japan Jaroslaw Kalisiak, Ph.D. Seiji Kamioka, Ph.D. Moumita Kar, Ph.D. Marcos Hernandez, Ph.D. Kwang Mi Kim, Ph.D. Par Holmberg, Ph.D. † Memorial Sloan Kettering Cancer Center New York, New York Wen-Xu Hong, Ph.D. Yu Fu, Ph.D. Ph.D. † Masaki Handa, Ph.D. † Sagami Chemical Research Center Ayase, Kanagawa, Japan Ph.D. † Jeffrey Culhane, Ph.D. Stephen Dalby, Ph.D. Tanja Gulder, Ph.D. Zhangyong Hong, Ph.D. Richard J. Hooley, Ph.D. † University of California Riverside, California Tamara Hopkins, Ph.D. † Boehringer Ingelheim Pharmaceuticals, Inc. Ridgefield, Connecticut Allen Horhota, Ph.D. Tony Horneff, Ph.D. F. Scott Kimball, Ph.D. Jeremy Kister, Ph.D. Keith Korthals, Ph.D. Larisa Krasnova, Ph.D. Arkady Krasovskiy, Ph.D. Luke Lairson, Ph.D. Jae Wook Lee, Ph.D. Jinq-Chyi Lee, Ph.D. † National Health Research Institutes Miaoli County, Taiwan Jong Seok Lee, Ph.D. Claas Hovelmann, Ph.D. Ki-Bum Lee, Ph.D. † Rutgers University Piscataway, New Jersey Fang Hu, Ph.D. † Department of Molecular Biology, Scripps Research Sejin Lee, Ph.D. † SK Drug Development Center Daejong, Korea Xiaoyi Hu, Ph.D. Alexandre Lemire, Ph.D.**** Zheng-Zheng Huang, Ph.D. † DuPont Central Research and Development Wilmington, Delaware Edward Lemke, Ph.D. Ben Hutchins, Ph.D. Chuang-Chuang Li, Ph.D. † Peking University Peking, China Jun-Li Hou, Ph.D. Der-ren Hwang, Ph.D. † Academia Sinica Taipei, Taiwan Christophe Letondor, Ph.D.**** Fangzheng Li, Ph.D.***** CHEMISTRY 2008 Hongming Li, Ph.D. † Schering-Plough Kenilworth, New Jersey Joonwoo Nam, Ph.D. † CytRx Corporation San Diego, California Ke Li, Ph.D. † DuPont Central Research and Development Wilmington, Delaware Tae-Gyu Nam, Ph.D. THE SCRIPPS RESEARCH INSTITUTE Troy Ryba, Ph.D. † Broad Institute of MIT and Harvard Cambridge, Massachusetts Ph.D. † Sebastian Steiniger, Ph.D. Antonia Stepan, Ph.D. James Stover, Ph.D. Youngha Ryu, Texas Christian University Fort Worth, Texas Bernhard Stump, Ph.D. Catherine Saccavini, Ph.D. Hui Kai Sun, Ph.D.***** Severin Odermatt, Ph.D.**** Nicholas Salzameda, Ph.D. Shinobu Takizawa, Ph.D. Yeon-Hee Lim, Ph.D. † Schering-Plough Kenilworth, New Jersey Christian Olsen, Ph.D. Antonio Sanchez-Ruiz, Ph.D. Yazmin Osornio, Ph.D.**** Yoshikazu Sasaki, Ph.D. Tongxiang Lin, Ph.D. Junguk Park, Ph.D. Stefan Schiller, Ph.D. Adam Talbot, Ph.D. † Institute of Chemical and Engineering Sciences Jurong Island, Singapore Troy Lister, Ph.D. † Novartis Cambridge, Massachusetts Nitin Patil, Ph.D. Niklas Schone, Ph.D. Annie Tam, Ph.D. Yefeng Tang, Ph.D. Christopher Liu, Ph.D. † Cubix Pharmaceuticals Lexington, Massachusetts Richard Payne, Ph.D. † University of Sydney Sydney, Australia Michael Schramm, Ph.D. † California State University Long Beach, California Wenshe Liu, Ph.D. † Texas A&M University College Station, Texas Xuemei Peng, Ph.D.**** Edward Sessions, Jr., Ph.D. Murali Peram Surakattula, Ph.D. † CytRx Corporation San Diego, California Shigeki Seto, Ph.D. Pi-Hui Liang, Ph.D. † Academia Sinica Taipei, Taiwan Michael Luzung, Ph.D. Utpal Majumder, Ph.D. Sreeman Mamidyala, Ph.D. Andrew Nguyen, M.D., Ph.D. Romain Noel, Ph.D. George Nora, Ph.D.***** Johan Paulsson, Ph.D. Alexander Mayorov, Ph.D. Charles Melancon, Ph.D. Lionel Moisan, Ph.D. † CEA Gif-Sur-Yvette, France Ana Montero, Ph.D.**** Miguel Morales, Ph.D. Shun Su, Ph.D. Mariola Tortosa, Ph.D. † Instituto de Quimica Organica, CSIC Madrid, Spain Craig Turner, Ph.D.**** Roshan Perera, Ph.D. † University of Texas Austin, Texas Takeshi Masuda, Ph.D. Michael Maue, Ph.D. † Bayer CropScience AG Monheim, Germany Young Jun Seo, Ph.D. 81 Mary Jo Sever, Ph.D. Alex Shaginian, Ph.D. † Ardea Biosciences San Diego, California David Shaw, Ph.D. Ramulu Poddutoori, Ph.D. Weijun Shen, Ph.D. Jonathan Pokorski, Ph.D. Xiao Shengxiong, Ph.D. Agustí Lledó Ponsati, Ph.D. Bingfeng Shi, Ph.D. Daniela Radu, Ph.D. † DuPont Central Research and Development Wilmington, Delaware Yan Shi, Ph.D. Vincent Trepanier, Ph.D. † Institute of Chemical and Engineering Sciences Jurong Island, Singapore Jonathan Tripp, Ph.D. Meng-Lin Tsao, Ph.D. † University of California Merced, California Andrew Udit, Ph.D. Hiroki Shigehisa, Ph.D. Hiroyuki Shimamura, Ph.D. Ronald Rahaim, Ph.D.***** Siddhartha Shenoy, Ph.D. Praveen Rao, Ph.D. † University of Waterloo Waterloo, Ontario, Canada Matthew Tremblay, Ph.D. Ryan Simkovsky, Ph.D. Chinnappan Sivasankar, Ph.D.**** Taiki Umezawa, Ph.D. † Hokkaido University Sapporo, Japan Kenji Usui, Ph.D. † Tokyo Institute of Technology Tokyo, Japan Carlos Valdez, Ph.D. † Rigel Pharmaceuticals, Inc. South San Francisco, California Adam Morgan, Ph.D. † Concert Pharmaceuticals, Inc. Lexington, Massachusetts Per Restorp, Ph.D. Ting-Wei Mu, Ph.D. Jin-Kyu Rhee, Ph.D. Michael Smolinski, Ph.D. † Kinex Pharmaceuticals Buffalo, New York S. Vasudeva Naidu, Ph.D. Fatima Rivas, Ph.D. Xinyi Song, Ph.D. Feng Wang, Ph.D. Yuya Nakai, Ph.D. Joshua Roth, Ph.D.***** Simon Stamm, Ph.D. Jian Wang, Ph.D. Kimberly Reynolds, Ph.D. Punna Venkateshwarlu, Ph.D. 82 CHEMISTRY Jiangyun Wang, Ph.D. † Institute of Biophysics Beijing, China Lin Wang, Ph.D. Sheng-Kai Wang, Ph.D. Weidong Wang, Ph.D. Xisheng Wang, Ph.D. Yajuan Wang, Ph.D. Yuanhua Wang, Ph.D. Timo Weide, Ph.D. Albert Willis, Ph.D. † Pharmagra Labs, Inc. Brevard, North Carolina Tao Wu, Ph.D. † Institute of Chemical and Engineering Sciences Jurong Island, Singapore Heiko Wurdak, Ph.D. Jian Xie, Ph.D. Wen Xiong, Ph.D. Yue Xu, Ph.D. Junichiro Yamaguchi, Ph.D. Ryu Yamasaki, Ph.D. † Tokyo University of Science Tokyo, Japan Ura Yasuyuki, Ph.D. † Nara Women’s University Nara, Japan Yan Yin, Ph.D. 2008 Heyue Zhou, Ph.D. Hongyan Zhou, Ph.D. Shoutian Zhu, Ph.D. Joerg Zimmermann, Ph.D. V I S I T I N G I N V E S T I G AT O R S Keisuke Fukuchi, Ph.D. Sankyo Co., Ltd. Tokyo, Japan Christine Hernandez, Ph.D. † University of Philippines Diliman, Philippines (Edmond) Shie-Liang Hsieh, Ph.D. National Yang-Ming University Taipei, Taiwan Masakazu Imamura, Ph.D. † Astellas Pharma Inc. Tsukuba, Ibaraki, Japan Kuniyuki Kishikawa, Ph.D. † Kyowa Hakko Kogyo Co., Ltd. Sunto-gun, Shizuoka, Japan Michael Meijler, Ph.D. † Ben-Gurion University of the Negev Be’er Sheva, Israel Takayoshi Suzuki, Ph.D. † Nagoya City University Nagoya, Japan Yoshiyuki Yoneda, Ph.D. † Daiichi Pharmaceutical Co., Ltd. Tokyo, Japan Ian Young, Ph.D. S C I E N T I F I C A S S O C I AT E Zhanqian Yu, Ph.D. Xu Yuan, Ph.D. Weiqiang Zhan, Ph.D. Hongjun Zhang, Ph.D. Xuejun Zhang, Ph.D. Yanghui Zhang, Ph.D. Yingchao Zhang, Ph.D. † Hoffmann-La Roche, Inc. Nutley, New Jersey Jon Ashley THE SCRIPPS RESEARCH INSTITUTE * Joint appointment in The Skaggs Institute for Chemical Biology ** Joint appointments in The Skaggs Institute for Chemical Biology and the Department of Immunology and Microbial Science *** Joint appointments in The Skaggs Institute for Chemical Biology and the Department of Molecular Biology **** Appointment completed ***** Scripps Florida † Appointment completed; new location shown CHEMISTRY 2008 Chairman’s Overview s the “central science,” chemistry stands between biology and medicine and between physics and materials science and provides the crucial bridge for drug discovery and development. But chemistry has a much more profound and useful role in science and society. It is the discipline that continually creates the myriad of new materials that we all encounter in our everyday lives: pharmaceuticals, high-tech materials, polymers and plastics, K.C. Nicolaou, Ph.D. insecticides and pesticides, fabrics and cosmetics, fertilizers, and vitamins—basically everything we can touch, feel, and smell. Chemistry at Scripps Research focuses on chemical synthesis and chemical biology, the areas most relevant to biomedical research and materials science. The members of our faculty are distinguished teacher-scholars who maintain highly visible and independent research programs in areas as diverse as biological and chemical catalysis, synthesis of natural products, combinatorial chemistry, molecular design, supramolecular chemistry, chemical evolution, materials science, and chemical biology. The chemistry graduate program attracts some of the bestqualified candidates from the United States and abroad. Our major research facilities, under the direction of Dee H. Huang (nuclear magnetic resonance), Gary Siuzdak (mass spectrometry), and Raj Chadha (x-ray crystallography), are second to none and continue to provide crucial support to our research programs. In addition, the Mabel and Arnold Beckman Center for the Chemical Sciences constantly receives high praise from visitors from around the world for its architectural design and operational aspects, both highly conducive to research. Research in the Department of Chemistry goes on unabated, establishing international visibility and attracting attention, as evidenced by numerous lecture invitations, visits by outside scholars, and headline news in the media. As of 2007, the Institute for Scientific Information ranked 2 members of the department as highly cited researchers (in the top 100 worldwide). Richard Lerner and his group continue their research on antibodies, chemical synthesis, and the biological A THE SCRIPPS RESEARCH INSTITUTE 83 role of polyoxygen species. Scientists in Albert Eschenmoser’s group continue to work on the chemical etiology of nucleic acids and the origins of life. Barry Sharpless and his group persist in their endeavors to discover and develop better catalysts for organic synthesis and to construct, through innovative chemistry and biology, libraries of novel compounds for biological screening. Their click chemistry, which has had a major impact in many areas of the molecular sciences, continues to be an important focus of their research. Members of my own group continue to explore chemical synthesis and chemical biology, with a focus on the total synthesis of new anticancer agents, antibiotics, marine-derived neurotoxins, antimalarial compounds, and other bioactive natural and designed molecules. Julius Rebek and his group devise biomimetic receptors, including molecules that bind neurotransmitters and membrane components, for studies in molecular recognition. Larger host receptors can surround 3 or more molecular guests and act as chambers in which the chemical reactions of the guests are accelerated. Scientists in the group also synthesize small molecules that act as protein helix mimetics for pharmaceutical applications. Peter Schultz and researchers in his laboratory are expanding the number of genetically encoded amino acids to include fluorescent, photocaged, metal-binding, chemically reactive, and posttranslationally modified amino acids. These scientists have also adapted this technology to mammalian cells and are applying these tools in basic and applied problems in cell biology. In addition, members of the group have used cell-based screens to identify small molecules that selectively differentiate and expand embryonic and adult stem cells and reprogram lineage-committed cells, as well as novel genes and small molecules that affect a number of physiologic and disease processes. Chi-Huey Wong and his group further advance the fields of chemoenzymatic organic synthesis, chemical glycobiology, and the development of enzyme inhibitors. A new strategy for the synthesis of glycoproteins based on sugar-assisted glycopeptide ligation has been developed. The programmable 1-pot synthesis of oligosaccharides developed by this group has been further used in the assembly of glycoarrays for study of saccharides that bind to proteins. Members of this group also developed new probes to study posttranslational glycosylation and identify glycoprotein biomarkers associated with cancer. Researchers in Dale Boger’s laboratory continue their work on chemical synthesis; combinatorial chemistry; het- 84 CHEMISTRY 2008 erocycle synthesis; anticancer agents, such as vinblastine, cyclostatin, chlorofusion, and yatakemycin; and antibiotics, such as vancomycin, teicoplanin, and ramoplanin. Scientists in Kim Janda’s laboratory conduct research grounded on organic chemistry as applied to specific biological systems. The targeted programs span a wide range of interests, from immunopharmacotherapy to biological and chemical warfare agents to filarial infections such as “river blindness” to quorum sensing in bacteria and new cancer therapeutic strategies. Recent achievements include in vivo detection of botulinum neurotoxin antagonists, the development of peptides and antibodies as drug delivery modules that home to cancer cells and active vaccines for nicotine addiction and obesity that are now in preclinical trials. M. Reza Ghadiri and his group are making important contributions in the design and study of a new generation of antimicrobial agents, based on self-assembling peptide nanotube architecture, to combat multidrug resistant infections. In addition, members of the group continue to make novel contributions in several ongoing basic research endeavors, such as designing biosensors, developing molecular computation, designing self-reproducing systems, understanding the origins of life, and creating emergent chemical systems. M.G. Finn and his group have pioneered the use of virus particles as chemical reagents and building blocks for nanochemical structures. This effort is directed toward the development of new diagnostics for disease and catalysts for organic reactions. Members of Dr. Finn’s laboratory also develop and investigate new organic and organometallic reactions and use these processes to synthesize biologically active compounds. Jeff Kelly and his group are exploring the interface between the chemistry, biology, and pathobiology of proteome maintenance. The aim of their projects is to understand the physical and biological basis of protein folding and the competitive misfolding and aggregation processes that lead to age-associated neurodegenerative diseases. Information on proteome maintenance is used to develop new small-molecule therapeutic strategies for a variety of diseases, including neurodegenerative diseases. Anita Wentworth and the researchers in her group are investigating the chemical basis of complex disease states and are synthesizing peptide- and small molecule–based therapeutic agents. These scientists focus on disease states in which inflammatory and reactive oxygen species are prominent, such as atherosclerosis, Alzheimer’s disease, and other diseases of ageing. THE SCRIPPS RESEARCH INSTITUTE Researchers in Floyd Romesberg’s laboratory are using diverse techniques ranging from bioorganic and biophysical chemistry to bacterial and yeast genetics to understand and manipulate the process of evolution. Major efforts include designing unnatural base pairs and using directed evolution of DNA polymerases to efficiently synthesize unnatural DNA containing the base pairs, using spectroscopy to understand biological function and how it evolves, and understanding how induced and adaptive mutations contribute to evolution in eukaryotic and prokaryotic cells. Phil Baran and his group are interested in how the general challenge of chemoselectivity in organic chemistry can be answered through the auspices of total synthesis. He and his coworkers have developed extremely concise chemical solutions to the synthetic challenges posed by numerous families of natural products. These syntheses systematically tackle the issue of chemoselectivity and are characterized by striking brevity, new biosynthetic postulates, the invention of new methods, and a minimum use or complete absence of protecting groups and superfluous oxidation state manipulations. The Frontiers in Chemistry Lecturers (19th Annual Symposium) for the 2007–2008 academic year were M. Christina White, University of Illinois; Ben L. Feringa, University of Groningen, the Netherlands; Ian Paterson, Cambridge University; and Harry Noller, University of California, Santa Cruz. In addition, we enjoyed hosting the following professors: Samir Zard, Ecole Polytechnique, France, as the Bristol-Myers Squibb Lecturer; E.J. Corey, Harvard University, as the Pfizer Lecturer; and Robert Bergman, University of California, Berkeley, as the Novartis Lecturer. CHEMISTRY 2008 THE SCRIPPS RESEARCH INSTITUTE 85 INVESTIGATOR’ S REPORT Synthesis of Natural Products, Development of Synthetic Methods, and Medicinal Chemistry W.R. Roush, R. Bates, D. Bykowski, M. Chen, E. Darout, A. DeBaillie, J. Dunetz, G. Halvorsen, M. Handa, J. Hicks, T. Hopkins, C.-W. Huh, F. Li, A. Legg, R. Lira, L. Martinez, C. Nguyen, G. Nora, R. Pragani, R. Rahaim, J. Roth, H. Sun, M. Tortosa, J. Whitaker, S. Winbush ur research has 2 major themes. One is the total synthesis of structurally complex, biologically active natural products such as those shown in Figure 1. In each of these syntheses, we emphasize the discovery, development, and/or illustration of new reactions and methods for achieving high levels of stereochemical control. These efforts are pursued in parallel with reaction design, stereochemical studies, and the development of new synthetic methods. We are particularly interested in stereochemical aspects of intramolecular and transannular Diels-Alder reactions, development of methods for the diastereoselective and enantioselective reactions of allylmetal compounds with carbonyl compounds, and nucleophilic phosphine-catalyzed organic reactions. Recent research has included stereochemical studies of transannular Diels-Alder reactions used in total syntheses of spinosyn A and superstolide A and development of new versions of the double allylboration reactions of aldehydes with γ-boryl-substituted allylboranes for stereocontrolled synthesis of 1,5-ene-diols, which are being used in several ongoing syntheses, including those of tetrafibricin, apoptolidin A, and peloruside. In addition, we have synthesized highly substituted tetrahydrofurans via [3+2]-annulation reactions of highly functionalized allylsilanes; this chemistry was recently applied to total syntheses of 10-hydroxytrilobacin and 3 stereoisomers. We have also developed phosphine-mediated organocatalytic reactions, and we recently completed the total synthesis of tedanolide. Our second area of major interest involves problems in bioorganic chemistry and medicinal chemistry. One long-term project is the design and synthesis of inhibitors of cysteine proteases isolated from tropical para- O F i g . 1 . Structures of recently synthesized natural products. sites, such as Trypanosoma cruzi, the causative agent of Chagas’ disease, and Plasmodium falciparum, the most virulent of the malaria parasites. This research is performed in collaboration with colleagues at the University of California, San Francisco. In collaboration with S. Reed, University of California, San Diego, we have developed a cysteine protease inhibitor with remarkable 86 CHEMISTRY 2008 ability to prevent Entamoeba histolytica from invading human intestinal tissue. Optimization of this inhibitor for in vivo applications is in progress. New projects involve discovery of small molecules that affect cancer and other disease-related biochemical targets (e.g., nuclear hormone receptors), studies of structure-activity relationships, and optimization of the pharmacologic profile of certain natural products. PUBLICATIONS Chen, Y.-T., Lira, R., Hansell, E., McKerrow, J.H., Roush, W.R. Synthesis of macrocyclic trypanosomal cysteine protease inhibitors. Bioorg. Med. Chem. Lett. 18:5860, 2008. Dunetz, J., Roush, W.R. Concerning the synthesis of the tedanolide C(13)-C(23) fragment via an anti-aldol reaction. Org. Lett. 10:2059, 2008. Handa, M., Scheidt, K.A., Bossart, M., Zheng, N., Roush, W.R. Studies on the synthesis of apoptolidin A, I: synthesis of the C(1)-C(11) fragment. J. Org. Chem. 73:1031, 2008. Handa, M., Smith, W.J. III, Roush, W.R. Studies on the synthesis of apoptolidin A, II: synthesis of the disaccharide unit. J. Org. Chem. 73:1036, 2008. Hicks, J.C., Huh, C.W., Legg, A.D., Roush, W.R. Concerning the selective protection of (Z)-1,4-syn-ene-diols and (E)-1,5-anti-ene-diols as allylic triethylsilyl ethers. Org. Lett. 9:5621, 2007. Hicks, J.D., Roush, W.R. Synthesis of the C(26)-C(42) and C(43)-C(67) pyrancontaining fragments of amphidinol 3 via a common pyran intermediate. Org. Lett. 10:681, 2008. Lira, R., Roush, W.R. Enantio- and diastereoselective synthesis of syn-β-hydroxyallylsilanes via a chiral (Z)-γ-silylallylboronate. Org. Lett. 9:4315, 2007. Methot, J.L., Roush, W.R. Applications of tricoordinated phosphorus compounds in organic catalysis. In: Organophosphorus Compounds. Trost, B.M. (Ed.). Thieme Chemistry, New York, in press. Vol. 42 in Science of Synthesis. Roth, J., Madoux, F., Hodder, P., Roush, W.R. Synthesis of small molecule inhibitors of the orphan nuclear receptor steroidogenic factor-1 (NR5A1) based on isoquinolinone scaffolds. Bioorg. Med. Chem. Lett. 18:2628, 2008. Roush, W.R. Total synthesis of biologically active natural products. J. Am. Chem. Soc. 130:6654, 2008. Tortosa, M., Yakelis, N.A., Roush, W.R. Total synthesis of (+)-superstolide A. J. Am. Chem. Soc. 130:2722, 2008. Winbush, S.M., Mergott, D.J., Roush, W.R. Total synthesis of (–)-spinosyn A: examination of structural features that govern the stereoselectivity of the key transannular Diels-Alder reaction. J. Org. Chem. 73:1818, 2008. Scheinost, J.C., Wang, H., Boldt, G.E., Offer, J., Wentworth, P., Jr. Cholesterol secosterol-induced aggregation of methylated amyloid-β peptides, insights into aldehydeinitiated fibrillization of amyloid-β. Angew. Chem. Int. Ed. 47:3919, 2008. Temperini, C., Cecchi, A., Boyle, N.A., Scozzafava, A., Cabeza, J.E., Wentworth, P., Jr., Blackburn, G.M., Supuran, C.T. Carbonic anhydrase inhibitors. Interaction of 2-N,N-dimethylamino-1,3,4-thiadiazole-5-methylsulfonamide with 12 mammalian isoforms: kinetic and x-ray crystallographic studies. Bioorg. Med. Chem. Lett. 18:999, 2008. Wentworth, P., Jr., Witter, D. Antibody-catalyzed water-oxidation pathway. Pure Appl. Chem. 80:1849, 2008. THE SCRIPPS RESEARCH INSTITUTE Infectology Distinguishing between prion strains 22L and Me7 with the mouse bioassay takes 6 months. The cerebellar Purkinje cell layer (immunostained for calbindin) remains intact in Me7-infected, terminally sick mice (top, arrow) but is obliterated by infection with 22L (bottom, arrow). With the cell panel assay (insets), the strains can be distinguished from each other in 2 weeks. 22L prions efficiently infect the 4 cell lines constituting the panel, whereas Me7 infects only LD9 and CAD cells. Stained sections and micrographs prepared by Sukhvir Mahal, Ph.D., staff scientist, and Alexsandra Sherman, research assistant; photomontage created by Christopher A. Baker, Ph.D., research associate. Work done in the laboratory of Charles Weissmann, Ph.D. BIOCHEMISTRY Corinne Lasmézas, D.V.M., Ph.D., Professor, and Paula Saá Prieto, Ph.D., Research Associate. INFECTOLOGY 2008 THE SCRIPPS RESEARCH INSTITUTE 195 DEPAR TMENT OF INFECTOLOGY S TA F F Charles Weissmann, M.D., Ph.D. Professor and Chairman Joaquin Castilla, Ph.D. Assistant Professor S E N I O R S TA F F R E S E A R C H A S S O C I AT E S Yervand Karapetyan, M.D. Ivan Angulo-Herrera, Ph.D. Minghai Zhou, Ph.D. Shawn Browning, Ph.D. SCIENTIST Corinne Lasmézas, D.V.M., Ph.D. Professor Sukhvir Mahal, Ph.D. Natalia Fernández-Borges, Ph.D. Maria Herva-Moyana, Ph.D. SENIOR RESEARCH Donny Strosberg, Ph.D. Professor Tim Tellinghuisen, Ph.D. Assistant Professor A S S O C I AT E S Paula Saá Prieto, Ph.D. Carlos Coito, Ph.D. Jiali Li, Ph.D. Chris Baker, Ph.D. Anja Oelschlegel, Ph.D. Chairman’s Overview he Department of Infectology focuses on prion diseases and hepatitis. Tim Tellinghuisen and his colleagues study hepatitis C virus (HCV) RNA replication and virion assembly. They identified the viral protein NS5A as an essential component of the viral replicase and mapped all amino acids in domains II and III essential for HCV RNA replication. They identified regions whose functions are required for Charles Weissmann, M.D., Ph.D. generating infectious virus but not for RNA replication, suggesting that NS5A regulates the switch between RNA replication and virus assembly. They are using high-throughput genetic screens to identify host components required for replicase activity. Donny Strosberg and his colleagues established for the first time a cell culture system for HCV genotype 1b. They identified peptides derived from the HCV core that inhibit (1) dimerization of core protein, the first step in viral assembly and (2) release of virions. F.V. Chisari and his group, Department of Molecular and Experimental Medicine, independently found that one such peptide inhibits HCV replication. These findings establish the HCV core as a target for the development of anti-HCV drugs. Three groups, those of Corinne Lasmézas and Joaquin Castilla and my own, study prion biology. Prions consist T of a multimeric assembly of PrPSc, a conformer of the normal host protein PrPC. The seeding hypothesis posits that prions replicate by recruiting host PrP C into the PrPSc assembly, a process that entails conformational rearrangement of the PrPC. Prions occur in the form of different strains, all associated with the same PrP Sc sequence but with distinct cell tropisms, both in brain and in cell culture. The cell-based assay for prions has been further streamlined by Emery Smith, and the cell panel assay, which allows rapid distinction between prion strains, was extended to more strains by Sukhvir Mahal. It has been proposed that “strain-ness” is encoded either by distinct conformations of PrP Sc or by the complex glycans present on the protein. This hypothesis has been negated by Shawn Browning and Dr. Mahal, who found that strain specificity is maintained when prions are propagated under conditions in which complex glycosylation is abrogated. Dr. Lasmézas and her colleagues determined that PrPSc in cultured prion-infected cells is undetectable at the outer cell surface and is therefore likely generated intracellularly. These investigators characterized a rapid animal model for prion disease based on a transgenic, PrP-overexpressing mouse strain and are researching the mechanism of pathogenesis. Dr. Castilla and his group have perfected the protein misfolding cyclic amplification procedure, which allows the cell-free replication of prions, and have shown that various prion strains can be propagated continuously without losing strain-specific properties. 196 INFECTOLOGY 2008 Investigators’ Reports Biology of Prion Strains THE SCRIPPS RESEARCH INSTITUTE Table 1. Susceptibility of cell lines to various prion strains.* Cell line Prion strain 22L 139A RML, 79A Me7 301C LD9 (EMEM) +++ +++ +++ +++ – CAD5* +++ +++ +++ – +++ PK1 +++ +++ +++ – – PK1/swa +++ ++ – – – ++ – – – – C. Weissmann, C.A. Baker, S. Browning, C. Demczyk, M. Herva-Moyana, J. Li, S.P. Mahal, A. Oelschlegel, A. Sherman, E. Smith, I. Suponitsky-Kroyter rions are thought to consist mainly or entirely of PrP Sc , an abnormal conformer of a normal host protein, PrPC, and to propagate via a PrPSccatalyzed conversion of PrPC. Intriguingly, distinct prion strains, which generate different disease phenotypes, are associated with the same PrP sequence, suggesting that the phenotypes are encoded posttranslationally. Our major interests are the mechanism of prion replication, the structural basis of strain specificity, and the mechanism of strain recognition by cells. Much of our research depends on a cell-based assay for prion infectivity, the standard scrapie cell assay. Therefore, considerable effort has gone into automating the procedure, reducing its cost, and improving its accuracy. We can now assay about 1000 samples per week in sextuplicate, orders of magnitude greater than the number that could be processed by using the classical mouse bioassay. The standard scrapie cell assay is the basis for the cell panel assay (CPA), which makes use of the finding that different cell lines have distinctive susceptibilities to various prion strains. Using the CPA, we can distinguish between various prion strains within 2 weeks, as opposed to the year or more required with the classical mouse bioassay. The panel originally consisted of 4 cell lines; however, when exposed to swainsonine (an α-mannosidase II inhibitor that modifies the glycans of N-glycosylated proteins), the PK1 cell line (but none of the others) becomes resistant to some prion strains but not to others. Therefore, swainsonine-treated PK1 cells provide an additional criterion for discriminating prion strains. We can currently distinguish 5 groups of strains (Table 1). We determined that the characteristic CPA responses of these strains are the same whether the strains are propagated in wild-type mice or in Tga20 mice, which overexpress PrP C and, conveniently, have a reduced incubation time. We have established a large collection of cell lines chronically infected with various prion strains. Interestingly, in many instances, the cell tropism of prions derived from such lines, as determined by using the P R33 * The number of pluses indicates the degree of susceptibility to infection. The minus sign indicates resistance to infection. CPA, differs from that of the prions derived from brain. To determine whether this change reflects a permanent modification of a strain, we inoculated cell-derived prions into mice; CPA analysis of the infected brains showed no differences between the original and the cell-passaged strains. We are currently considering the possibility that the cell tropism of a prion strain may depend on the host cell in which the strain is generated, reflecting some host-imparted property (the “cytotype”), for example, the glycosylation pattern of the PrP. It has been proposed that the N-linked complex glycans attached to PrP might encode the “strain-ness” of prions. To address this question, we propagated RML, 22L, and Me7 prions in CAD5 cells in the presence of the glycosylation inhibitors deoxymannojirimycin and swainsonine. These inhibitors prevent processing of the precursor of N-linked glycans and result in PrP with [mannose]9[N-acetylglucosamine]2 as the only glycan, rather than a multiplicity of complex sugars. We injected cell lysates of the drug-treated, prion-infected cells into mice and used the CPA to analyze the infected brains. In all instances, the strain-specific properties had been retained, proving that complex glycosylation was not the strain-determining element and adding weight to the hypothesis that strain-ness is encoded by the conformation of PrPSc. In another project, we are exploring why certain subclones derived from the same cell line, occurring with a frequency of 1 in 5000 to 1 in 10,000, can vary by 100 to 1000 times in their susceptibility to prions. We tagged highly susceptible and resistant subclones with 2 different markers of antibiotic resistance and fused the cells. Currently, we are determining whether susceptibility, as measured in heterokaryons, INFECTOLOGY 2008 is a dominant or a recessive property. In the next step, we will generate microcells containing single, tagged chromosomes from one of the cell lines and fuse the microcells to cells from the other line to determine which chromosome encodes the critical trait. PUBLICATIONS Julius, C., Hutter, G., Wagner, U., Seeger, H., Kana, V., Kranich, J., Klöhn, P., Weissmann, C., Miele, G., Aguzzi, A. Transcriptional stability of cultured cells upon prion infection. J. Mol. Biol. 375:1222, 2008. Mahal, S.P., Baker, C.A., Demczyk, C.A., Smith, E.W. Julius, C., Weissmann, C. Prion strain discrimination in cell culture: the cell panel assay. Proc. Natl. Acad. Sci. U. S. A. 104:20908, 2007. Mahal, S.P., Demczyk, C.A., Smith, E.W., Jr., Klöhn, P.-C., Weissmann, C. Assaying prions in cell culture: the standard scrapie cell assay (SSCA) and the scrapie cell assay in end point format (SCEPA). Methods Mol. Biol. 459:49, 2008. Pathogenesis of Transmissible Spongiform Encephalopathies C.I. Lasmézas, N. Salès, P. Saá Prieto, M. Zhou, Y. Karapetyan, F. Sferrazza, G. Ottenberg rions, the transmissible agents responsible for prion diseases, are thought to consist mainly of PrPSc, an abnormally folded isoform of the ubiquitous prion protein PrP. Prions are thought to replicate by an autocatalytic process of template-induced conformational change. In collaboration with C. Weissmann, Department of Infectology, we are devising a new method for propagating and characterizing different strains of prions. Dr. Weissmann and his group have developed a cellbased infectivity assay, the cell panel assay, in which prion strains are distinguished on the basis of cell tropism. We have studied the fate of prions in the Tga20 mouse model. Compared with wild-type mice, Tga20 mice express 8-fold higher levels of PrP C in the brain, and clinical disease occurs more quickly after inoculation of prions. Many aspects of prion replication in Tga20 mice were unknown, for instance, the response of these animals to different prion strains. We found that PrPC from the brains of Tga20 mice has a higher intrinsic resistance to proteolytic digestion by protease than does PrPC from the brains of wild-type mice. We also discovered that the levels of PrPC overexpression in different regions of the brain vary and hence could influence the pathologic lesions and PrPSc distribution that occur after prion infection. Vacuolation and PrPSc deposition pro- P THE SCRIPPS RESEARCH INSTITUTE 197 files in brains of Tga20 mice differed from those in C57BL/6 mice for the 3 scrapie strain studied. Each strain had a characteristic profile in Tga20 mice and could be readily distinguished by neuropathologic analysis, showing that the level of PrPC expression is not the main determinant of brain tropism. Importantly, despite generating different neuropathologic phenotypes in C57BL/6 and Tga20 mice, all 3 prion strains retained their intrinsic identity after being replicated in either mouse line, as determined by the cell panel assay. This study provides a new reliable method for rapid propagation and characterization of prion strains by using a combination of Tga20 mice and the cell panel assay. Fundamentally, our results indicate that despite different biochemical characteristics of PrPC, different expression patterns of PrPC in the 2 murine hosts, and different genetic background of the 2 mouse strains, prions are propagated faithfully in both types of mice. This finding raises once more the question of the molecular basis of prion-strain properties. Previously, we found that oligomeric assemblies of recombinant prion protein are toxic to primary cultures of cortical neurons and in mice when injected via a stereotaxic method. We are now devising new intervention strategies to block the toxic/infectious PrP species. A first goal is to determine the cellular location of PrP aggregates, in order to know which cellular compartment to target. The many reports of the presence of protease-resistant PrP (PrPres) at the cell membrane are contradictory. Using a cell-surface biotinylation strategy and comprehensive controls to account for the presence of dead cells and for the intrinsic capacity of PrPres to be biotinylated, we found no detectable PrPres at the cell membrane. This finding has major implications for the development of diagnostic molecules. We are continuing our efforts to locate the cellular compartment that must be targeted for therapeutic purposes, and we are setting up new models for PrPinduced toxic effects in cell lines. Inhibitors of Protein-Protein Interactions in Hepatitis C A.D. Strosberg, C. Coito, R. Henderson, S. Kota, G. Mousseau S everal small-molecule drugs are in advanced clinical development for the treatment of hepatitis C, a situation that may affect the 170 mil- 198 INFECTOLOGY 2008 lion carriers of hepatitis C virus (HCV) worldwide, including more than 3 million in the United States. Because of its high mutability, HCV likely will become resistant to these potential drugs, which are mostly inhibitors of a viral protease and the viral polymerase. This past year, we continued our studies of protein interactions involving HCV proteins to better understand the respective roles of the proteins and to identify novel target proteins that would not induce resistance. The core HCV protein, which is highly conserved across all 6 major HCV genotypes, is a good candidate for such studies. H C V C O R E P R O T E I N A S A TA R G E T The HCV core protein functions primarily as the structural element of the virus. The core contains several residues essential for HCV production. Most of these residues are located in the N-terminal two-thirds of the core protein and mediate core dimerization and most interactions with intracellular proteins. Core dimerization is the first step in nucleocapsid assembly; its inhibition should block formation and release of infectious HCV particles. THE SCRIPPS RESEARCH INSTITUTE tive of SL-175 bound to core106 with a dissociation constant of 1.9 µM and was displaced by the uncoupled peptide, with a 50% inhibitory concentration of 18.7 µM. In a collaborative surface plasmon resonance study with J.-P. Lavergne, Centre National de la Recherche Scientifique, Lyon, France, SL-175 bound core169 with a dissociation constant of 7.2 µM. PEPTIDE INHIBITORS OF HCV RELEASE FROM H E PAT O M A C E L L S When added to Huh-7.5 hepatoma cells infected with the HCV genotype 2a J6/JFH-1 or the 1b CG strain, peptides SL-173 and SL-175 prevented release of newly formed infectious HCV into the medium. Using a different approach, F.V. Chisari and his group, Department of Molecular and Experimental Medicine, independently found that SL-173 inhibits HCV focus formation in vitro by more than 90% and viral RNA synthesis 11-fold, 72 hours after infection. The combined results of our 2 groups thus establish that the HCV core is a useful novel target for development of anti-HCV drugs. H E PAT O M A C E L L C U LT U R E S Y S T E M F O R H C V A S S AY S F O R H C V C O R E D I M E R I Z AT I O N Using a pair of domains that consist of the first 106 residues of the HCV core protein (core106) tagged with oligonucleotides encoding the octapeptide Flag or glutathione-S-transferase (GST), we developed an assay based on the use of an antibody to Flag that binds to Flag-tagged core106 interacting with GST-tagged core106 adsorbed on a glutathione-coated microtiter plate. We designed a 384 well–based sensitive and high-throughput time-resolved fluorescence assay with fluorescent antibodies to Flag and GST. Untagged core106 completely inhibits core106 dimerization. One of our industrial partners will use this assay to screen 2 million chemically diversified small molecules to identify novel nonpeptidic inhibitors of HCV production. P E P T I D E I N H I B I T O R S O F C O R E D I M E R I Z AT I O N We also designed an amplified luminescent proximity homogeneous assay to monitor core-core interactions on the basis of donor and acceptor beads that respond to the specific tags on the proteins. Using this assay, which has a high signal-to-background ratio, we screened 14 18-residue-long peptides derived from the HCV core and identified 2 partially overlapping peptides, SL-173 and SL-174, which caused 68% and 63% inhibition, respectively, of core106 dimerization. SL-175, a 3-residue shorter version of SL-173, inhibited core106 dimerization by 50%, with a 50% inhibitory concentration of 22 µM. Using fluorescence polarization, we found that a fluorophore-coupled deriva- GENOTYPE 1B The culture system for HCV used routinely in laboratories worldwide was derived from strain JFH-1 of an HCV of genotype 2a. In Western countries, however, the most prevalent infections are caused by HCV strains of genotype 1. To understand differences between HCV of different genotypes and further evaluate potential inhibitors of protein-protein interactions in HCV or between HCV and human host proteins, we developed and characterized a unique culture system for the HCV genotype 1b CG strain. Two different protocols have been developed: the first one is based on the coculture of infected, virus-releasing cells with uninfected cells in a 2-chamber system; the second is a direct incubation of uninfected cells with supernatant from cells electroporated with the RNA from strain CG or from infected cells. Using either system, we have confirmed transfer of infectivity in several passages. Furthermore, we showed that cyclosporine A has comparable inhibitory effects on J6/JFH-1 and CG strains of HCV and that antibodies to CD81, a coreceptor for HCV, block infectious particles released into the supernatant of cells infected by the 1b CG strain. Initial results also suggest that the 2 strains are diversely affected by IFN-α; J6/JFH-1 appears to be sensitive, and CG appears to be resistant. This finding, if confirmed, would reflect the situation in patients; patients infected with genotype 2 HCV generally respond better to interferon treatment than do patients infected with genotype 1 HCV. INFECTOLOGY 2008 PUBLICATIONS Strosberg, A.D., Nahmias, C. G-protein-coupled receptor signaling through protein networks. Biochem. Soc. Trans. 35:23, 2007. Hepatitis C Virus RNA Replication and Virion Assembly T.L. Tellinghuisen, J.C. Treadaway, K.L. Foss, THE SCRIPPS RESEARCH INSTITUTE 199 work. We have also begun applying high-throughput genetic screens to identify required host components of the replicase. Our ultimate goal is to understand, at the molecular level, the assembly, activity, and regulation of the HCV RNA replication machinery. Greater insight into the poorly understood replicase components, such as NS5A, will provide a more complete view of the replicase complex and will fuel the design of new drugs. I. Angulo-Herrera epatitis C virus (HCV) is a human pathogen of global importance; according to some estimates, nearly 3% of the world’s population are chronically infected. Long-term viral replication in these individuals leads to severe liver disease, including cirrhosis and, often, hepatocellular carcinoma. The current treatment with agents nonspecific for HCV is poorly tolerated and is ineffective in about half of the patients, emphasizing the need for effective antiviral drugs specific for the virus. The HCV replicase, the multicomponent machine that replicates the viral RNA, is an ideal drug target. The core replicase consists of 5 HCV proteins associated with well-characterized polymerase, protease, and helicase activities. Some HCV replicase proteins, such as NS5A, are essential for HCV replication; however, their specific functions remain enigmatic. We have been characterizing NS5A. Our goal is to understand the role of this protein in replication and, more generally, the replicase itself. We have defined NS5A as an essential, 3-domain metalloprotein component of the replicase. Our crystal structure of domain I of NS5A has provided a glimpse of the potential interactions of NS5A in the viral replicase. We recently identified all of the amino acids in the poorly understood domains II and III that are required for HCV RNA replication. Additionally, we have discovered an interaction between the membrane anchor of NS5A and the protein NS4B, another component of the replicase. This interaction appears to localize NS5A to the replicase and is essential for RNA replication. We are identifying regions of NS5A whose functions are required for the production of infectious virus but not for RNA replication. Our findings suggest that NS5A may function as a regulator of the switch between RNA replication and virus production. We are conducting biochemical, genetic, and structural experiments to evaluate the potential interaction surfaces and activities of NS5A observed in our previous structural and genetic H PUBLICATIONS Lindenbach, B.D., Tellinghuisen, T.L. Insights into hepatitis C virus RNA replication. In: Viral Genome Replication. Götte, M., Cameron, C., Raney, K. (Eds.). Springer, New York, in press. Tellinghuisen, T.L., Evans, M.J., Von Hahn, T., You, S., Rice, C.M. Studying hepatitis C virus: making the best of a bad virus. J. Virol. 81:8853, 2007. Tellinghuisen, T.L., Foss, K.L., Treadaway, J. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032, 2008. Tellinghuisen, T.L., Foss, K.L., Treadaway, J.C., Rice, C.M. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073, 2008. Tellinghuisen, T.L., Lindenbach, B.D. Reverse transcription PCR based sequence analysis of hepatitis C virus replicon RNA. In: Hepatitis C: Methods and Protocols, 2nd ed. Tang, J. (Ed.). Humana Press, Totowa, NJ, in press. Vol. 510 in Methods in Molecular Biology. Walker, J. (Series Ed.). Tellinghuisen, T.L., Marcotrigiano, J. Preparation of hepatitis C virus NS5A protein for structural studies. In: Hepatitis C: Methods and Protocols, 2nd ed. Tang, J. (Ed.). Humana Press, Totowa, NJ, in press. Vol. 510 in Methods in Molecular Biology. Walker, J. (Series Ed.). Strain and Species Barrier Phenomena in a Cell-Free System J. Castilla, N. Fernández-Borges, J. de Castro ransmissible spongiform encephalopathies are fatal neurodegenerative disorders that affect both humans and animals. The disorders can be classified as genetic, sporadic (putatively spontaneous), or infectious. The infectious agent associated with these encephalopathies, the prion, appears to consist of the single protein PrPSc, an abnormal conformer of the natural host protein PrPC. Prions propagate by converting host PrPC into PrPSc. One characteristic of prions is their ability to infect some species and not others. This phenomenon is known as the transmission barrier. Interestingly, prions occur in the form of different strains with distinct biological and physicochemical properties, even though all the strains have the same PrP amino acid sequence, albeit in presumably different conformations. In general, the T 200 INFECTOLOGY 2008 transmission barrier is manifested as an incomplete attack rate (i.e., the percentage of animals in a group in which disease develops after inoculation with prions is less than 100) and long incubation times (time from inoculation to the onset of the clinical signs of disease), which become shorter after serial passages of the prion strain in animals. Compelling evidence indicates that the transmission barriers are closely related to differences in PrP amino acid sequences between the donor and recipients of the infectious prions and the prion strain conformation. Unfortunately, the molecular basis of the transmission barrier and its relationship to prion-strain conformations are currently unknown, and we cannot predict the degree of a species barrier simply by comparing the prion proteins from 2 species. We have conducted a series of experiments in which we used protein misfolding cyclic amplification, a technique that mimics in vitro some of the fundamental steps involved in prion replication in vivo, albeit with accelerated kinetics. The in vitro generated prions have key prion features: they are infectious in vivo and maintain their strain specificity. We have used this technique to efficiently replicate a variety of prion strains from, among others, mice, hamsters, bank voles, deer, cattle, sheep, and humans. The correlation between in vivo data and our in vitro results suggests that protein misfolding cyclic amplification is a valuable tool for assessing the strength of the transmission barriers between diverse species and for different prion strains. We are using the method to determine which amino acids in the PrPC sequence contribute to the strength of the transmission barrier. These studies are useful in evaluating the potential risks to humans and animals not only of established prion strains but also of new (atypical) strains. For example, although the prion strain that causes classical sheep scrapie cannot cross the human transmission barrier in vitro, the strain that causes bovine spongiform encephalopathy can cross the human transmission barrier efficiently after propagation in sheep. In addition, we have generated prions that are infectious to species hitherto considered resistant to prion diseases. PUBLICATIONS Hetz, C., Lee, A.H., González-Romero, D., Thielen, P., Castilla, J., Soto, C., Glimcher, L.H. Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 105:757, 2008. Morales, R., González, D., Soto, C., Castilla, J. Advances in prion detection. In: Microbial Food Contamination. Wilson, C.L. (Ed.). CRC Press, Boca Raton, FL, 2007, p. 255. THE SCRIPPS RESEARCH INSTITUTE Molecular and Integrative Neurosciences A, Circadian rhythm profile for wild-type (WT) littermates and EP3R–/– mice. Although diurnal distribution of motor activity follows the light-dark cycle, EP3R–/– mice have bouts of increased activity during the light cycle that are associated with grooming and eating behavior. Those bouts of activity are irregular and are better detected during the resting phase (see carets). B, Continuous recording of core body temperature (CBT) and motor activity (MA) during 5 days at normothermic conditions (room temperature, 30°C) confirms that EP3R–/– and WT mice are nocturnal and that they follow the low activity–high resting (light cycle) and high activity–low resting (dark cycle) pattern. C, Averaged data indicate that EP3R–/– mice have an increase in motor activity characterized by bouts of activity that increases the core body temperature (see arrows). The increase in motor activity is associated with grooming and eating behavior. D, Cumulative data confirm that EP3R–/– mice are more active during the light period. *P = .03. Work done in the laboratory of Tamas Bartfai, Ph.D., professor. Reprinted from Sánchez-Alavez, M., Klein, I., Brownell, S.E., et al. Night eating and obesity in the EP3R-deficient mouse. Proc. Natl. Acad. Sci. U. S. A. 104:3009, 2007. Copyright 2007 National Academy of Sciences U.S.A. Cindy Ehlers, Ph.D., Professor, Gina Stouffer, Research Assistant, and José Criado, Jr., Ph.D., Staff Scientist MOLECUL AR AND INTEGRATIVE NEUROSCIENCES MOLECULAR AND I N T E G R AT I V E NEUROSCIENCES DEPAR TMENT S TA F F Tamas Bartfai, Ph.D. Chairman and Professor Director, Harold L. Dorris Neurological Research Institute Serge Ahmed, Ph.D. Adjunct Assistant Professor Etienne Baulieu, Ph.D. Adjunct Professor Floyd Bloom, M.D. Professor Emeritus Executive Director, Science Communication 2008 THE SCRIPPS RESEARCH INSTITUTE Steven J. Henriksen, Ph.D. Adjunct Professor Amanda Roberts, Ph.D. Associate Professor Paul L. Herrling, Ph.D. Adjunct Professor Michael G. Rosenfeld, M.D. Adjunct Professor Tomas Hokfelt, M.D., Ph.D. Adjunct Professor Pietro P. Sanna, M.D. Associate Professor Danny Hoyer, Ph.D. Adjunct Professor George R. Siggins, Ph.D. Professor Koki Inoue, Ph.D. Adjunct Associate Professor Iustin Tabarean, Ph.D. Assistant Professor Harvey Karten, M.D. Adjunct Professor Antoine Tabarin, Ph.D. Adjunct Associate Professor Henri Korn, M.D., Ph.D. Adjunct Professor Lars Terenius, Ph.D. Adjunct Professor Thomas Krucker, Ph.D. Adjunct Assistant Professor Claes Wahlestedt, M.D., Ph.D.* Professor Stefan Kunz, Ph.D. Adjunct Professor Mehrdad Alirezaei, Ph.D. Michal Bajo, M.D., Ph.D. Hilda Bajova, D.V.M. Fulvia Berton, Ph.D. Vez Repunte Canonigo, Ph.D. Kazuki Hagihara, Ph.D. Izabella Klein, Ph.D. Kayo Mitsukawa, Ph.D. Olivia Osborn, Ph.D. Covadonga Paneda, Ph.D. Gurudutt Pendyala, Ph.D. Jerry Pinghwa Pian, Ph.D. Tammy Wall, Ph.D. Adjunct Associate Professor Jilla Sabeti, Ph.D. Friedbert Weiss, Ph.D. Professor V I S I T I N G I N V E S T I G AT O R S Cary Lai, Ph.D. Associate Professor Karen T. Britton, M.D., Ph.D. Adjunct Associate Professor Ulo Langel, Ph.D. Adjunct Professor Michael Buchmeier, Ph.D. Adjunct Professor Xiaoying Lu, Ph.D. Assistant Professor Iain L. Campbell, Ph.D. Adjunct Professor Jan O. Lundstrom, Ph.D. Adjunct Professor Zhen Chai, Ph.D. Adjunct Assistant Professor Athina Markou, Ph.D. Adjunct Professor Jerold Chun, M.D., Ph.D. Adjunct Professor Madis Metsis, Ph.D. Adjunct Associate Professor Bruno Conti, Ph.D. Associate Professor Benjamin Neuman, Ph.D. Adjunct Assistant Professor Cindy L. Ehlers, Ph.D. Professor Shirley M. Otis, M.D. Adjunct Professor Ralph Feuer, Ph.D. Adjunct Assistant Professor Tommy Phillips, Ph.D. Adjunct Assistant Professor Howard S. Fox, M.D., Ph.D. Associate Professor John Polich, Ph.D. Associate Professor Brendan Walker, Ph.D. Hermann H. Gram, Ph.D. Adjunct Associate Professor Luigi Pulvirenti, M.D. Adjunct Associate Professor S C I E N C E A S S O C I AT E S S TA F F S C I E N T I S T S Roberto Ciccocioppo, Ph.D. José Criado, Ph.D. Walter Francesconi, Ph.D. David Gilder, M.D. Salvador Huitrón-Reséndiz, Ph.D. M. Cecilia Marcondes, Ph.D. Teresa Reyes, Ph.D. Adjunct Assistant Professor R E S E A R C H A S S O C I AT E S Zhifeng Chen, Ph.D. Jason Botten, Ph.D. Assistant Professor Donna L. Gruol, Ph.D. Associate Professor 327 Remi Martin-Fardon, Ph.D. Tom Nelson, Ph.D. Manuel Sánchez-Alavaz, M.D., Ph.D. Mitra Rebek, Ph.D. Caroline Lanigan, Ph.D. Sam Madamba Hedieh Badie, Ph.D. Genomics Institute of the Novartis Research Foundation San Diego, California Persephone Borrow, Ph.D. Edward Jenner Institute for Vaccine Research Compton, England Urs Christen, Ph.D. La Jolla Institute for Allergy and Immunology La Jolla, California Jean E. Gairin, Ph.D. CNRS Toulouse, France Karine Guillem, Ph.D. University of Pennsylvania Philadelphia, Pennsylvania Katsuro Hagiwara, Ph.D. Rakuno Gakuen University Ebetsu, Japan Dirk Homann, M.D., Ph.D. University of Colorado Health Sciences Center Denver, Colorado 328 MOLECUL AR AND INTEGRATIVE NEUROSCIENCES Shinchi Iwasaki, M.D., Ph.D. Osaka City University Medical School Osaka, Japan Rolf Kiessling, Ph.D. Karolinska Institutet Stockholm, Sweden Denise Naniche, Ph.D., M.P.H. Universitat de Barcelona Barcelona, Spain Noemi Sevilla, Ph.D. Universidad Autonoma de Madrid Madrid, Spain Christina Spiropoulou, Ph.D. Centers for Disease Control and Prevention Atlanta, Georgia Elina Zuniga, Ph.D. University of California San Diego, California * Scripps Florida 2008 THE SCRIPPS RESEARCH INSTITUTE MOLECUL AR AND INTEGRATIVE NEUROSCIENCES Chairman’s Overview I n the past year, we experienced scientific successes as well as organizational and policy changes in the Molecular and Integrative Neurosciences Department. The scientific work of several faculty members resulted in high-significance, high-visibility publications and important new research grants and renewals of earlier grants from the National Institutes of Health. Tamas Bartfai, Ph.D. Particularly noteworthy because of their immediate clinical usefulness are the findings of George Siggins and his collaborators in the Committee on the Neurobiology of Addictive Disorders that the widely used antiepileptic compound gabapentin may be useful in treating alcohol addiction. Pietro Sanna published important findings on the molecular mechanisms of alcohol-induced adaptation of nerve cells. Friedbert Weiss expanded our knowledge of the pharmacologic potential of the subtype-selective antagonists that can block the endogenous anxiogenic stress signal corticotropin-releasing factor. Research by Cindy Ehlers in pharmacogenomics led to new conclusions about the genetic basis of vulnerability of Native Americans to alcohol addiction, and Donna Gruol added new data on the effects of the proinflammatory cytokine IL-6 in the brain. John Polich expanded his noninvasive studies on the human brain by using attentional tasks. Bruno Conti made important findings about the role of the cytokine IL-18 in the regulation of feeding behavior and energy efficiency and through these mechanisms, the control of body weight. He also collaborated with Manuel Sánchez-Alavez and Iustin Tabarean, who uncovered a previously undetected night-eating phenotype in the commonly studied strain of mice that lack the gene for prostanoid receptor 3. These mice may be good models of night bingeing. Xiaoying Lu, Amanda Roberts, and I have 2008 THE SCRIPPS RESEARCH INSTITUTE 329 added to the studies on galanin and galanin receptors in anxiety and in depressive behaviors. The scientists of the department have engaged in many intradepartmental and interdepartmental collaborations. Numerous high-impact invited lectures and seminars were presented by the faculty nationally and internationally. For example, I was the keynote speaker at the largest drug development meeting (12,000 attendees) in Shanghai in June 2007. Despite a difficult economic climate, scientific progress in the department was good, and our educational goals for our graduate students and postdoctoral fellows were all successfully met. Several faculty and students received prestigous stipends. 330 MOLECUL AR AND INTEGRATIVE NEUROSCIENCES Investigator’s Reports Neuroscience Discovery and Pharmacogenomics C. Wahlestedt, M.A. Faghihi, J. Huang, J. Kocerha, A.M. Khalil, S. Brothers, F. Modarresi ur research involves aspects of Alzheimer’s disease, schizophrenia, alcohol addiction, fragile X syndrome, autism, and aging. In addition to drug discovery efforts, we focus on basic aspects of mammalian genomics, genetics, and transcriptomics (RNA research). O 2008 THE SCRIPPS RESEARCH INSTITUTE mals, can be used to elucidate gene functions by rapidly silencing expression of a target gene. Today, siRNAs are widely used and have potential for becoming therapeutic agents. We have built up a powerful and versatile portfolio of siRNA technology. Moreover, we have introduced the use of locked nucleic acids as components of siRNAs (and antisense oligonucleotides) and have shown a range of beneficial properties of these modified agents. G PROTEIN–COUPLED RECEPTORS AS DRUG TA R G E T S F O R C N S D I S O R D E R S I D E N T I F I C AT I O N A N D F U N C T I O N A L A N A LY S I S O F More than half of known drugs bind to G protein– coupled receptors. We have continued our long-standing work on these receptors. Currently, we are focusing on neuropeptide Y and nociceptin receptors. This research involves ultra-high-throughput screening. R E G U L AT O R Y R N A T R A N S C R I P T S HUMAN GENETICS AND PHARMACOGENOMICS We are among the few neuroscientists who have been and continue to be involved in high-throughput sequencing of transcriptomes (i.e., all the RNA transcripts in a cell) of humans and mice. Such efforts have provided strong evidence that in contrast to earlier understanding, in mammalian cells, a majority of the genome is transcribed. Analysis of such data sets has indicated that most mammalian RNA transcripts are noncoding. Thus, conventional protein-coding genes appear to account for only a minority of human RNA transcripts. A substantial component of the full-length mouse and human cDNA sets that we and others have analyzed does not contain an annotated protein- coding sequence and likely corresponds to noncoding RNA. In addition to small RNAs, many of the noncoding RNAs constitute natural antisense RNA transcripts. We have shown that many noncoding RNAs identified to date have substantial conservation across species. We have also shown that many small noncoding RNAs and antisense transcripts have differential expression under various conditions and can affect conventional gene expression. These novel RNA transcripts also likely are affected by a range of disease processes in humans. A fruitful study during the past year has been the investigation of RNA transcripts in the FMR1 locus, which is related to fragile X syndrome and to autism spectrum disorders. We are also involved in several genotyping and genome-wide association studies related to human CNS disorders, including schizophrenia. We wish to understand what makes certain individuals susceptible to disease and how their responses to drug treatment may differ (pharmacogenomics). One of our goals is to identify biomarkers associated with human disorders, including Alzheimer’s disease. RNA INTERFERENCE AND DEVELOPMENT OF HIGH-THROUGHPUT GENOMICS TECHNOLOGY RNA interference has become one of the most important gene manipulation technologies. Short interfering RNA (siRNA), the inducer of RNA interference in mam- PUBLICATIONS Dahlgren, C., Zhang, H.Y., Du, Q., Grahn, M., Norstedt, G., Wahlestedt, C., Liang, Z. Analysis of siRNA specificity on targets with double-nucleotide mismatches. Nucleic Acids Res. 36:e53, 2008. Faghihi, M.A., Modarresi, F., Khalil, A.M., Wood, D.E., Sahagan, B.E., Morgan, T.E., Finch, C.E., St-Laurent, G. III, Kenny, P.J., Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 14:723, 2008. Hong, J., Wei, N., Chalk, A., Wang, J., Song, Y., Yi, F., Qiao, R.P., Sonnhammer, E.L., Wahlestedt, C., Liang, Z., Du, Q. Focusing on RISC assembly in mammalian cells. Biochem. Biophys. Res. Commun. 368:703, 2008. Huang, J., Young, B., Pletcher, M.T., Heilig, M., Wahlestedt, C. Association between the nociceptin receptor gene (OPRL1) single nucleotide polymorphisms and alcohol dependence. Addict. Biol. 13:88, 2008. Kemmer, D., Podowski, R.M., Yusuf, D., Brumm, J., Cheung, W., Wahlestedt, C., Lenhard, B., Wasserman, W.W. Gene characterization index: assessing the depth of gene annotation. PLoS ONE 3:e1440, 2008. Khalil, A.M., Faghihi, M.A., Modarresi, F., Brothers, S.P., Wahlestedt, C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS ONE 3:e1486, 2008. Khalil, A.M., Wahlestedt, C. Epigenetic mechanisms of gene regulation during mammalian spermatogenesis. Epigenetics 3:21, 2008. Scheele, C., Nielsen, A.R., Walden, T.B., Sewell, D.A., Fischer, C.P., Brogan, R.J., Petrovic, N., Larsson, O., Tesch, P.A., Wennmalm, K., Hutchinson, D.S., Cannon, B., Wahlestedt, C., Pedersen, B.K., Timmons, J.A. Altered regulation of the PINK1 locus: a link between type 2 diabetes and neurodegeneration? FASEB J. 21:3653, 2007. St-Laurent, G. III, Wahlestedt, C. Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 30:612, 2007. MOLECULAR THERAPEUTICS 2008 THE SCRIPPS RESEARCH INSTITUTE Molecular Therapeutics Identification of posttranslational modifications on peptides by using high-resolution mass spectrometry and MS3 scanning for absolute assignment of site of modification. Work done in the laboratory of Jennifer Caldwell Busby, Ph.D., assistant professor. 347 Jennifer Caldwell Busby, Ph.D., Assistant Professor, and Kristie Rose, Ph.D., Staff Scientist MOLECULAR THERAPEUTICS 2008 THE SCRIPPS RESEARCH INSTITUTE 349 DEPAR TMENT OF MOLECULAR THERAPEUTICS S TA F F Patrick Griffin, Ph.D. Professor and Chairman Director, Translational Research Institute Jennifer Caldwell-Busby, Ph.D.* Assistant Professor Gregg Fields, Ph.D. Adjunct Professor Philip LoGrasso, Ph.D.* Associate Professor Mathew T. Pletcher, Ph.D.** Assistant Professor S TA F F S C I E N T I S T S Monica Istrate, Ph.D. Lisa Cherry, Ph.D. Brook Miller, Ph.D. Kristie Rose, Ph.D. Jun Zhang, Ph.D. * Joint appointment in the Translational Research Institute SENIOR SCIENTISTS R E S E A R C H A S S O C I AT E S Scott Busby, Ph.D. Brian Ember, Ph.D. Michael Chalmers, Ph.D. Christie Fowler, Ph.D. ** Joint appointments in the Department of Biochemistry and the Translational Research Institute Kevin Hayes, Ph.D. Paul J. Kenny, Ph.D. Assistant Professor Chairman’s Overview he Department of Molecular Therapeutics was established on the Florida campus of Scripps Research in 2007. Faculty in the department use chemical biology approaches to dissect signaling pathways and transcriptional programs. We rely on state-of-theart multidisciplinary technology and methods and a variety of model systems for target identification, validation, and preclinical studies. Currently, the department Patrick R. Griffin, Ph.D. has 5 tenure-track faculty members and several non–tenure track members who oversee key functional cores on the Florida campus. These investigators have created strong research programs that take advantage of the unique high-throughput core facilities at the Florida campus, including genomics, cell-based screening, and proteomics. Research activities include discovery and development of therapeutic agents for unmet medical needs in neurodegeneration, Parkinson’s disease, acute respiratory distress syndrome, spinal cord injury, cardiovascu- T Jonathan Hollander, Ph.D. lar disease, cancer, addiction, and metabolic disorders, including insulin resistance, obesity, and type 2 diabetes. Paul Kenny and his group focus on the neuropharmacology of addiction and on establishing the role of several G protein–coupled receptors in addictive behavior. Phil LoGrasso and members of his laboratory are involved in the discovery of small-molecule therapeutic agents to be used as neuroprotective agents in diseases such as Parkinson’s and are determining the role of rho kinase in vascular bed modulation and glaucoma. Thomas Burris and his group are studying the role of orphan nuclear receptors in circadian rhythms and metabolic disorders such as obesity. Scientists in Jennifer Caldwel Busby’s laboratory use state-of-the-art mass spectrometry to identify, quantify, and characterize proteins and protein modifications to map the signaling pathways related to diabetes and cancer. Peter Hodder and coworkers focus on technology and assay development and novel chemical approaches to expand compound libraries. Michael Cameron and his group are involved in mechanistic studies of P450s and drug biotransformation mechanisms. Researchers in my group are dissecting the mechanism of ligand-dependent activation of orphan nuclear receptors implicated in cancer and metabolic disorders. 350 MOLECULAR THERAPEUTICS 2008 Investigators’ Reports Probing Protein Dynamics With Hydrogen-Deuterium Exchange Mass Spectrometry P.R. Griffin, S.A. Busby, M.J. Chalmers, S.Y. Dai, J. Zhang, M. Istrate, R. Garcia-Ordonez, S. Novick, B. Pascal, J. Conkright, G. Zastrow-Hayes, K. Hayes, T. Schröter, F. Madoux, D. Minond, P.S. Hodder e use a wide range of technologies to study ligand activation of nuclear receptors. During the past few years, we focused on the ligandbinding domains of the well-characterized nuclear receptors peroxisome proliferator–activated receptor γ (PPARγ) and the α and β estrogen receptors. Recently, we have focused on developing hydrogen-deuterium exchange (HDX) technology for probing the mechanism of activation of several orphan nuclear receptors. In addition, in collaboration with scientists at Xencor, Monrovia, California, we are studying the dynamics of TNF-α. W L I G A N D A C T I VAT I O N O F P PA R γ PPARγ is a multidomain ligand-dependent transcription factor. Ligands regulate PPARγ activation by binding to the receptor’s ligand-binding domain, inducing a change in the conformational dynamics of the domain that leads to dissociation of corepressor molecules and formation of suitable neoepitopes for the binding of coactivator molecules. We used structural, biochemical, and cell-based techniques to examine the mechanism of ligand regulation of PPARγ transcriptional activity. We found that the magnitude of PPARγ agonism is regulated by coactivator recruitment selectivity of p160 coactivators. In mutagenesis studies, we determined the key residues on the receptor that facilitate these selective coactivator interactions. In other studies, we are using coactivators as chemical tools to generate desired functional responses and to differentiate pharmacologically beneficial function from adverse function, a novel unexploited therapeutic avenue for treating insulin resistance. Our goals are to determine the structure-activity relationships between PPARγ ligands and their coactivator recruitment selectivity and to obtain PPARγ ligands with specific coactivator preferences by screening for agonists that favor specifically the association of a given cofactor. For a large-scale THE SCRIPPS RESEARCH INSTITUTE high-throughput screening to identify coactivator-selective agonists of the receptor, we have developed a validated time-resolved fluorescence resonance energy transfer assay for ligand-dependent recruitment of the coactivator to PPARγ. Scientists at the Scripps Research Institute Molecular Library Screening Center used these assays to examine the National Institute of Health small-molecule library. The results obtained from this research are providing molecular insight into coactivator recruitment and receptor activation and will result in chemical tools to dissect the biological role of specific coactivators in modulating PPARγ. L I G A N D A C T I VAT I O N O F T H E V I TA M I N D R E C E P T O R In collaboration with scientists at Eli Lilly and Company, Indianapolis, Indiana, we are using HDX to characterize activation of the full-length heterodimer complex composed of the vitamin D receptor and its coreceptor retinoid X receptor α. This project is promoting further development of our HDX platform to facilitate the analysis of large transcriptional complexes. Although this research is in an early stage, we have data that suggest HDX is useful for probing dynamics of large transcriptional complexes. PROBING G PROTEIN–COUPLED RECEPTORS G protein–coupled receptors are an important family of transmembrane signaling proteins. Characterization of the structure and dynamics of these proteins is an analytical challenge because their transmembrane domains are hydrophobic. We have begun to expand the application of HDX to probe the dynamics of these receptors. This work is being done in collaboration with H. Rosen, Department of Chemical Physiology, and R.C. Stevens, Department of Molecular Biology. PUBLICATIONS Bruning, J., Chalmers, M.J., Prasad, S., Busby, S.A., Kamenecka, T., He, Y., Nettles, K.W., Griffin, P.R. Partial agonists activate PPARγ using a helix 12 independent mechanism. Structure 15:1258, 2007. Chalmers, M.J., Busby, S.A., Pascal, B.D., Southern, M.R., Griffin, P.R. A twostage differential hydrogen deuterium exchange method for the rapid characterization of protein/ligand interactions. J. Biomol. Tech. 18:194, 2007. Dai, S.Y., Chalmers, M.J., Bruning, J., Bramlett, K.S., Osborne, H.E., MontroseRafizadeh, C., Barr, R.J., Wang, M., Burris, T.P., Dodge, J.A., Griffin, P.R. Prediction of the tissue-specificity of selective estrogen receptor modulators using a single biochemical method. Proc. Natl. Acad. Sci. U. S. A. 105:7171, 2008. Madoux, F., Li, X., Chase, P., Zastrow, G., Cameron, M.D., Conkright, J.J., Griffin, P.R., Thacher, S., Hodder, P.S. Potent, selective and cell penetrant inhibitors of SF-1 by functional ultra-high-throughput screening. Mol. Pharmacol. 73:1776, 2008. MOLECULAR THERAPEUTICS 2008 Mass Spectrometry for Identification of Proteins THE SCRIPPS RESEARCH INSTITUTE 351 modifications associated with sites of transcription can be used as the basis for further experiments to determine the biological roles of the proteins in gene regulation and activation. J.A. Caldwell Busby, V. Cavett ur general focus is the use of cutting-edge separation and mass spectrometry techniques to identify proteins involved in biological events. The biological applications are determined by the research needs of a large group of collaborators in various disciplines, with a wide variety of questions to be answered. We provide these collaborators access to powerful and novel approaches to examine posttranslational modifications and measure protein levels in multiple samples. In addition to these collaborative efforts, we are developing a method to identify and temporally map chromatin proteins involved in transcriptional regulation. Gene regulation is a fundamental biological process that is studied from a variety of perspectives with a variety of methods; however, research to date has been highly gene centric, and only a few reports have been published on the proteomics of gene regulation. We target these missing proteomics components, particularly the components of the supermolecular complex of chromatin, including nucleosome substructure and regulatory and transcription complexes. Methods for whole-system approaches are difficult to implement because traditional technologies tend to focus on the isolation and analysis of individual parts of the whole—DNA, RNA, or protein. Our techniques combine advances in molecular biology with the power of mass spectrometry to identify novel biomolecules involved in multibiopolymer complexes. In particular, we are modifying and combining techniques such as protein-protein and protein-DNA cross-linking, immunoprecipitation methods, chromatin immunoprecipitation, mass spectrometry, and liquid chromatography to determine the larger regulatory mechanisms involved in the fate of cells. The keystone of this method is a modified chromatin immunoprecipitation protocol that maintains the integrity of the DNA while allowing for the isolation, recovery, and analysis of the protein components of the nucleosome complex. This advanced method targets proteins that regulate chromatin function and correlates those proteins with histone modification states and gene occupancy. Incorporating proteomics into a traditionally DNA-based experimental protocol provides a new perspective and a novel approach to genetic regulation. Newly identified proteins and novel protein O PUBLICATIONS Amelio, A.L., Miraglia, L.J., Conkright, J.J., Mercer, B.A., Batalov, S., Cavett, V., Orth, A.P., Busby, J., Hogenesch, J.B., Conkright, M.D. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 104:20314, 2007. Neurobiology of Addiction P.J. Kenny, P. Bali, C.D. Fowler, J.A. Hollander, H.-I. Im, P.M. Johnson,* Q. Lu, B.H. Miller * Kellogg School of Science and Technology, Scripps Research e focus on understanding the neurobiological mechanisms of addiction. This knowledge will be used to develop novel therapeutic agents for treatment of substance abuse. We use a multidisciplinary approach that includes mouse behavioral genetics, virus-mediated gene expression, RNA and protein analyses, and in vivo behavioral testing. W NICOTINE ADDICTION We seek to identify the subtypes of nicotinic acetylcholine receptors and downstream signaling cascades through which nicotine promotes tobacco addiction. Currently, we are assessing the reinforcing effects of nicotine in mice with null mutations in various subunits of the receptors. In addition, we are testing the effects on nicotine reinforcement of using lentivirusbased short hairpin RNAs to silence the genes of targeted nicotinic acetylcholine receptor subunits in brain reward circuitries. Finally, we are using a proteomics approach to identify the intracellular proteins coupled to nicotinic acetylcholine receptors in the brains of mice. Our goal is to identify novel scaffold and signaling proteins involved in transducing the addictive actions of nicotine. These studies promise to yield significant new insights into the neurobiological mechanisms of nicotine addiction, with direct relevance for the treatment of the tobacco habit in humans. BRAIN SYSTEMS INVOLVED IN ADDICTION In collaboration with other scientists at Scripps Research, we found that the neuropeptide orexin (hypocretin) plays a critical role in drug reward. In ongoing studies, we are identifying the mechanisms through which orexin-mediated transmission regulates drug reward. We are also investigating the roles of 352 MOLECULAR THERAPEUTICS 2008 novel constitutive mechanisms of gene regulation in the neuroplasticity induced by drugs of abuse that may promote addiction. Further, we are testing the hypothesis that drug addiction and obesity share common reward and motivational mechanisms. These studies may identify novel targets for the development of therapeutics against addiction and obesity. DEVELOPMENT OF NOVEL ANTIADDICTION M E D I C AT I O N S In collaborations with scientists in the Translational Research Institute, Scripps Florida, we are developing small-molecule drugs that may be useful as novel therapeutic agents for treatment of substance abuse disorders. The targets for these drugs are G protein–coupled receptors that we previously showed play a role in drug dependence. PUBLICATIONS Faghihi, M.A., Modarresi, F., Khalil, A.M., Wood, D.E., Sahagan, B.G., Morgan, T.E., Finch, C.E., St-Laurent, G. III, Kenny, P.J., Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 14:723, 2008. Johnson, P.M., Hollander, J.A., Kenny, P.J. Decreased brain reward function during nicotine withdrawal in C57BL6 mice: evidence from intracranial self-stimulation (ICSS) studies. Pharmacol. Biochem. Behav. 90:409, 2008. Kenny, P.J., Chartoff, E., Roberto, M., Carlezon, W.A., Jr., Markou, A. NMDA receptors regulate nicotine-enhanced brain reward function and intravenous nicotine self-administration: role of the ventral tegmental area and central nucleus of the amygdala. Neuropsychopharmacology 14:723, 2008. Inhibition of Jun N-Terminal Kinase 2/3 for the Treatment of Parkinson’s Disease P. LoGrasso, M. Cameron, W. Chen, S. Clapp, D. Duckett, B. Ember, J. Habel, R. Jiang, T. Kamenecka, S. Khan, L. Ling, Y.-Y. Ling, M. Lopez, A. Pachori, C. Ruiz, Y. Shin, X. Song, T. Vojkovsky, D. Zadory poptosis, or programmed cell death, plays a vital role in the normal development of the nervous system and is also thought to contribute to the aberrant neuronal cell death that characterizes many neurodegenerative diseases. Therefore, blocking neuronal apoptosis could be an approach for treating neurodegenerative diseases. A major pathway implicated in neuronal cell death and survival is the MAP kinase pathway, which controls cell proliferation and cell death in response to many extracellular stimuli. Recent studies have linked Jun N-terminal kinase (JNK) activity with A THE SCRIPPS RESEARCH INSTITUTE the cell death associated with Parkinson’s disease and Alzheimer’s disease. JNK is linked to many of the hallmark pathophysiologic components of Parkinson’s disease, such as oxidative stress, programmed cell death, and microglial activation. Many pieces of evidence support JNK as a target for treatment of the pathologic changes that underlie Parkinson’s disease. One attractive feature of JNK3 as a selective drug target is that this kinase is almost exclusively expressed in the brain. In contrast, JNK1 and JNK2 are ubiquitously expressed. Despite the ubiquitous expression of JNK2, we are developing a therapy to prevent degeneration of dopaminergic neurons and halt the progression of Parkinson’s disease by targeting JNK2/3. Our strategy for inhibiting JNK2/3 is based on the results of experiments with mice in which the gene for JNK3 or JNK2 was deleted and mice in which the genes for both JNK2 and JNK3 or both JNK1 and JNK2 were deleted. In contrast to mice lacking the gene for JNK1 alone, which had defective T-cell differentiation, mice lacking the gene for JNK2 alone had normal T- and B-cell development and normal T-cell proliferation. Moreover, mice lacking the gene for JNK2 alone and mice lacking the gene for JNK3 alone were protected against the effects of 1-methyl-4-phenyl-1,2,3,5-tetrahydropyridine (MPTP), a compound used to induce parkinsonian signs in animal models of Parkinson’s disease, whereas both wild-type mice and mice lacking the gene for JNK1 were not. In other research, compared with wild-type mice, mice lacking the genes for both JNK2 and JNK3 were dramatically protected against acute MPTP-induced injury of the nigrostriatal pathway. This protective effect resulted in a 3-fold increase in the number of neurons positive for tyrosine hydroxylase, an indication of the increase in survival of dopaminergic neurons. On the basis of these in vitro and in vivo data, we are synthesizing potent, selective JNK 2/3 inhibitors that we will test for efficacy in MPTP animal models of Parkinson’s disease. We have established homogenous time-resolved fluorescence biochemical assays for JNK3 and counterscreens for JNK1 and p38. We have generated more than 1000 compounds from 3 different structural classes; many of the compounds are inhibitory for JNK3 in nanomolar concentrations. Some of the compounds have a cellular potency of 40–60 nM and in vitro efficacy in promoting primary survival of dopaminergic neurons. We have tested com- MOLECULAR THERAPEUTICS 2008 pounds in vivo in rats and mice for drug metabolism and pharmacokinetic properties. Many of the JNK3 inhibitors have had good oral absorption, good brain penetration, and good pharmacokinetic properties that enable efficacy studies. We have also solved the crystal structure of 10 complexes of JNK3 with inhibitor at approximately 2.2-Å resolution. This information is being used in structurebased drug design to help guide medicinal chemistry studies and optimize compounds for potency, selectivity, brain penetration, oral absorption, half-life, clearance, and efficacy. We have also begun investigating the role of JNK in myocardial infarction. We have set up animal models to test for the ability of JNK inhibitors to decrease infarct size and preserve cell function in these models. Finally, we have determined the kinetic mechanism for JNK3 and have shown that it is a random sequential mechanism. We are investigating the kinetic mechanism of JNK1 and are examining differences substrate specificity that may exist between the isoforms. We plan to investigate the role played by different JNK isoforms and, more specifically, different splice variants in various apoptosis scenarios in different cell types. The purposes of these basic mechanistic studies is to understand structure-function relationships at the molecular level and to design specific inhibitors that may be selective for one isoform or splice variant. PUBLICATIONS Ember, B., Kamenecka, T., LoGrasso, P. Kinetic mechanism and inhibitor characterization for c-jun-N-terminal kinase 3α1. Biochemistry 47:3076, 2008. Jiang, R., Duckett, D., Chen, W., Habel, J., Ling, Y.-Y., LoGrasso, P., Kamenecka, T.M. 3,5-Disubstituted quinolines as novel c-Jun N-terminal kinase inhibitors. Bioorg. Med. Chem. Lett. 17:6378, 2007. Schröter, T., Minod, D., Weiser, A., Dao, C., Habel, J., Spicer, T., Chase, P., Baillargeon, P., Scampavia, L., Schürer, S., Chung, C., Mader, C., Southern, M., Tsinoremas, N., LoGrasso, P., Hodder, P. Comparison of miniaturized time-resolved fluorescence energy transfer and enzyme-coupled luciferase high-throughput screening assays to discover inhibitors of Rho-kinase II (ROCK-II). J. Biomol. Screen. 13:17, 2008. THE SCRIPPS RESEARCH INSTITUTE 353 Translational Research Institute Neuronal differentiation is essential for the formation of the mammalian nervous system. Activation of the Rho Kinase (ROCK) pathway by lysophosphatidic acid (LPA) causes neurite retraction. In order to examine the effect of ROCK inhibitors on the prevention of LPAinduced neurite retraction, PC12 cells were allowed to differentiate for 4 days in the presence of nerve growth factor before treatment with ROCK inhibitors and then stimulation with LPA. Cells were fixed and stained for α-tubulin, and nuclei were visualized by using Hoechst 33342 dye. Cells were imaged in a 96well format with the IN Cell 1000 platform. Images were analyzed for neurite length (red) and cell count (green) by using the Developer Toolbox. Work done in the laboratory of Thomas Schröter, Ph.D., senior scientist. Thomas Schröter, Ph.D., Senior Scientist TRANSL ATIONAL RESEARCH INSTITUTE T R A N S L AT I O N A L RESEARCH INSTITUTE S TA F F Patrick Griffin, Ph.D.* Director 2008 William Roush, Ph.D.** Executive Director, Medicinal Chemistry Associate Dean, Kellogg School of Science and Technology THE SCRIPPS RESEARCH INSTITUTE Romain Noel, Ph.D. Sanjay Saldanha, Ph.D. E. Hamp Sessions, Ph.D. Anthony Smith, Ph.D. Xinyi Song, Ph.D. Thomas D. Bannister, Ph.D. Associate Scientific Director, Medicinal Chemistry SENIOR SCIENTISTS Prem Subramaniam, Ph.D. Yenting Chen, Ph.D. Dusica Vidovic, Ph.D. Jennifer Caldwell Busby, Ph.D.* Associate Scientific Director, Proteomics Michael Cameron, Ph.D.* Associate Scientific Director, Drug Metabolism and Pharmacokinetics Derek R. Duckett, Ph.D. Associate Scientific Director, Discovery Biology Rong Jiang, Ph.D. Kristen Clarke Ware, Ph.D. Marcel Koenig, Ph.D. Yan Yin, Ph.D. Jiuxiang Ni, Ph.D. Alok Pachori, Ph.D. Louis Scampavia, Ph.D. Thomas Schröter, Ph.D. Peter Hodder, Ph.D. Scientific Director, Lead Identification Ted Kamenecka, Ph.D. Associate Scientific Director, Medicinal Chemistry Dmitriy Minond Timothy Spicer Youseung Shin, Ph.D. Tomas Vojkovsky, Ph.D. Yangbo Feng, Ph.D. Associate Scientific Director, Medicinal Chemistry S C I E N T I F I C A S S O C I AT E S HTS ROBOTICS ENGINEERS S TA F F S C I E N T I S T S Pierre Baillargeon Lisa Cherry, Ph.D. Peter Chase Juliana Conkright, Ph.D. Lina Deluca Dympna Harmey, Ph.D. Sahba Tabrizifard, Ph.D. I N F O R M AT I C S S TA F F Congxin Liang, Ph.D. Scientific Director, Medicinal Chemistry SENIOR RESEARCH A S S O C I AT E Philip LoGrasso, Ph.D.* Senior Director, Discovery Biology Franck Madoux, Ph.D. Caty Chung Yasel Cruz Kashif Hoda Bruce Pascal Patricia McDonald, Ph.D. Associate Scientific Director, Discovery Biology R E S E A R C H A S S O C I AT E S Becky Mercer, Ph.D. Associate Scientific Project Manager, Lead Identification Melissa Crisp, Ph.D. Mathew T. Pletcher, Ph.D.** Assistant Professor, RNA Core Stephan Schuerer Sarwat Chowdhury, Ph.D. Mark Southern Brian Ember, Ph.D. Xingang Fang, Ph.D. Yuanjun He, Ph.D. Xiaohai Li, Ph.D. * Joint appointment in the Department of Molecular Therapeutics ** Joint appointment in the Department of Chemistry 367 368 TRANSL ATIONAL RESEARCH INSTITUTE 2008 Director’s Overview he Translational Research Institute merges drug discovery efforts at the Scripps Research Florida campus with advanced technology platforms to rapidly identify and validate biological pathways that can be targeted for therapeutic intervention. The goal of the drug discovery operation is to discover and develop small-molecule therapeutic agents for unmet medical needs in neurodegeneration, Parkinson’s disease, acute respiratory distress syndrome, glaucoma, Patrick R. Griffin, Ph.D. spinal cord injury, cancer, and metabolic disorders, including insulin resistance, type 2 diabetes, and obesity, by targeting G protein– coupled receptors, proteases, ion channels, and kinases. The drug discovery component of the Translational Research Institute is fully integrated with the following groups: Lead Identification and High-Throughput Screening, headed by Peter Hodder, Department of Molecular Therapeutics; Medicinal Chemistry, headed by William Roush, Department of Chemistry; Discovery Biology, headed by Phil LoGrasso, Department of Molecular Therapeutics; Drug Metabolism and Pharmacokinetics, headed by Mike Cameron, Department of Molecular Therapeutics; and Informatics, headed by Mark Southern. The Lead Identification team enables drug-target lead identification via ultra-high-throughput screening technology. Using state-of-the-art automation and instrumentation, members in this group are responsible for developing and executing biochemical or cell-based high-throughput screening assays in a miniaturized 1536-well microtiter plate format. In addition to its support of internal Scripps Research objectives, the group participates in the National Institutes of Health Molecular Libraries Probe Production Centers Network (MLPCN), in which qualified assays are screened against the network’s high-throughput screening compound library. Several internal and external investigators have accessed the group’s expertise via collaborative or core-charge mechanisms. The genomics core is headed by Brandon Young. Scientists in this core oversee genotyping and gene expression profiling. The services provided by the core T THE SCRIPPS RESEARCH INSTITUTE allow Scripps Research investigators to examine the genome at both the genetic and the transcriptional level for the genes that underlie common diseases. In collaboration with colleagues on the Florida campus, members of the core have been involved in projects to identify the genes responsible for pathologic conditions, such as addiction and alcoholism, systemic lupus erythematosus, autism, obsessive-compulsive disorder, diabetes, obesity, and prion diseases. The cell-based screening platform is headed by Julie Conkright, Department of Molecular Therapeutics. The faculty advisor to the core is Michael Conkright, Department of Cancer Biology. In this group, high-throughput technologies are used to provide a systematic description of the function of genes encoded by the human genome and a more comprehensive understanding of the genetic basis for human disease. Members of the group provide investigators access to genome-wide collections of cDNAs and short interfering RNAs that can be used to examine cellular models of signal transduction pathways and phenotypes. In addition, the cell-based screening platform participates in one of the center-based initiatives of the Scripps Research MLPCN center. The proteomics platform is headed by Jennifer Caldwell Busby, Department of Molecular Therapeutics. The focus of this core is using liquid chromatography and state-of-the-art mass spectrometry technology to identify, quantify, and characterize proteins and protein modifications. Researchers in the core are involved in scientific collaborations in which novel technologies are used to identify biologically important proteins and protein modifications. Large-scale differential analysis is being used to map the pathways related to insulin sensitization and adipogenesis. In other projects, chromatographic enrichment techniques are used to identify sites of phosphorylation and other posttranslational modifications. Researchers in the proteomics core collaborate with other scientists to create experiments that will provide meaningful mass spectrometric results. TRANSL ATIONAL RESEARCH INSTITUTE 2008 Investigators’ Reports Drug Discovery: Medicinal Chemistry Efforts T.D. Bannister, Y. Feng, T.M. Kamenecka, C. Liang, W.R. Roush, Y. Chen, S. Chowdhury, X. Fang, Y. He, R. Jiang, M. Koenig, R. Noel, E.H. Sessions, Y. Shin, X. Song, T. Vojkovsky, Y. Yin e seek to discover new compounds to treat diseases for which current therapies are inadequate. In our major programs during the past year, we targeted glaucoma, Parkinson’s disease, and breast cancer. In each program, we have attempted to block the action of a specific protein kinase that is overactive or overabundant in affected patients and that hastens the progression of disease. In 2 of the programs, we began by identifying chemical leads from a highthroughput biochemical screen of the Scripps collection of more than 700,000 compounds. The structural information from these screens, in combination with computational and biological analysis of compounds made in other laboratories targeting the same enzymes, provides insights for modifying the structures to obtain unique and patentable leads with the required druglike biochemical, physical, and pharmacologic properties. All of these are evaluated internally by Scripps scientists in the biology, pharmacology, and drug metabolism and pharmacokinetics groups of the Translation Research Institute, who work closely with the medicinal chemists on fully integrated interdisciplinary project teams. W GLAUCOMA We are designing inhibitors of the serine-threonine kinase ROCK, or Rho kinase, which regulates intraocular pressure by controlling the outflow of aqueous humor. Excess ROCK activity is associated with high intraocular pressure, which is a primary risk factor for glaucoma, and with retinal damage. Application of a ROCK inhibitor increases outflow, lowers intraocular pressure, and preserves retinal neurons. Current antiglaucoma drugs have limited efficacy or cause side effects, including discomfort, hyperemia (red eye), and/or undesired changes in cardiovascular function. No glaucoma drugs on the market act by directly altering the Rho kinase pathway, but ROCK inhibitors have strong pressurelowering and neuroprotective effects and thus could be a valuable new treatment. An ideal ROCK inhibi- THE SCRIPPS RESEARCH INSTITUTE 369 tor applied topically to the eye must simultaneously have many properties, including high ROCK affinity, aqueous solubility, excellent corneal permeability, high cellular penetration, and low ocular clearance, to provide a long-lasting effect. Most importantly, the inhibitor must be selective for ROCK over other enzymes and receptors so that no serious side effects occur. We have synthesized thousands of new ROCK inhibitors in multiple chemical classes; many have low nanomolar potency in both biochemical and cell-based assays, high selectivity, and a profile of properties appropriate for preclinical development. For example, SR-3677 was tested in an animal model for glaucoma by our collaborator, V. Rao, Duke University, Durham, North Carolina. The inhibitor lowered intraocular pressure more than 30% within 1 hour, an efficacy comparable to that of antiglaucoma drugs in current use. The reduction in pressure waned after 2 hours, however, so we are designing other compounds intended to have a similarly powerful yet more sustained effect. As expected, the reduction in pressure was due to an increased rate of fluid outflow. We have also made compounds that distinguish between the enzyme isoforms ROCK-I and ROCK-II to test their precise roles. An inhibitor selective for ROCK-II, for example, would lack any unwanted side effects due to ROCK-I inhibition. Such effects are unclear, because no other isoform-selective ROCK inhibitors targeting glaucoma are known. PARKINSON’S DISEASE In collaboration with the National Institute of Neurological Disorders and Stroke, we are developing a therapy to interrupt the loss of dopamine-containing neurons in the midbrain that is a hallmark of Parkinson’s disease. Activation of the transcription factor c-Jun by c-Jun N-terminal kinase (JNK) promotes neurodegeneration. Inhibitors of JNK, which exist in 3 isoforms, JNK1, JNK2, and JNK3, are neuroprotective in animal models of Parkinson’s disease. Our approach, using inhibitors selective for JNK2 and JNK3, would be a quantum leap in the clinical treatment of Parkinson’s disease for several reasons. All current therapies merely treat the symptoms of the disease rather than address the underlying pathologic changes, they tend to lose therapeutic efficacy over time, and they typically elicit undesired side effects. Our challenge is to develop a compound that is a potent, selective, and cell-permeable JNK2/3 inhibitor; has the pharmacokinetic properties for oral dosing (ideally once a day); has good brain penetration; and has a benign toxicology profile. 370 TRANSL ATIONAL RESEARCH INSTITUTE 2008 We have synthesized thousands of new JNK inhibitors in multiple chemical scaffolds and are evaluating compounds with the best combination of properties in several preclinical animal models for Parkinson’s disease. For example, in a pilot study, the Scripps JNK inhibitor SR-3306 delivered systemically to rodents via osmotic minipump at 10 mg/kg reduced CNS-mediated behaviors that occur after a chemically induced brain lesion used to mimic the parkinsonian condition. Newer generation compounds, including SR-3562, will soon be evaluated in animal models and are particularly promising because of improved properties, including high oral bioavailability (45%), high cell-based potency (0.06 µM), and excellent distribution to the brain of rodents after oral dosing. CANCER In collaboration with Poniard Pharmaceuticals, South San Francisco, California, we have synthesized many potent and selective novel inhibitors of focal adhesion kinase (FAK). FAK inhibitors could be an important new means of treating solid tumors, including breast cancer. FAK has been implicated in promoting detachment of tumor cells and metastasis, characteristics of almost all advanced-stage solid tumors that are responsible for most of the suffering and death related to cancer. By blocking FAK and thus stopping the first step of metastasis, the detachment of cancer cells from their primary site, we hope to halt this process and thereby interrupt progression of the disease. We have recently completed our FAK chemistry efforts after identifying highly potent and selective FAK inhibitors, including SR2516. This lead compound is effective in in vitro tumor metastasis models, is efficacious in animal models of tumor progression, has desirable pharmaceutical properties suitable for convenient once-a-day oral dosing, and is being licensed for further development. FUTURE DIRECTIONS We are continuing our research on glaucoma and Parkinson’s disease and have smaller or exploratory efforts in other areas, including methods for treating diabetes, for curbing drug addiction, and for targeting cancer progression by other mechanisms. We hope to expand these efforts. Many of the compounds identified in the ROCK and JNK inhibitor programs are also likely to be useful in the treatment of other diseases. For example, animal data suggest that ROCK inhibitors might be an effective treatment for multiple sclerosis. Strong preclinical evidence shows that JNK inhibitors, in addition to treating Parkinson’s disease, also may THE SCRIPPS RESEARCH INSTITUTE prevent neuronal damage in a host of other disorders including stroke, Alzheimer’s disease, and amyotrophic lateral sclerosis. We anticipate that in each of our research programs we can continue to synthesize novel compounds with the right combination of properties that would permit development of the compounds as safe and effective agents for stopping the progression of important diseases. PUBLICATIONS Chen, Y.T., Bannister, T.D., Weiser, A., Griffin, E., Lin, L., Ruiz, C., Cameron, M.D., Schürer, S., Duckett, D., Schröter, T., Lograsso, P., Feng, Y. Chroman-3amides as potent Rho kinase inhibitors. Bioorg. Med. Chem. Lett., in press. Feng, Y., Cameron, M.D., Frackowiak, B., Griffin, E., Lin, L., Ruiz, C., Schröter, T., LoGrasso, P. Structure-activity relationships and drug metabolism and pharmacokinetic properties for indazole piperazine and indazole piperidine inhibitors of ROCK-II. Bioorg. Med. Chem. Lett. 17:2355, 2007. Feng, Y., Yin, Y., Weiser, A., Griffin, E., Cameron, M.D., Lin, L., Ruiz, C., Schürer, S.C., Inoue, T., Rao, P.E., Schröter, T., LoGrasso, P. Discovery of substituted 4(pyrazol-4-yl)-phenylbenzodioxane-2-carboxamides as potent and highly selective Rho kinase (ROCK-II) inhibitors. J. Med. Chem. 51:6642, 2008. Jiang, R., Duckett, D., Chen, W., Habel, J., Ling, Y.Y., LoGrasso, P., Kamenecka, T.M. 3,5-Disubstituted quinolines as novel c-Jun N-terminal kinase inhibitors. Bioorg. Med. Chem. Lett. 17:6378, 2007. LoGrasso, P., Kamenecka, T. Inhibitors of c-jun-N-Terminal Kinase (JNK). Mini Rev. Med. Chem. 8:755, 2008. Sessions, E.H., Yan, Y., Bannister, T.D., Pocas, J., Cameron, M.D., Ruiz, C., Schürer, S.C., Schröter, T., LoGrasso, P., Feng, Y. Benzimidazole- and benzoxazole-based inhibitors of Rho kinase. Bioorg. Med. Chem. Lett., in press. Proteomics Laboratory J.A. Caldwell Busby, V. Cavett he Proteomics Laboratory at Scripps Florida provides proteomics services and expertise to scientific collaborators at Scripps Research facilities in both Florida and California, universities within the state of Florida, and other educational institutions. We use cutting-edge mass spectrometry technology to identify proteins, map modifications that occur after translation, and do relative quantitation experiments with a variety of sample types. In its lifetime, a protein can have several locations and functions within a cell. Location, function, and 3-dimensional structures of proteins are all influenced by static and dynamic chemical modifications that occur after translation. These modifications vary from small methyl and acetyl groups, which are part of the histone codes, to large lipid and glycosylation modifications, which act as cellular markers and signaling molecules. With mass spectrometry, we can detect both the small T TRANSL ATIONAL RESEARCH INSTITUTE 2008 and the large changes in mass that occur in proteins because of these modifications, and we can identify the specific amino acids modified. Relative changes in protein levels or level of posttranslational modification between multiple samples provide biologically relevant information about cellular pathways and proteins of interest. Large-scale studies of this type require rigorous sample preparation and highly tuned algorithms for comparing different mass spectrometric analyses. We are currently validating methods for both sample fractionation and data analysis for these types of large-scale differential protein experiments. Mass spectrometers at the facility include an ion trap spectrometer, which is used mostly to identify proteins and peptides, and a triple quadrupole mass spectrometer, which is used for relative quantitation experiments. A new addition is a mass spectrometer that can be used to perform accurate mass and high-resolution experiments. Each mass spectrometer is interfaced to nano-flow electrospray ionization sources and capillary high-performance liquid chromatography columns. Data analysis is performed primarily via automated workflow on a cluster maintained by the bioinformatics group. Automation of the front-end processing allows a more thorough review of the data and more time for development of innovative software in collaboration with information technology groups at Scripps Research and beyond. Drug Metabolism and Pharmacokinetics Laboratory M.D. Cameron, L. Lin, C. Ruiz, S. Khan, Z. Li he Drug Metabolism and Pharmacokinetics Laboratory at Scripps Florida provides in vitro and in vivo evaluation of the pharmacokinetic and pharmacodynamic properties of new chemical entities. We work on project teams within the drug discovery group of the Department of Molecular Therapeutics and support chemistry efforts within the Scripps Research Institute Molecular Screening Center. We help bridge medicinal chemistry and pharmacology by evaluating the metabolic fate and identifying the liabilities of compounds. Pharmacokinetic studies provide basic parameters, including peak plasma concentration, bioavailability, exposure, half-life, clearance, volume of distribution, and tissue distribution. Research interests T THE SCRIPPS RESEARCH INSTITUTE 371 include P450 structure-function relationships and the formation of reactive intermediates during metabolism. The laboratory is equipped with a liquid chromatography–tandem mass spectrometry system and a Q-trap hybrid triple quadrupole/linear ion-trap mass spectrometer. PUBLICATIONS Cameron, M.D., Wen, B., Roberts, A.G., Atkins, W.M., Campbell, A.P., Nelson, S.D. Cooperative binding of acetaminophen and caffeine within the P450 3A4 active site. Chem. Res. Toxicol. 20:1434, 2007. Cameron, M.D., Wright, J., Black, C.B., Ye, N. In vitro prediction and in vivo verification of enantioselective human tofisopam metabolite profiles. Drug Metab. Dispos. 35:1894, 2007. Madoux, F., Li, X., Chase, P., Zastrow, G., Cameron, M.D., Conkright. J.J., Griffin, P.R., Thacher, S., Hodder, P. Potent, selective and cell penetrant inhibitors of SF-1 by functional ultra-high-throughput screening. Mol. Pharmacol. 73:1776, 2008. Miller, B.H., Schultz, L.E., Gulati, A., Cameron, M.D., Pletcher, M.T. Genetic regulation of behavioral and neuronal responses to fluoxetine. Neuropsychopharmacology 33:1312, 2008. Cell-Based Screening Core J.J. Conkright, G. Zastrow, J. Cartzendafner, M. Morris he Cell-Based Screening Core provides highthroughput screening of functional genomic platforms and consults with researchers from Scripps, both in California and Florida; universities in Florida; and other outside academic institutions to perform these screens. We curate 2 large libraries: the Mammalian Genome Collection cDNA library and the Qiagen Druggable siRNA library. Screening these libraries allows investigators to determine if overexpression of a single gene (Mammalian Genome Collection cDNA library) or reduction in expression levels of a single gene (Qiagen siRNA library) positively or negatively influences their particular biological readout. These libraries provide investigators a unique tool to identify novel factors and pathways involved in biological systems. The findings can lead to new areas of research and novel targets for drug development. In addition to our large libraries, we have 2 small libraries that we built: a transcription factor library and a nuclear receptor library. These libraries are important new tools for investigators who study the effects of proteins and signaling pathways on gene expression. These libraries are also a mechanism for studies of the specificity of new potential drugs and chemical probes that modulate gene expression. A third area of expertise we provide is the generation of mutagenesis screens. Determining the regions T 372 TRANSL ATIONAL RESEARCH INSTITUTE 2008 or residues in a protein that are important for its biological function can be a key component in dissecting how the protein interacts with other factors. Chemical mutagenesis of a gene permits an unbiased approach to identifying these biologically critical residues of the protein. We perform the mutagenesis and provide screening sets for investigators to examine the effect the mutation has on a chosen biological end point. Last, we counsel researchers on how to validate their screens and counterscreen the rank-order hits. These tasks are extremely important to prove the statistical significance of a finding and to ascertain the specificity of the finding for that precise biological function or pathway. PUBLICATIONS Amelio, A.L., Miraglia, L.J., Conkright, J.J., Mercer, B.A., Batalov, S., Cavett, V., Orth, A.P., Busby, J., Hogenesch, J.B., Conkright, M.D. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 104:20314, 2007. Madoux, F., Li, X., Chase, P., Zastrow, G., Cameron, M.D., Conkright, J.J., Griffin, P.R., Thacher, S., Hodder, P. Potent, selective and cell penetrant inhibitors of SF-1 by functional ultra-high-throughput screening. Mol. Pharmacol. 73:1776, 2008. Discovery Biology: Kinases THE SCRIPPS RESEARCH INSTITUTE United States. The prognosis for these patients is poor, and treatment options are limited. We will focus on defining the role Jun N-terminal kinase signaling plays in tumor maintenance and cell dispersal and whether inhibition of this kinase has therapeutic potential in this devastating disease. In addition, we are involved in the Scripps-Pfizer collaboration that was started in 2007. Since then, several assays have been designed for high-throughput screening of targets of therapeutic interest to Pfizer. PUBLICATIONS Jiang, R., Duckett, D., Chen, W., Habel, J., Ling, Y.Y., LoGrasso, P., Kamenecka, T.M. 3,5-Disubstituted quinolines as novel c-Jun N-terminal kinase inhibitors. Bioorg. Med. Chem. Lett. 17:6378, 2007. Lansing, T.J., McConnell, R.T., Duckett, D.R., Sephar, G.R., Knick, V.B., Hassler, D.F., Noro, N., Furuta, M., Emmitte, K.A., Gilmer, T.M., Mook, R.A., Jr., Cheung, M. In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol. Cancer Ther. 6:450, 2007. Rech, J.C., Yato, M., Duckett, D., Ember, B., LoGrasso, P.V., Bergman, R.G., Ellman, J.R. Synthesis of potent bicyclic bisarylimidazole c-jun N-terminal kinase inhibitors by cyclic C-H bond activation. J. Am. Chem. Soc. 129:490, 2007. Rhodes, N., Heerding, D.A., Duckett, D.R., Eberwein, D., Knick, V.B., Lansing, T.J., McConnell, R.J., Gilmer, T.M., Zhang, S.Y., Robell, K., Kahana, J., Geske, R.S., Kleymenova, E.V., Choudhry, A.E., Lai, Z., Leber, J.D., Minthorn, E.A., Strum, S.L., Wood, E.R., Huang, P.S., Copeland, R.A., Kumar, R. Characterization of an Akt kinase inhibitor with potent pharmacodynamic and antitumor activity. Cancer Res. 68:2366, 2008. D.R. Duckett, J. Anderson, W. Chen, D. Harmey, Y.Y. Ling e are investigating the use of small-molecule kinase inhibitors of biologic interest and therapeutic potential. Protein kinases are important components of signal transduction pathways, and deregulation of kinase activity in humans can lead to disease. Kinases have become one of the most important target classes for drug development. We are optimizing a novel class of kinase inhibitors for treatment of Parkinson’s disease. Although the cause of Parkinson’s disease is unknown, a strong correlation exists between loss of primary dopaminergic neurons within the substantia nigra and progression to the diseased state. Our current goal is to develop an inhibitor of the Jun N-terminal family of kinases; our aim is to protect the primary dopaminergic neurons from cell death, thus slowing or halting the progression of the disease. Working closely with scientists in other disciplines necessary for lead optimization (chemistry, pharmacology, and drug metabolism), we were successful in securing funding from the National Institute of Neurological Disorders and Stroke for this research. We are also investigating the role of MAP kinases in primary brain cancers. In 2008, brain tumors will be diagnosed in approximately 20,000 patients in the W Probe and Drug Discovery: The Lead Identification Department P. Hodder, A. Abovich, P. Baillargeon, P. Chase, M. Crisp, L. DeLuca, R. Einsteder, K. Emery, F. Madoux, B. Mercer, D. Minond, M. Petrillo, A. Porto, S. Saldanha, L. Scampavia, M. Spaargaren, T. Spicer, V. Fernandez-Vega he Lead Identification Department is responsible for developing and executing high-throughput screening (HTS) assays and for supporting downstream medicinal chemistry and related “hit-to-lead” efforts (Fig. 1). The anchors of the department are 2 fully automated robotic platforms. One supports screening of 384- and 1536-well microtiter plates in a variety of biochemical and cell-based assay formats. The other is used to manage and distribute the more than 600,000 compounds used for drug discovery at Scripps Research and 300,000 compounds for the Molecular Libraries Probe Production Centers Network. The facility also contains an assay development laboratory, equipped with bacterial culture, protein purification, compound characterization, and tissue culture laboratories as well as semi-automated equipment for liquid handling and T TRANSL ATIONAL RESEARCH INSTITUTE 2008 THE SCRIPPS RESEARCH INSTITUTE 373 F i g . 1 . The uHTS laboratory of the Lead Identification Department houses equipment and instrumentation necessary to develop and support a uHTS campaign and medicinal chemistry follow-up efforts. The anchor of the department is a fully automated uHTS platform (center right), which is used to screen libraries of compounds for biological activity in a variety of pharmacologically relevant assays, including cell-based, protein, RNA, and DNA targets. Flanking the uHTS platform are an assay development laboratory (left and center) containing equipment and instrumentation necessary to develop an HTS assay and a mammalian tissue culture suite (upper right). Behind the uHTS platform (not shown) is a fully automated compound management platform capable of storing, retrieving, and aliquoting desirable compounds from the screening file and a liquid chromatography–mass spectrometry platform (bottom right) used to perform routine compound quality assurance/quality control. Not shown are fully equipped protein expression/purification and microbiology laboratories. detection. Supporting this operation is an integrated laboratory information management system, which tracks HTS assay data and compound usage and quality. Additionally, we are involved in developing metallo-β-lactamase class B1 chemical probes. related collaborations (Table 1) and have contributed to the discovery more than 10 novel leads (chemical probes) of G protein–coupled receptors, metalloproteinases, nuclear receptors/transcription factors, and kinases (http://molscreen.florida.scripps.edu/). THE SCRIPPS RESEARCH INSTITUTE MOLECULAR DISCOVERY AND DEVELOPMENT OF CLASS B SCREENING CENTER M E TA L L O - β- L A C TA M A S E I N H I B I T O R S Established in July 2005, the Scripps Research Institute Molecular Screening Center is a national resource for small-molecule screening and the development of chemical probes. It is 1 of 9 members in the Molecular Libraries Probe Production Centers Network, a translational research initiative sponsored by National Institutes of Health (NIH) and part of the NIH Roadmap. The mission of the Scripps center is to screen the NIH library of more than 300,000 individual compounds against peer-reviewed targets; the goal is to discover proof-of-concept probes. The results are available to the scientific community through the PubChem Web site of the National Center for Biotechnology Information: http://pubchem.ncbi.nlm.nih.gov. Currently, the Lead Identification department serves as the HTS core within the Scripps screening center; our responsibilities are to develop biological and biochemical assays, perform HTS campaigns, manage the resulting data, act as steward of the NIH screening library, and provide assay support for the development of probes. The diversity of bacterial β-lactamases continues to outpace the development of useful β-lactam–based antibiotics. Although the development of class B β-lactamase inhibitors has been an active area of past research, an array of potent, class-specific small-molecule inhibitors has yet to be fully characterized in the clinically relevant VIM-2 metallo-β-lactamase system. Additionally, VIM2 inhibitors that are effective inhibitors of other class B β-lactamases will be of great interest. Such compounds will be useful as tools for characterizing gram-negative pathogens or as adjuvant in antibiotic therapy. One of our goals is to develop HTS-ready assays suitable for rapid identification of compounds that modulate the activity of Ambler molecular class B (Bush-JakobyMedeiros group 3) metallo-β-lactamases, specifically VIM-2 and IMP-1 enzymes. In preliminary research efforts, we have developed HTS-ready fluorescence- and absorbance-based VIM-2 and IMP-1 inhibition assays. In collaboration with K.B. Sharpless, Department of Chemistry, we have screened a diverse click-chemistry library or compounds designed specifically to inhibit metallo-βlactamases. Currently we are developing several novel scaffolds that appear to be specific inhibitors. OTHER SCREENING ACTIVITIES Since the inauguration of the ultra-HTS (uHTS) operation in November of 2005, we have also been actively screening the Scripps collection of compounds against drug discovery targets not only from the MLPCN but also from scientists at Scripps Research and from outside partners. So far, members of the department have initiated and successfully completed more that 50 uHTS- PUBLICATIONS Chung, C.C., Ohwaki, K., Schneeweis, J.E., Stec, E., Varnerin, J.P., Goudreau, P.N., Chang, A., Cassaday, J., Yang, L., Yamakawa, T., Kornienko, O., Hodder, P., Inglese, J., Ferrer, M., Strulovici, B., Kusunoki, J., Tota, M.R., Takagi, T. A fluorescence-based thiol quantification assay for ultra-high-throughput screening for inhibitors of coenzyme a production. Assay Drug Dev. Technol. 6:361, 2008. 374 TRANSL ATIONAL RESEARCH INSTITUTE 2008 THE SCRIPPS RESEARCH INSTITUTE T a b l e 1 . Summary of collaborations in the development of HTS and HTS assays. Target class Antibacterial ATPase Target name (Abbreviation) β-Lactamase VIM-2 Pseudomonas aeruginosa p97 IMP-1 5HT1a 5HT1e GalR2 GPR7 S1P1 S1P2 G protein–coupled receptor S1P3 GLP-1 GPR119 m Opioid heterodimers NPY- Y1 NPY- Y2 RBBP9 b-gluc Aquaporins (AQP) Ion channel TRPML3 TRPN1 JAK2 JNK3 Kinase P. Hodder, Scripps Research, Jupiter, Florida M. Teitler, Albany Medical College, Albany, New York S. Brown, Scripps Research, La Jolla, California O. Civelli, University of California, Irvine, California H. Rosen, Scripps Research, La Jolla, California S1P4 AGTRL-1 (APJ) Hydrolase Collaborator, affiliation R. Miller, Pfizer, Groton, Connecticut R. Deshaies, California Institute of Technology, Pasadena, California ROCK2 PKA FAK L. Smith, Burnham Institute for Medical Research, Orlando, Florida P. LoGrasso, Scripps Research, Jupiter, Florida P. McDonald. Scripps Research, Jupiter, Florida L. Devi, Mt. Sinai School of Medicine, New York, New York C. Wahlestedt, Scripps Research, Jupiter, Florida B. Cravatt, Scripps Research, La Jolla, California J. Kelly, Scripps Research, La Jolla, California M. Yeager, Scripps Research, La Jolla, California S. Heller, Stanford University, Stanford, California R. Levine, G. Gilliland, Sloan Kettering, New York, New York P. LoGrasso, Scripps Research, Jupiter, Florida T. Schröter, Scripps Research, Jupiter, Florida P. Hodder, Scripps Research, Jupiter, Florida ADAMTS4 MMP13 Metalloproteinase Falciparum M18 metalloprotease IDE NADPH oxidase Nox-1 SHP-1 Estrogen receptor Nuclear receptor RAR SF1 (NR5A1) RORa (NR1F1) Phosphotransferase Proliferation/viability TPT1 Jurkat E6.1 cells EphB4-ephrinB2 Protein/protein HCV core homodimer NS5B/CYPB Hsp70 Protein misfolding AL-09 PERK Protein/RNA Reductase Stem cell proliferation G. Fields, Florida Atlantic University, Boca Raton, Florida MMP8 HIV Rev-RRE RNA msrA Notch D. Gardiner, Queensland Institute of Medical Research, Queensland, Australia M. Leissring, Mayo Clinic, Jacksonville, Florida G. Bokoch, Scripps Research, La Jolla, California P. Griffin, Scripps Research, Jupiter, Florida K. Nettles, Scripps Research, Jupiter, Florida P. Griffin, Scripps Research, Jupiter, Florida X. Li, Orphagen Pharmaceuticals, San Diego, California H. Harding, New York University, New York, New York P. Hodder, Scripps Research, Jupiter, Florida P. Kuhn, Scripps Research, La Jolla, California D. Strosberg, Scripps Research, Jupiter, Florida R. Morimoto, Northwestern University, Chicago, Illinois M. Ramirez-Alvarado, Mayo Clinic, Rochester, Minnesota D. Ron, New York University, New York, New York J. Williamson, Scripps Research, La Jolla, California H. Weissbach, Florida Atlantic University, Boca Raton, Florida H. Petrie, Scripps Research, Jupiter, Florida PPARg/Src1 PPARg/Src2 P. Griffin, Scripps Research, Jupiter, Florida PPARg/Src3 Transcription factor NF-kB STAT1 STAT3 KLF5 AHR Ubiquitin proteolysis WEE1 J. Reed, Burnham Institute for Medical Research, La Jolla, California D. Frank, Dana-Farber Cancer Institute, Boston, Massachusetts V. Yang, Emory University, Atlanta, Georgia M. Denison, University of California, Davis, California N. Ayad, Scripps Research, Jupiter, Florida TRANSL ATIONAL RESEARCH INSTITUTE 2008 Lauer-Fields, J.L., Minond, D., Chase, P.S., Baillargeon, P.E., Saldanha, S.A., Stawikowska R., Hodder, P., Fields, G.B. High throughput screening of potentially selective MMP-13 exosite inhibitors utilizing a triple-helical FRET substrate. Bioorg. Med. Chem. Lett., in press. Lauer-Fields, J.L., Spicer, T.P., Chase, P.S., Cudic, M., Burstein, G.D., Nagase, H., Hodder, P., Fields, G.B. Screening of potential a disintegrin and metalloproteinase with thrombospondin motifs-4 inhibitors using a collagen model fluorescence resonance energy transfer substrate. Anal. Biochem. 373:43, 2008. Madoux, F., Li, X., Chase, P., Zastrow, G., Cameron, M.D., Conkright, J.J., Griffin, P.R., Thacher, S., Hodder, P. Potent, selective and cell penetrant inhibitors of SF-1 by functional ultra-high-throughput screening. Mol. Pharmacol. 73:1776, 2008. Roth, J., Madoux, F., Hodder, P., Roush, W.R. Synthesis of small molecule inhibitors of the orphan nuclear receptor steroidogenic factor-1 (NR5A1) based on isoquinolinone scaffolds. Bioorg. Med. Chem. Lett. 18:2628, 2008. Schröter, T., Minond, D., Weiser, A., Dao, C., Habel, J., Spicer, T., Chase, P., Baillargeon, P., Scampavia, L., Schürer, S., Chung, C., Mader, C., Southern, M., Tsinoremas, N., LoGrasso, P., Hodder, P. Comparison of miniaturized time-resolved fluorescence resonance energy transfer and enzyme-coupled luciferase high-throughput screening assays to discover inhibitors of Rho-kinase II (ROCK-II). J. Biomol. Screen. 13:17, 2008. Schürer, S.C., Brown, S.J., Gonzales-Cabrera P.J., Schaeffer, M.T., Chapman, J., Jo, E., Chase, P., Spicer, T., Hodder, P., Rosen, H. Ligand-binding pocket shape differences between sphingosine 1-phosphate (S1P) receptors S1P1 and S1P3 determine efficiency of chemical probe identification by ultrahigh-throughput screening. ACS Chem. Biol. 3:486, 2008. Discovery Biology: G Protein–Coupled Receptors P. McDonald, D. Obradovich, A. Smith, E. Sturchler, S. Tabrizifard protein–coupled receptors (GPCRs) are the largest and most versatile family of cell-surface receptors. The ubiquitous cell-surface distribution and involvement of these proteins in almost all biological processes explain why the largest percentage of currently marketed therapeutic drugs target these receptors. We focus on developing biochemical and cell-based functional assays to monitor GPCR activity that involve high-throughput and high-content technologies. Using a multidisciplinary approach that involves collaborations with disciplines such as lead identification, chemistry, drug metabolism and pharmacokinetics, and in vivo pharmacology, we aim to identify and develop small-molecule modulators of GPCRs for the treatment of metabolic diseases such as type 2 diabetes mellitus and obesity. We have developed a series of novel cell-based assays for the glucagon-like peptide 1 receptor (GLP-1R) to promote a drug discovery program for this clinically validated target of type 2 diabetes. In parallel with the GLP-1R assays, we have developed similar assays for 2 other closely related receptors, GLP-2R and glucagon receptor, which serve as counterscreens for selectivity against GLP-1R. We are also working on an orphan G THE SCRIPPS RESEARCH INSTITUTE 375 GPCR that has been implicated in type 2 diabetes and obesity and that signals and functions in an analogous manner to GLP-1R. In collaboration with P. Kenny, Molecular Therapeutics, we are also developing smallmolecule inhibitors of a GPCR previously shown to be involved in drug dependence that may lead to a novel therapy for substance abuse. As part of the collaboration between Scripps and Pfizer, Inc., that was initiated in 2007, we are designing and developing 3–4 assays per year for GPCR targets of therapeutic interest to Pfizer. In Vivo Pharmacology A.S. Pachori, M. Ganno, S. Khan, S. Clapp, D. Hansen he In Vivo Pharmacology group at Scripps Florida is an integrated group of investigators involved in preclinical studies in support of drug discovery efforts at Scripps Research in both Florida and California. We develop appropriate animal models of diseases for ongoing projects such as studies in hypertension, glaucoma, Parkinson’s disease, diabetes, and heart failure. These models are then used to test the efficacy of new compounds for a particular therapeutic area. An important aspect of establishing the efficacy of a compound is to determine if the compound is altering its intended target. We initially evaluate the role of a particular target in primary cell culture and then evaluate the target in vivo. The cell culture experiments are also used to screen novel compounds for the effects of the compounds on targets. In addition, we evaluate the pharmacodynamic properties of new chemical entities in vivo by doing dose-response and time-course studies to determine the effects of the compounds on the intended target. For these evaluations, we use standard techniques such as Western blotting, enzyme-linked immunosorbent assays and immunohistochemistry. We also collaborate with the drug metabolism and pharmacokinetics laboratory to monitor plasma and tissue concentrations of chemical entities; the results help us further refine and develop the disease models. Finally, we evaluate the compounds for toxicity. In the past year, we have successfully developed animal models of hypertension to test the efficacy of Rho kinase inhibitors as novel antihypertensive agents. We have screened several novel Rho kinase inhibitors for their efficacy and toxicity and established the need for isoform-selective inhibitors to avoid toxicity issues. T 376 TRANSL ATIONAL RESEARCH INSTITUTE 2008 We have also established the use of primary dopaminergic neurons in an in vitro target modulation assay to screen compounds for treatment of Parkinson’s disease. In addition, we have adopted a strategy to explore and seek alternative therapeutic uses for novel compounds currently under development. For example, we have expanded the use for novel Jun N-terminal kinase inhibitors for Parkinson’s disease to include their use as cytoprotective agents against heart failure induced by ischemia-reperfusion injury. Our preliminary data indicate that in rodents, these inhibitors can successfully reduce tissue damage in a dose-dependent manner. We are also exploring the use of Rho kinase inhibitors, which were originally developed as antihypertensive agents, in the treatment of glaucoma. This strategy will help us not only expand our portfolio but also discover new approaches for some of the biggest unmet needs of patients. Omics Informatics B.D. Pascal he Omics Informatics group at Scripps Florida addresses support and software needs of various laboratories. Our goals are to identify and integrate existing solutions where possible and to build new solutions only when necessary. A primary specialization is the analysis and management of mass spectrometric data and development of proteomics software research tools that enable proteomics researchers to validate, visualize, and share their data. T HD DESKTOP Scientists at Scripps Florida are using hydrogendeuterium exchange mass spectrometry to characterize protein dynamics and protein-protein or protein-ligand interactions. The Deuterator software, released last year, addresses some of the data analysis bottlenecks by providing a platform to automate and visualize centroid calculations. Despite these efforts, the task of assembling and visualizing the resulting data is still a manual operation left to the end user. The current software, HD Desktop, leverages the existing code base and stores all data in a relational database. Novel rendering and analysis tools have been presented in an integrated user interface (Fig. 1). M A S S S P E C T R O M E T R Y C A L I B R AT I O N S O F T WA R E Although every mass spectrometry laboratory will vary in instrument manufacturer and experimental designs, most proteomics laboratories have a common THE SCRIPPS RESEARCH INSTITUTE F i g . 1 . HD Desktop experiment view. need to measure and validate the calibration of the instruments. An automated method to process and display the mass calibration shift at various time points allows the maximization of instrument run time and contributes to the standardization and validation of proteomics data. We have developed an automated method for determining calibrated mass drift on high-resolution instruments. On a daily basis, the quality of the data collection is monitored by routine analysis of a tryptic digest of a β-casein standard. The resulting binary files are collected, moved into the laboratory information management system, and then processed through an automated workflow, which conducts file conversions and peak quality assessments and sends the files to a compute cluster for peptide identification search. The results are then parsed, and the difference between the observed and calculated mass is stored in a database; the data are made available through a Web-based interface (Fig. 2, page 377). PUBLICATIONS Chalmers, M.J., Busby, S.A., Pascal, B.D., Southern, M.R., Griffin, P.R. A twostage differential hydrogen deuterium exchange method for the rapid characterization of protein/ligand interactions. J. Biomol. Tech. 18:194, 2007. Pascal, B.D., Chalmers, M.J., Busby, S.A., Mader, C.C., Southern, M.R., Tsinoremas, N.F., Griffin, P.R. The Deuterator: software for the determination of backbone amide deuterium levels from H/D exchange MS data. BMC Bioinformatics 8:156, 2007. Drug Discovery Biology: Cell Biology T. Schröter, A.M.W. Handy, E. Griffin, J.R. Pocas, K. Clarke, C. Hahmann W ith more than 500 members, protein kinases are important drug discovery targets for a wide variety of therapeutic indications. These TRANSL ATIONAL RESEARCH INSTITUTE 2008 THE SCRIPPS RESEARCH INSTITUTE 377 For the cancer program, in 2007, we successfully finished a collaboration with scientists at Poniard Pharmaceuticals, Inc., South San Francisco, California, to discover novel inhibitors of focal adhesion kinase. This kinase has been implicated in tumor cell detachment and metastasis. We supported the program by developing biochemical and cell-based assays to monitor the effect of newly discovered small molecules on biochemical inhibition of focal adhesion kinase and on cellular growth, migration, and invasion. We are also collaborating with researchers at Pfizer, Inc., in developing biochemical and cell-based high-throughput screening assays for a diverse set of novel disease targets, including protease inhibitors, hydrolases, and membrane transporters. PUBLICATIONS Feng, Y., Cameron, M.D., Frackowiak, B., Griffin, E., Lin, L., Ruiz, C., Schröter, T., LoGrasso, P. Structure-activity relationships and drug metabolism and pharmacokinetic properties for indazole piperazine and indazole piperidine inhibitors of ROCK-II. Bioorg. Med. Chem. Lett. 17:2355, 2007. F i g . 2 . Data flow of mass spectrometry calibration software. kinases control signal transduction pathways, and in humans, deregulation of their activity can lead to diseases such as glaucoma and cancer. The serine-threonine kinase Rho kinase (ROCK) regulates intraocular pressure by controlling the outflow of aqueous humor. In glaucoma, increased intraocular pressure leads to loss of retinal ganglion cells and, ultimately, loss of vision. Inhibition of ROCK activity increases outflow, lowers intraocular pressure, and preserves retinal neurons. We concentrate on developing biochemical and cell-based functional assays to monitor ROCK activity via both highcontent and high-throughput screening technologies. Working closely with researchers in high-throughput screening, medicinal chemistry, pharmacology, and drug metabolism and pharmacokinetics, we identified small-molecule lead compounds from a high-throughput screening of the Scripps collection of more than 700,000 compounds. During lead optimization, we screened thousands of new ROCK inhibitors for the biochemical activity against the enzyme and the close family members protein kinase A, Akt1, and MRCKα. Hundreds of these compounds were chosen, and their cell-based activity against ROCK was tested by using target modulation and functional assays. Changes in myosin light-chain phosphorylation were measured by using a 96-well immunocytochemical assay and infrared imaging, and changes in the formation of stress fibers and neurite protection were evaluated by using a highcontent imaging system. Schröter, T., Minond, D., Weiser, A., Dao, C., Habel, J., Spicer, T., Chase, P., Baillargeon, P., Scampavia, L., Schürer, S., Chung, C., Mader, C., Southern, M., Tsinoremas, N., Lograsso, P., Hodder, P. Comparison of miniaturized time-resolved fluorescence resonance energy transfer and enzyme-coupled luciferase highthroughput screening assays to discover inhibitors of Rho-kinase II (ROCK-II). J. Biomol. Screen. 13:17, 2008. Chemical Informatics Program S.C. Schürer, D. Vidović, C. Chung e collaborate with scientists in the Scripps Research Institute Molecular Screening Center, and we also received a grant from the Columbia University Molecular Library Screening Center. Both of these centers are part of the national Molecular Libraries Screening Centers Network. We are also involved in drug discovery efforts within the Translational Research Institute. We have developed a platform of industry-standard software tools for analysis, visualization, hypothesis building, and modeling of large and focused experimental screening data sets. Our platform enables us to generate and evaluate a large variety of structural, pharmacophore, and physicochemical 2- and 3-dimensional descriptors. The platform includes computational chemistry tools for 3-dimensional pharmacophore-alignment ligand-based quantitative structure-activity relationships, ligand-protein docking (in a variety of approaches and scoring functions), homology modeling, molecular modeling and dynamics, and statistical tools. We have also developed various interactive reporting and visualization protocols that are used in collaborative research. Our platform provides broad chemical informat- W 378 TRANSL ATIONAL RESEARCH INSTITUTE 2008 ics and computational chemistry capabilities. Examples of research projects in which these technologies play a key role include analysis of data on toxic effects in cells and animals; development of small-molecule modulators of a broad variety of targets, including metalloproteases, phosphatases, kinases, nuclear receptors, and sphingosine lipid receptors (for which we also modeled the receptor structures); and image-based high-content assays, including probing the inflammatory pathway at the stage of NF-κB translocation and expression of E-selectin or vascular adhesion molecule 1 and targeting the aggregation of the protein huntingtin. In collaboration with the screening informatics team, we play a key role in implementing work flow, procedures, and business rules and in integrating discovery informatics with the operational informatics infrastructure to facilitate discovery processes. Examples include the Scripps Research compound registration system; integration of the Molecular Libraries Screening Centers Network chemoinformatics server, which is hosted at Scripps Research, with PubChem; integration of images from image-based screening with the operational infrastructure; and publication of screening data to PubChem. To date, more than 5.5 million data points and more than 120 assays obtained by using these protocols have been published. Other chemoinformatics research efforts are focused on structure-based comprehensive analyses of target-similarity relationships in the phosphatase and kinase gene families, ligand-based and target “fishing,” and the development of integrative applied chemoinformatics methods. PUBLICATIONS Guha, R., Schürer, S.C. Utilizing high throughput screening data for predictive toxicology models: protocols and application to MLSCN assays. J. Comput. Aided Mol. Des. 22:367, 2008. Schröter, T., Minond, D., Weiser, A., Dao, C., Habel, J., Spicer, T., Chase, P., Baillargeon, P., Scampavia, L., Schürer, S., Chung, C., Mader, C., Southern, M., Tsinoremas, N., Lograsso, P., Hodder, P. Comparison of miniaturized time-resolved fluorescence resonance energy transfer and enzyme-coupled luciferase highthroughput screening assays to discover inhibitors of Rho-kinase II (ROCK-II). J. Biomol. Screen. 13:17, 2008. Xie, Y., Deng, S., Thomas, C.J., Liu, Y., Zhang, Y.Q., Rinderspacher, A., Huang, W., Gong, G., Wyler, M., Cayanis, E., Aulner, N., Többen, U., Chung, C., Pampou, S., Southall, N., Vidović, D., Schürer, S., Branden, L., Davis, R.E., Staudt, L.M., Inglese, J., Austin, C.P., Landry, D.W., Smith, D.H., Auld, D.S. Identification of N-(quinolin-8-yl)benzenesulfonamides as agents capable of down-regulating NFκB activity within two separate high-throughput screens of NFκB activation. Bioorg. Med. Chem. Lett. 18:329, 2008. Xie, Y., Liu, Y., Gong, G., Rinderspacher, A., Deng, S.X., Smith, D.H., Többen, U., Tzilianos, E., Branden, L., Vidović, D., Chung, C., Schürer, S., Tautz, L., Landry, D.W. Discovery of a novel submicromolar inhibitor of the lymphoid specific tyrosine phosphatase. Bioorg. Med. Chem. Lett. 18:2840, 2008. THE SCRIPPS RESEARCH INSTITUTE Screening Informatics Program M.R. Southern, K. Hoda, Y. Cruz cientists in the Screening Informatics Program at Scripps Florida collaborate broadly with researchers in the Scripps Research Institute Molecular Screening Center, part of the national Molecular Libraries Screening Centers Network, and in drug discovery efforts within the Translational Research Institute. Our responsibilities include high- and lowthroughput screening assays and downstream drug metabolism and pharmacology, medicinal chemistry, and probe development. We have an operational environment for data management and quality assurance and a knowledge environment that facilitates efficient optimization of probes. These activities take place at both the Florida and the California sites. The software systems have been built primarily by using the MDL Discovery Experiment Management Framework from Symyx Technologies, Inc., Santa Clara, California, and support specific work flows involving tasks such as chemical compound registration, plate and sample registration, assay development, and entire screening campaigns. On top of these systems, we have developed in-house software that is tightly coupled to provide additional functionality and to improve our efficiency. Examples include robotic automation, plate mapping operations, and structure search. A key component is our Assay Exploration Data Warehouse, which along with its Web-based front end is known to end users as ChemInfo. ChemInfo is assay metric and structure centric, enabling exploration of assay data by compound, target, or assays. It integrates chemical descriptors, physical properties, and data on drug metabolism and pharmacokinetics to facilitate probe optimization. Complicated Venn-like queries are possible. ChemInfo contains individual and aggregated assay data from our internal assays as well as from PubChem. The database has up to 30 users within Scripps Research. S PUBLICATIONS Schröter, T., Minond, D., Weiser, A., Dao, C., Habel, J., Spicer, T., Chase, P., Baillargeon, P., Scampavia, L., Schürer, S., Chung, C., Mader, C., Southern, M., Tsinoremas, N., Lograsso, P., Hodder, P. Comparison of miniaturized time-resolved fluorescence resonance energy transfer and enzyme-coupled luciferase high-throughput screening assays to discover inhibitors of Rho-kinase II (ROCK-II). J. Biomol. Screen. 13:17, 2008.