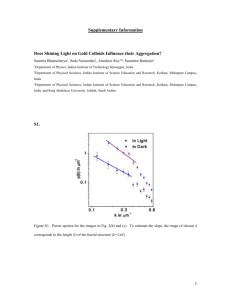
Document 12617765
advertisement
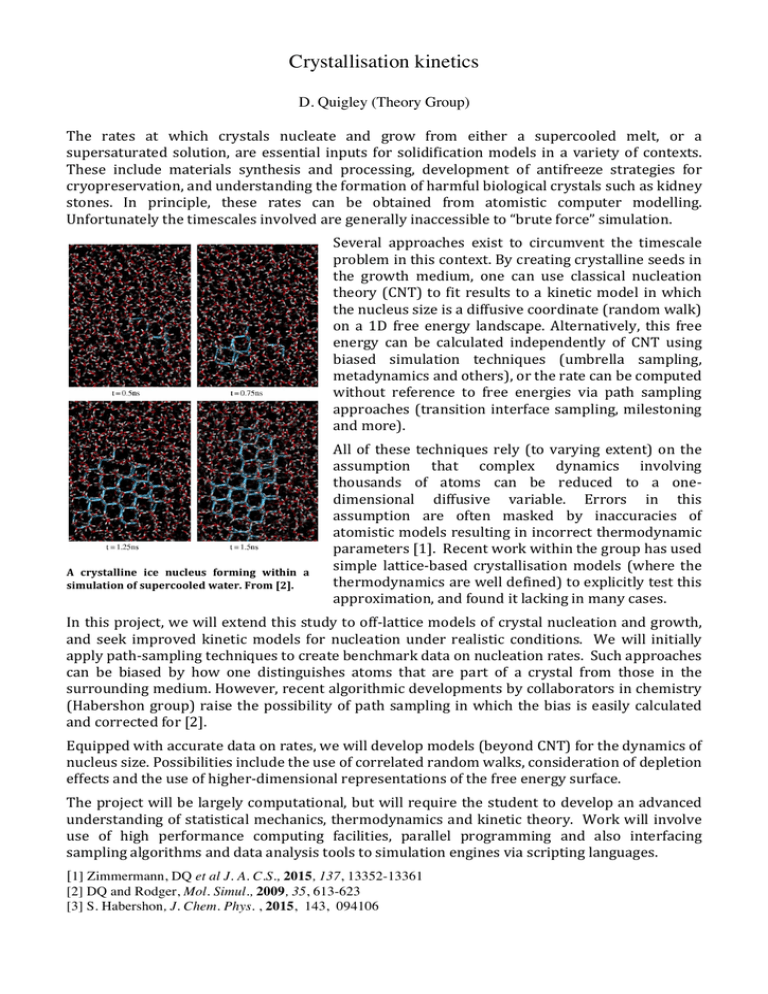
Nsolid – Neq lde et al. together al orders ugh this ledge. n for xc disorder n easily multiple herefore gest that e greatly contrast, eation of biasing we have of liquid an those ow the ccording we plot mber of he first is 2496 d order energy. thin the crease in e largest multiple 200 Crystallisation kinetics 0 0 0.5 1 D. Quigley (Theory Group) Figure 3. Total number of solid water molecules, and number The rates at which crystals nucleate and grow from either a supercooled melt, or a within the largest cluster as a function of time minus the induction time (t ) to the first nucleation event in a metadynamics i supersaturated solution, are essential inputs for solidification models in a variety of contexts. simulation of 2496 TIP4P water molecules freezing at 180 K. These materials synthesis and processing, development of antifreeze strategies for Each value isinclude plotted minus the average equilibrium (unbiased) background value up to the formation of a critical cluster. The cryopreservation, and understanding the formation of harmful biological crystals such as kidney total number of molecules in the simulation is 2496. stones. In principle, these rates can be obtained from atomistic computer modelling. Unfortunately the timescales involved are generally inaccessible to “brute force” simulation. t–ti (ns) Several approaches exist to circumvent the timescale problem in this context. By creating crystalline seeds in the growth medium, one can use classical nucleation theory (CNT) to fit results to a kinetic model in which the nucleus size is a diffusive coordinate (random walk) on a 1D free energy landscape. Alternatively, this free energy can be calculated independently of CNT using biased simulation techniques (umbrella sampling, metadynamics and others), or the rate can be computed without reference to free energies via path sampling approaches (transition interface sampling, milestoning and more). 0.12 a critical fluid at Figure 4. Growth of a critical nucleus in a metadynamics simulation of 2496 ice TIP4P water molecules freezing at 180 A crystalline nucleus forming within a K. Hydrogen bonds molecules identified solid are simulation of connecting supercooled water. From as [2]. highlighted. Solid molecules are identified by the same method used in [7]. Times correspond to the x-axis in figure 3. All of these techniques rely (to varying extent) on the assumption that complex dynamics involving thousands of atoms can be reduced to a one-­‐ dimensional diffusive variable. Errors in this assumption are often masked by inaccuracies of atomistic models resulting in incorrect thermodynamic parameters [1]. Recent work within the group has used simple lattice-­‐based crystallisation models (where the thermodynamics are well defined) to explicitly test this approximation, and found it lacking in many cases. In this project, we will extend this study to off-­‐lattice models of crystal nucleation and growth, and seek improved kinetic models for nucleation under realistic conditions. We will initially apply path-­‐sampling techniques to create benchmark data on nucleation rates. Such approaches can be biased by how one distinguishes atoms that are part of a crystal from those in the surrounding medium. However, recent algorithmic developments by collaborators in chemistry (Habershon group) raise the possibility of path sampling in which the bias is easily calculated and corrected for [2]. Equipped with accurate data on rates, we will develop models (beyond CNT) for the dynamics of nucleus size. Possibilities include the use of correlated random walks, consideration of depletion effects and the use of higher-­‐dimensional representations of the free energy surface. The project will be largely computational, but will require the student to develop an advanced understanding of statistical mechanics, thermodynamics and kinetic theory. Work will involve use of high performance computing facilities, parallel programming and also interfacing sampling algorithms and data analysis tools to simulation engines via scripting languages. [1] Zimmermann, DQ et al J. A. C.S., 2015, 137, 13352-13361 [2] DQ and Rodger, Mol. Simul., 2009, 35, 613-623 [3] S. Habershon, J. Chem. Phys. , 2015, 143, 094106