Exploiting Next Generation Sequencing to investigate the genetics of
advertisement

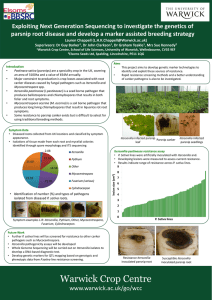
Exploiting Next Generation Sequencing to investigate the genetics of parsnip root disease and develop a marker assisted breeding strategy Lauren 1 Chappell 1 Barker , 1 Clarkson , 1 Teakle (L.H.K.Chappell@Warwick.ac.uk) Supervisors: Dr Guy Dr John Dr Graham & Mrs Sue 1Warwick Crop Centre, School of Life Sciences, University of Warwick, Wellesboune, Warwick CV35 9EF 2Elsoms Seeds Ltd, Spalding, Lincolnshire, PE11 1QG 2 Kennedy Introduction Pastinaca sativa (parsnips) are a speciality crop within the UK, covering an area of 3100 hectares with a value of £64M annually. Currently the major constraint to production is up to 80% losses associated with root canker diseases caused by fungal pathogens such as Itersonilia and Mycocentrospora spp. Some resistance to parsnip canker exists but is difficult to select for using traditional phenotype screening methods. This project will use new technologies to identify and exploit these sources of resistance in addition to developing a variety of methodologies to further understand the epidemiology of the pathogens. To develop tools to facilitate breeding for quantitative resistance to parsnip root canker diseases caused by Itersonilia and Mycocentrospora spp. Objectives 1. 2. 3. 4. 5. To determine the range of fungal pathogens causing root cankers on parsnip To obtain Itersonilia pastinacae isolates from different sources and locations and characterise them using molecular and biological approaches To develop screening tests to identify I. pastinacae resistance in parsnip breeding lines To determine the pathogenicity of different I. pastinacae isolates To develop genetic markers linked to resistance through QTL mapping and Next Generation Sequence analysis. Symptom Data Root Assay A Parsnip root assay was develop to determine disease resistance within a P. sativa population. Breeding and Parent lines from Elsoms parsnip breeding program were tested. Pot grown roots were inoculated with an agar plug of Itersonilia or Mycocentrospora and assessed for disease. Final Lesion Size (cm2) Aims Whole genome sequencing of I. pastinacae isolates. Root and seedling screening for pathogen resistance. QTL mapping from genotypic and phenotypic data from Elsoms Breeding Lines. 2.08 12.50 6.0 25.00 5.0 14.58 4.0 Itersonilia Pythium Other Mycocentrospora 22.92 3.0 2.0 22.92 Fusarium (various) Cylindrocarpon 1.0 Figure 2: Identification of number (%) and types of pathogens isolated from diseased roots harvested from various locations around the UK 0.0 Future Work Diseased roots obtained from locations around the UK including a disease tunnel and field trial (Elsoms). Symptoms were photographed and classified with subsequent isolations of tissue made from each root, and plated onto agar. Plates were incubated at 20°C for 10 days; once mycelial growth was identified colonies were subcultured. Provisional identification was through colony and spore morphology with confirmation through ITS sequencing. Parsnip Parent Line Figure 1: Final lesion size for 10 Parsnip Parent line roots inoculated with I. pastinacae. Showing resistance and susceptibility within the population. Figure 3: Examples of symptoms L-R Itersonilia, Pythium, Other, Mycocentrospora, Fusarium, Cylindrocarpon