Document 12559212
advertisement

Identiation of horizontally aquired DNA
using genome signature analysis
1
1
2
Stephen A. Jarvis , Jason S. Mirsky , John F. Peden
and Nigel J. Saunders
3
y
1
Programming Researh Group, Oxford University Computing Laboratory ,
University of Oxford, Oxford OX1 3QD, UK.
2
3
MRC Moleular Haematology Group , Moleular Infetious Diseases Group ,
Institute of Moleular Mediine,
University of Oxford, Oxford OX3 9DS, UK.
September 1, 2000
Abstrat
Introdution
Identiation of regions with untypial
perentage G+C omposition and dinuleotide signatures are two genome analysis tehniques used in the
identiation of horizontally aquired DNA. We desribe a generi program framework for performing
both types of analysis in linear time. The approah
is extended for length > 2 oligonuleotide signatures.
Using the derived program we test some of the onlusions of Karlin and Burge, the primary exponents
of these tehniques.
We demonstrate that no single method of
signature analysis is suÆient for the omplete identiation of horizontally aquired DNA. Consequently
we produe a fast program - and a robust methodology - for the prodution and employment genome signatures in the identiation of horizontally aquired
DNA.
Software available from rst author.
DNA; genome; signature; algorithms.
In addition to the vertial transmission of adaptive
mutations, horizontal transfer of genes between unrelated speies is inreasingly reognised as an important omponent of evolution [Doolittle, 2000℄. Most
if not all speies inlude material whih has been aquired in this way. The sope for this type of geneti
exhange is limited in higher organisms in whih the
germ ells are sequestered and are not normally exposed to foreign DNA. However, in bateria, in whih
the ells that form the origins of new populations are
exposed to DNA in the environment, horizontal exhange of DNA is an ongoing and important proess
[Arber, 1993, Lawrene et al., 1998, Maddox, 1998,
Woese, 1998, Brown and Doolittle, 1999, Doolittle,
1999, Forterre and Philippe, 1999, Jain et al., 1999,
Martin, 1999, Nelson et al., 1999, Faguy and Doolittle, 2000, Moreire, 2000℄. For example, it has been
estimated that 18% of the genes in E. oli have been
aquired by horizontal exhange sine its divergene
from Salmonella spp.approximately 100 million years
ago [Lawrene et al., 1998℄.
Horizontal exhange is reognised as being important in the evolution of baterial virulene, the
most widely aknowledged examples of whih are
so-alled `pathogeniity islands'. The typial har-
Motivation:
Results:
Availability:
Keywords:
now
versity
at:
of
Department
Warwik,
of
Coventry
stephen.jarvisds.warwik.a.uk.
Computer
CV4
7AL,
Siene,
UK.
UniEmail:
To whom orrespondene
should be addressed
y urrent
address:
Sir William Dunn Shool of Pathology,
University of Oxford, Oxford. OX1 3RE
1
ateristis of these regions of DNA are: a %G+C
omposition whih diers from that of the rest of
the genome; terminal inverted repeats; proximity to
tRNA genes [Hou, 1999℄ and assoiation with transposases whih may be intat or present as remnants. These islands often ontain groups of genes
with funtions whih an readily be invoked in making a speies more virulent, suh as toxin prodution and seretion [Groisman and Ohman, 1996,
Haker et al., 1997, Alfano et al., 2000℄. Usually the
origin of the transferred DNA is not known and
the soure of these genes - so signiant in virulene - remains an interesting enigma. In a
small number of instanes, however, the likely origin of horizontally transferred genes an be inferred with some ertainty [Vasquez et al., 1995,
Kroll et al., 1998, Saunders et al., 1999℄.
In order to beome established within a baterial
population, an aquired gene (or other gene with
whih it is assoiated) must lead to an inrease in baterial tness. This takes plae through one or more of
the following fators: inreasing transmissibility, inreasing the size of the olonising population, establishing new nihes in whih the organism an grow,
or evading host immune responses. This inrease in
tness must then be suÆient for the sequene to
beome prevalent in the population to whih it has
been transferred. This ours either through lonal
replaement of the previous population that did not
have the gene or by further horizontal transfer of the
gene within the new speies to whih it has been
transferred. The proesses whih might be expeted
to be involved in inreased tness are broadly similar
to those involved in virulene. It is therefore important to be able to identify sequenes whih have
been aquired by horizontal transfer in order to understand the evolutionary history of a speies and, in
the ontext of baterial pathogens, to identify genes
potentially assoiated with virulene.
DNA from dierent geneti bakgrounds (e.g., baterial speies) has dierent harateristis. These are
a produt of the odons whih are preferentially used
by the partiular speies and the environmental onditions in whih it grows, as well as the types of error to whih the DNA repliation enzymes are prone
and dierenes in the ability of eah enzyme to or-
ret mistakes. This makes it possible to identify setions of DNA whih are derived from dierent geneti bakgrounds by means of their untypial base
omposition. DNA whih is inorporated following
horizontal transfer will be exposed to the same pressures whih gave the reipient its harateristi omposition. As a result, over time the aquired DNA
will begin to onform to the average DNA omposition of the reipient by a proess alled amelioration [Lawrene and Ohman, 1997℄. However, the
rate of amelioration is suÆiently slow that it is possible to identify regions of DNA whih have been horizontally aquired. The extent to whih this sequene
is untypial therefore depends on both the extent of
the dierene between the donor and reipient speies
and the period of time whih has elapsed sine aquisition.
The rst method for the identiation of regions of
potentially horizontally transferred DNA is based on
%G+C harateristis of the sequene. This is useful beause the %G+C within prokaryoti genomes
is relatively homogeneous within a speies but differs between unrelated speies [Karlin et al., 1998℄.
Signiantly dierent %G+C ontent in setions of
DNA therefore indiates possible horizontal aquisition [Karlin et al., 1998℄.
Experimental studies showed that the dinuleotide odds ratios were similar in most organisms for the bulk of their DNA and essentially the same for dierent parts of the same
genome [Josse et al., 1961℄. Looking at sequenes derived from dierent sequene bakgrounds revealed
that more losely related speies tended to have more
similar dinuleotide ompositions than unrelated
speies [Karlin et al., 1994.a, Karlin et al., 1994.b,
Karlin et al., 1994.℄.
Karlin and his olleagues noted that the speies
dierenes in base usage were more omplex and
that, in addition to dierenes in %G+C omposition, dierent speies used harateristi patterns of
dinuleotide pairs within their genomes. This observation provided the basis for the seond approah
in whih the DNA is seen not as a string of single bases but as a sequene of pairs of dinuleotides:
the sequene ACGTG, for example, would be onsidered as a sequene of dinuleotide pairs AC, CG,
2
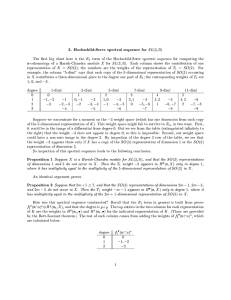
Number of steps Estimated exeution
GT and TG. The perentage of eah of the 16 din(bounded by O) time
uleotide pairs was found to dier between speies
O(log 2 n)
0:1 seonds
and although the reasons for these dierenes are not
p
O( n)
5 seonds
fully understood, this provides a seond method for
identifying horizontally aquired DNA. The method
O(n)
1 hour 15 minutes
is known as dinuleotide signature (DNS) analyO(n2 )
450 years
sis [Karlin and Burge, 1995℄.
In this paper:
Figure 1: Length of time n omputations take when
= 5; 000; 000 and a single omputation takes 1 mil Karlin's approah to %G+C ontent and DNS nliseond.
Table adapted from [Kaldewaij, 1990℄.
analysis is examined and extensions to his methods are proposed.
alulation. The hromosomes of ba A generi program for the linear omputation of mathematial
teria are typially omposed of 0.5 to 5 million base
n-oligonuleotide signatures is developed.
pairs (Mb). Performing alulations on the DNA
(of length n) requires a large amount of
Karlin and Burge's observation that n- sequene
omputation:
rst the omplete sequene statistis
oligonuleotide signatures are highly implied
are
obtained,
O(n); then individual segments (winby the DNS is onrmed. However, it is also
and ompared with
demonstrated that if dierent window sizes and dows) are statistially analysed
the
omplete
sequene,
O(n2 ). A straightforward reword lengths are used, n-oligonuleotide signaan implementation
tures point to dierent, potentially relevant, nement would therefore produe
whih
was
bounded
by
O(n2 ) steps.
areas from the DNS graphs.
Figure 1 shows the relationship between asymptoti
analysis and omputation time.
A omparison is made of the dierent methods
Optimisations
to programs an of ourse be made
for identifying horizontally aquired DNA; onand
omputers
work
at a far greater rate than the
lusions are drawn and supporting results, whih
example
suggests.
However,
the table demonstrates
desribe how the methods an be used, are prethat
a
dierene
in
the
order
of
omplexity of an algosented.
rithm is diretly proportional to the runtime. Reduing an algorithm from an O(n2 ) solution to a O(n)
solution will make a onsiderable dierene to the
number (and variety) of results whih an feasibly be
obtained.
Systems and methods
Computational analysis
Asymptoti analysis is a way of omparing the performane of dierent omputational algorithms. This
omparison is used to selet one omputational approah, whih may provide a more eÆient solution,
over another.
The big-O notation desribes the upper bound of
the asymptoti growth of a funtion. For instane, if
an algorithm takes 31 n2 2n + 4 steps, it is of O(n2 ),
sine it is bounded from above by a funtion whih
takes n2 steps times some onstant [Kaldewaij, 1990℄.
Karlin's methods for nding horizontally aquired
DNA rely on omparing DNA with itself by means of
Methods of genome analysis
%G+C ontent
Calulating the %G+C ontent of an organism's
DNA is a simple method for nding regions of geneti interest. %G+C ontent sores are alulated
for segments (windows) of DNA by adding the frequeny (number divided by window length) of G to
the frequeny of C. A plot of %G+C ontent is derived by sliding the window over the omplete genome
sequene and plotting the results.
3
Dinuleotide signatures
nally the sequene is ompletely overed by windows. Graphing the results shows the regions of
Dinuleotide signatures an be used to show areas largest dierene, whih are andidates for horizontal
where pairs of bases are lustered more frequently aquisition.
than if they were distributed aross the sequene by
hane. A DNS graph is based on a alulation of
odds ratios for eah of the 16 dinuleotides. This
odds ratio (probability) is alulated by taking the
frequeny of a dinuleotide and dividing it by the ex- Just as greater information and resolution is obtained
peted probability of nding the dinuleotide within when dinuleotide sequene omposition is onsidered rather than single base omposition, analyza partiular sequene:
ing sequenes on the basis of even longer motifs
may provide additional useful information. Trinufxy
pxy =
(1) leotide omposition addresses odon usage in exfx fy
pressed portions of open reading frames. However, it
fx is the frequeny of nuleotide x within the se- may be that preferenes for partiular pairs or more
quene and fxy is the frequeny of the dinuleotide of odons, mutational proesses whih favour or sexy within the (irular) sequene of DNA:
let against longer sequenes, or proesses whih affet the omposition of the non-transribed strand
#xy
will
inuene the sequene omposition in additional
fxy =
(2)
#dinuleotides
ways. The method has therefore been extended so
that signatures an be determined for longer mowhere #dinuleotides = SequeneLength 1
tifs
[Karlin et al., 1997, Mirsky, 1999℄.
pxy represents the odds ratio for dinuleotides
Dinuleotide analysis is easily extendible to
in single-stranded DNA. To alulate the odds ra
oligonuleotide
signatures of length n.
tio for double-stranded DNA, written pxy , the sequene and its reverse omplement are onate4l
1
nated [Burge et al., 1992℄. The alulation remains
j p i (f ) p i (g) j
(4)
Æ (f; g ) = ( l )
the same, but with a sequene twie the length of the
4 i :s
original.
To nd areas of possible horizontally aquired where l is the length of oligonuleotides whih omDNA, a window (f ) (of some hosen size { 50 Kb for prise the signature, s is the set of all permutations of
example [Karlin and Burge, 1995℄) is reated. The length l and i is one suh permutation.
In addition, the variane v and standard deviation
dinuleotide probabilities of this window are then
ompared with the dinuleotide probabilities of the sd an be determined for the absolute dierenes of
overall sequene (g ). This is done for eah of the 16 the p values at eah point and for the Æ values aross
dinuleotides and the normalised results are added. the entire sequene.
The result is an overall dinuleotide dierene ben
x2 ( x)2
tween the window f and the sequene g .
v=
(5)
2
Oligonuleotide signatures
X
P
1 X p
j
Æ (f; g ) = ( )
16
16
xy
xy (f )
P
n
pxy (g )
j
(3)
To alulate the variane for Æ , eah Æ value or
responds to a x and the number of Æ values (the sequene length) orresponds to n. To alulate the
variane of the p values, x = p and n = 4l . The
standard deviation
p is simply the sum of the variane
values, sd = v .
Using the same mehanis as those used in %G+C
ontent signatures, Karlin and Burge suggest repeatedly sliding the window one position along the sequene and repeating the dierene alulation until
4
When the entire population is not known, that is
if the window slides by more than one base between
alulations, an estimate of the variane is alulated:
v
=
n
P x2
n(n
P
( x)2
1)
A.
Sequence
Movement of sliding window
(6)
Windows wrap round
before termination because
of circularity of bacterial DNA
Window 1 2 3
Implementation
B.
Window
The same proedure is used to implement %G+C
ontent, dinuleotide and n-oligonuleotide signatures. The sliding window implementation (or similar) is mehanially equivalent for eah; the primary
dierene between eah signature is simply the formula used to alulate the points.
A generi implementation is explored.
%G+C ontent requires the smallest speiation
and is therefore most appropriate to illustrate the
derivation of the program. An O(n2 ) solution is derived and, by a method known as loop attening, this
is onverted into an O(n) solution.
The O(n) implementation provides the basis for a
omparative study of these methods for the identiation of horizontally transferred DNA.
ws
0 <= ws < WS
C.
Sequence
Old window
C
A
New window
A
Nucleotide leaving window
If ( n = C or n = G )
GC(window’) = GC(window) - 1
T
G
Nucleotide entering window
If ( n = C or n = G )
GC(window’) = GC(window) + 1
Figure 2: Derivation of a generi DNA signature algorithm: A. Generate a window for every position
in the sequene by sliding the window through the
sequene, the outer loop; B. Calulate signatures for
eah window, the inner loop; C. Flatten the inner
loop for O(N ) solution.
Generi genome signatures
The preondition states that the sequene must be
The speiation of a program for %G+C ontent
of
at least length 1 (for %G+C ontent this is approanalysis (GCprog ) is presented in the guarded ompriate;
for larger oligonuleotides this minimum must
mand language [Kaldewaij, 1990℄.
be the length of the oligonuleotide).
P = fN 1g
The frame ontains the information relevant to the
[ on N: int fN 1g; A: array[0..N) of DNA;
development of the program: the onstant N is the
on WS: int f1 WS Ng;
length of the DNA sequene A; W S is the window
var n: int;
size; n is a variable used for traversing the sequene
GCprog
and GCprog is simply a marker for the program itself.
℄.
Q = fr = 8p : 0 p < N : 8q : 0 q < W S :
The postondition desribes the result r, a list of
g = (#i : p i (p + q ) :
%G+C ontent dierenes, one for eah of the winA:(i mod N ) = C
dows in the sequene. The modulo (mod) is ontained
_ A:(i mod N ) = G)=W S )g
within the speiation to aount for the irularity
of the baterial DNA. It is also noted that the results
The speiation ontains three parts: the preon- list r is written to a le as the program proeeds dition P; the part in the frame, denoted by square this saves on RAM.
brakets; and the post ondition Q, found after the
The program derivation ontains three stages:
frame.
5
Derivation: stage 1
window and adding the items whih enter the window
eah time the window slides. See Figure 2-C.
To alulate the new inner loop we now split the
loop into two parts: the rst alulates the ontent
for the rst window, O(N ) one only; the seond
alulates the remaining N 1 windows, onstant
2 O(N ) = O(N ). As we are adding terms rather
than multiplying them, the new algorithm is O(N ).
To alulate a window from every position in the sequene, the window slides through the sequene (f.
Karlin). This outer loop is determined by replaing
the onstant N by the variable n and using the postondition Qfn = N g. This suggests starting n at 0
and setting the loop guard to n 6= N . This oers a
solution to the outer loop of O(N ). See Figure 2-A.
Computing other signature types
Derivation: stage 2
The derived algorithm is generi in the sense
that it provides the neessary O(N ) framework for
whihever type of signature one is trying to program. The inner loop alulation (for %G+C ontent) an simply be replaed by the formula for noligonuleotide signatures [Mirsky, 1999℄.
One interesting observation should be noted for noligonuleotide signatures.
When nuleotides enter and leave a window, some
adjustment needs to be made to the tallies for the
oligonuleotides in the new window. In Figure 2 part
C, for example, a ount of the dinuleotides in the
window requires 1 to be subtrated from the CA
ount and 1 to be added to the TG ount.
This is an easy alulation to make, yet one must
be areful about storing and aessing a list of
oligonuleotide ourrenes. If a linear searh is used
then the omplexity of the overall algorithm may
hange; if 4l > N , the generi signature program will
not maintain O(N ).
To overome this problem a hash funtion is used
to alulate the array position of any permutation.
DNA is oded as an enumerated type, where A=0,
T=1, C=2, G=3. The hash funtion is alulated as
follows: if the oligonuleotide being searhed for in
the hash table is ATCA, the hash table oset is as
follows:
The inner loop is now alulated. The inner loop
orresponds to the alulation for one window. Sine
the inner loop will be plaed within the outer loop,
p an be replaed by n in the inner loop speiation.
[ var ws, g : int;
ws, g = 0, 0;
℄.
Qi
Wprog
= ft = 8q : 0 q < ws :
= ((#i : n i < (n + q ) :
A:(i mod N ) = C
_ A:(i mod N ) = G)=W S )g
g
The inner loop is derived by replaing onstant W S
with variable ws and splitting the invariant into two
hunks, one for alulating G ontent and the other
for alulating C ontent. The inner blok is also
O(N ). See Figure 2-B.
Sine the windows range from size 1 to N 1, a
mathematial average for the window size is N=2.
This gives O(W S )[W S n(N=2)℄, i.e., O(N ) for the inner loop. As this ours O(N ) times, one for eah
iteration of the outer loop, the omplexity of the algorithm is O(N 2 ).
Derivation: stage 3
0 43 + 1 42 + 2 41 + 0 40 = 0 + 16 + 8 + 0 = 24
It is possible to atten the inner loop from an average
The ourrene ount for this oligonuleotide is
size of N=2 to a onstant of 2. Instead of simply
ounting and disarding the number of Gs and Cs therefore found at position 24 in the hash table.
The alulation is always unique, so searhing the
in eah window, it is possible to use the information
from the previous window to alulate the next. This hash table is never neessary. It is also onstant, so
is possible by subtrating the items whih leave the the lookup is always very quik.
6
Figure 3: N-oligonuleotide signatures for H. pylori. Figure 4. Signature analyses of H. pylori, window
Using large windows the HNS (part b, bottom) is size 1 Kb. The rst graph shows the DNS; the
highly implied from the DNS (part a, top).
seond shows the HNS.
Disussion
This method also has the advantage of not requiring any of the oligonuleotide permutations to be
stored textually, and thus wasting memory. Eah
oligonuleotide permutation is simply a formula
based on the nuleotide ontent and the enumerated
type values.
One last optimisation an be made. This imple
mentation is for Æ and not Æ , as the alulation is for
single-stranded DNA. It is possible to write a funtion whih derives the omplement of the sequene
and onatenates it onto the end, as suggested by
Karlin and Burge (1995) and originally implemented
by Burge et al. (1992). This turns N into 2 N .
We prove that Æ atually equals Æ , even though p 6=p.
The proof of this equivalene is illustrated by Mirsky
(1999). The 50% gain redues the omputation time
of a derived program still further.
Comparative results
Karlin and Burge (1995) present a DNS graph of H.
pylori; this graph is reprodued by our derived program and is shown in Figure 3-a. Karlin and Burge
doument that signatures reated from longer permutations are \highly implied" by the DNS graph.
The length 6-oligonuleotide signature graph (HNS)
for window size 50,000 (Figure 3-b) is indeed implied by the DNS results, although the results are
not the same. The observation of impliation does
not hold uniformly for other window sizes. Dierent
areas of interest are highlighted as the window-length
and permutation-length variables are modied (Figure 4).
The results also show that Æ ats as an averag7
ing funtion, removing a large amount of potentially
valuable data. By studying Pxy pxy (dinuleotide
probability for the whole sequene minus the probability for an individual window) for all xy permutations of length 2, regions of importane appear whih
are averaged out from the Æ graphs.
The graphs for dierent permutation lengths an
be ompared. This is beause signatures are based
on odds ratios and so the number of permutations
is absorbed into the ratio yielding absolute results.
Signiant results are identied by seletion based
on standard deviation thresholds from the mean. The
most appropriate thresholds should be determined on
an organism-by-organism basis and on the nature of
the study being performed, and remain dependent on
window size and permutation length.
DNS analysis using a 50,000 base window has
previously been used to identify regions that have
been horizontally aquired in N. meningitidis, using a threshold for detetion of 3 standard deviations [Tettelin et al., 2000℄. Analysis of the H.
pylori sequene, using the same parameters the
DNS analysis, identied two regions inluding reading frames HP0412 to HP0465 and HP0497 to
HP0573. The seond region ontains the ag
pathogeniity island previously identied using this
method [Karlin and Burge, 1995℄.
Using the same threshold of 3 standard deviations
the HNS identied only a single region (from HP0499
to HP0578) ontaining the seond region identied by
the DNS. Using a redued threshold of greater than
2 standard deviations the HNS also identied a region inluding the rst identied by the DNS (from
HP0409 to HP0480), and an additional region inluding reading frames HP0974 to HP1030.
One diÆulty with this approah is that the large
size of the windows means that this method only gives
a general indiation of where the atypial sequene
is loated. In the ase of the ag pathogeniity island the region whih had been horizontally aquired
was dened by a judgement based upon interpretation of the oding regions [Karlin and Burge, 1995℄.
In the ase of N. meningitidis, in whih a similar
analysis was performed, the limits of the regions
that had been horizontally aquired were determined
by omparison with an unrelated seond sequened
strain [Tettelin et al., 2000℄.
Using a window length of 1000, DNS analysis identied 177 windows of > 3 standard deviations above
the mean. These orrespond to 25 regions with ontiguous ORFs and 2 regions whih did not ontain annotated oding regions. The HNS analysis identied
a broadly similar number of regions, with 222 windows > 3 standard deviations above the mean. These
orrespond to 32 regions with ontiguous ORFs (and
none not ontaining any annotated oding regions).
However, although the analyses based upon dierent
length motifs identied 20 ommon reading frames
(40% of DNS and 25.3% of HNS nds) the majority of the reading frames identied were unique to
one or other analysis. The unique genes identied in
both analyses inluded those whih are known to be
the subjet to horizontal transfer suh as restrition
modiation system genes (2 identied by DNS and
7 by HNS). It should also be noted that genes whih
have atypial sequene omposition for reasons other
than horizontal aquisition are also identied by this
method, inluding in this analysis the `poly E rih
protein' (HP0322) and the `histidine and glutaminerih protein' (HP1432).
Reduing the window size inreases the granularity of the results. This is assoiated with an inrease in the speiity of this analysis, although
some genes adjaent to those that generate divergent
results will still be inluded due to the sliding window nature of this method. The results onerning
the ag pathogeniity island proved interesting. Both
1000 bp window size analyses identied HP0527 and
HP0528 (oding for ag pathogeniity island proteins
7 and 8). However, the DNS also identied HP0522,
HP0523, HP0531, HP0532, HP0533 and HP0534 (enoding ag pathogeniity island proteins 3, 4, 11, 12,
13 and a hypothetial protein, respetively); while
the HNS identied HP0527 and HP0528 (enoding
ag pathogeniity island proteins 15 and 16) as well
as two other reading frames within the region of divergene but not part of the reognized pathogeniity
island. These results indiate that although the overall patterns of the longer window analyses are similar
this an be the onsequene of the presene of different signals within the sanned areas and that the
8
speiity and sensitivity of the dierent word length
analyses dier.
Karlin and Burge state that the area of
most signiane in the 50 Kb window dinuleotide signature points to a pathogeniity island [Karlin and Burge, 1995℄. This is supported by
the 50 Kb-window DNS and %G+C ontent results.
However, the analysis using shorter windows indiates that some of the genes within this region, even
within the ag pathogeniity island itself, are not divergent by 1 standard deviation (see Figure 4). On
the basis of the signature analysis it annot be onluded that all of the genes within this region have
features that suggest that they have been horizontally
aquired. While a 1 Kb window implies the results
of the 50 Kb window, sine the larger window merely
averages the results of the smaller windows it omprises, the reverse is not true. Examination of the
region identied by the 50 Kb DNS using the smaller
window analysis allows us to onlude that the large
peak surrounding the pathogeniity island is in part
identifying regions of atypial assoiated DNA, and
that the abnormal regions do not omprise all the
reognised genes that ompose the ag pathogeniity
island. Signature analysis with a large window (e.g.
50 Kb) annot be relied upon for nding short regions
of atypial sequene (e.g. 2 Kb) in a genome.
Reently, Lawrene and Ohman mined the
DNA of E. oli for horizontally aquired regions [Lawrene et al., 1998℄. Using a odon bias
tehnique, they determined that 18% of the genes
in E. oli (approximately 10% of the sequene) have
been horizontally aquired sine its divergene from
Salmonellae. A similar proportion of untypial sequene in E. oli is identied using signature analysis.
A length-2 oligonuleotide signature and a length-6
oligonuleotide signature showed that 14% and 12%
respetively of the windows in eah were at least 1
standard deviation above the mean.
ods to plot individual probabilities and to alulate
standard deviations and means for use in ondene
estimates. The development of a generi O(n) algorithm for these tasks is doumented. This has onsiderable benet over an O(n2 ) solution. The speed
at whih the resulting program exeutes means that
methods of signature analysis an be ompared using
large datasets and previous onlusions an be tested.
Testing these signature analysis methods on
genome sequene data has revealed that %G+C ontent alone is not a reliable method for identifying
horizontally aquired DNA. The variane that implies horizontal transfer is not always reeted in
%G+C ontent and therefore we reommend the use
of %G+C ontent only to support data from one of
the other doumented methods.
DNS analysis does identify sequenes onsistent
with those thought to be horizontally transferred.
The generation of length-n>2 oligonuleotide signatures also provides useful results. Karlin's results
suggesting that the n-oligonuleotide signatures are
highly implied by the dinuleotide signature are onrmed for a 50 Kb window. However, with dierent
window sizes, the n-oligonuleotide signatures often
point to other, potentially relevant, areas from the
dinuleotide signature graphs. In addition, we nd
that plotting individual dierenes in probabilities,
before averaging, results in a ner granularity of results. Often an average annot distinguish between
a high variation in an individual permutation whih
is oset by low variations in the other permutations,
on the one hand, and moderate dierene in all permutations on the other.
Our omparison of methods leads us to onlude
that no single method of signature analysis is suÆient for the omplete identiation of horizontally
aquired DNA. There is some overlap between the
results, but dierent approahes ontribute valuable
additional results that any one method would miss.
Aknowledgements
Conlusions
Nigel Saunders is supported by a Welome Trust felThis paper desribes new software for the identia- lowship in medial mirobiology.
tion of horizontally aquired DNA. Programs have
been developed whih perform dinuleotide and noligonuleotide signatures and whih inlude meth9
Referenes
Alfano,J.R., Charkowski,A.O., Deng,W.L., Badel,
J.L., Petniki-Owieja,T., van Dijk,K. and
Collmer, A. (2000) The Pseudomonas syringae
hrp pathogeniity island has a tripartite mosai
struture omposed of a luster of type III seretion genes bounded by exhangeable eetor and
onserved eetor loi that ontribute to parasiti
tness and pathogeniity in plants. Pro. Natl.
Aad. Si. USA.
4856-61.
97
genomes: The omplexity hypothesis. Pro. Natl.
Aad. Si. USA.
3801-?.
96
Josse,K., Kaiser,A.D. and Kornberg,A. (1961) Enzymati synthesis of deoxyribonulei aid VIII. Frequenies of nearest neighbor base sequenes in deoxyribonulei aid. J. Biol. Chem.
864-875.
263
Kaldewaij,A. (1990) Programming: The Derivation
of Algorithms. Prentie Hall Int. Series in Comp.
Siene, Prentie Hall, New York.
Karlin,S. and Burge,C. (1995) Dinuleotide relative
Arber,W. (1993) Evolution of prokaryoti genomes. abundane extremes: a genomi signature. Tren.
Genet.
283-290.
Gene
49-56.
11
135
Brown,J.R. and Doolittle,W.F. (1999) Gene desent,
dupliation, and horizontal transfer in the evolution
of glutamyl- and glutaminyl-tRNA synthetases. J.
Mol. Evol.
485-95.
49
Burge,C., Campbell,A.M., Karlin,S. (1992) Overand under-representation of short oligonuleotides in
DNA sequenes. Pro. Natl. Aad. Si. USA
1358-1362.
89
Karlin,S., Campbell,A.M. and Mrazek,J. (1998)
Comparative DNA analysis aross diverse genomes.
Annual Rev. Genet.
185-225.
32
Karlin,S., Ladunga,I., Blaisdell,B.E. (1994) Heterogeneity of genomes: measures and values. Pro. Natl.
Aad. Si. USA
12837-12841.
91
Karlin,S. and Ladunga,I. (1994) Comparisons of eukaryoti genomi sequenes. Pro. Natl. Aad. Si.
12832-12836.
Doolittle,W,F. (1999) Lateral genomis. Trends Cell USA
Biol. M5-8.
Karlin,S., Moarski,E.S. and Shahtel,G.A. (1994)
Doolittle,W.F. (2000) Uprooting the tree of life. Si. Moleular evolution of herpesviruses: genomi and
protein sequene omparisons. J. Virol.
1886Am.
90-5.
1902.
Faguy,D.M. and Doolittle,W.F. (2000) Horizontal
transfer of atalase-peroxidase genes between arhaea Karlin,S., Mrazek,J. and Campbell,A.M. (1997)
Compositional biases of baterial genomes and evoand pathogeni bateria. Trends Genet.
196-7.
lutionary impliations. J. Bat.
3899-3913.
Forterre,P. and Philippe,H. (1999) Where is the root
Kroll,J.S., Wilks,K.E., Farrant,J.L. and Langof the universal tree of life? Bioessays
871-9.
ford,P.R. (1998) Natural geneti exhange between
Groisman,E.A. and Ohman,H. (1996) Pathogeniity Haemophilus and Neisseria: intergeneri transfer of
islands: baterial evolution in quantum leaps. Cell hromosomal genes between major human pathogens.
791-4.
Pro. Natl. Aad. Si. USA
12381-5.
Haker.J., Blum-Oehler,G., Muhldorfer,I. and Lawrene,J.G. and Ohman,H. (1997) Amelioration
Tshape,H. (1997) Pathogeniity islands of viru- of baterial genomes: rates of hange and exhange.
lent bateria: struture, funtion and impat on J. Mol. Evol.
383-397.
mirobial evolution. Mol. Mirobiol.
1089-1097.
Lawrene,J.G., Ohman,H. (1998) Moleular arhaeHou,Y.M. (1999) Transfer RNAs and pathogeniity ology of the Esherihia oli genome. Pro. Natl.
islands. Trends Biohem. Si.
295-8.
Aad. Si. USA
9413-7.
9
20
68
282
16
179
21
87
95
44
23
24
95
Jain et al. (1999) Horizontal gene transfer among Maddox,J. (1998) What Remains to Be Disovered.
10
TheFree Press, Simon & Shuster, New York.
us that has aquired a gonooal PIB porin. Mol.
Mirobiol.
1001-1007.
15
Martin,W. (1999) Mosai baterial hromosomes: a
Woese,C. (1998) The universal anestor. Pro. Natl.
hallenge enroute to a tree of genomes. BioEssays
Aad. Si. USA
6854-9.
99-104.
21
Mirsky,J. (1999) Genome Analysis Methodologies for
the Identiation of Horizontally Aquired DNA. Oxford University Computing Laboratory M.S. thesis.
Moriere (2000) Multiple independent horizontal
transfers of informational genes from bateria to plasmids and phages: impliations for the origin of baterial repliation mahinery. Mol. Mirobiol.
1-5.
35
Nelson,K.E., Clayton,R.A., Gill,S.R., Gwinn,M.L.,
Dodson,R.J., Haft,D.H., Hikey,E.K., Peterson,J.D.,
Nelson,W.C., Kethum,K.A., MDonald,L., Utterbak, T.R., Malek,J.A., Linher,K.D., Garrett,M.M., Stewart,A.M., Cotton,M.D., Pratt,M.S.,
Phillips,C.A., Rihardson,D., Heidelberg,J., Sutton,
G.G., Fleishmann,R.D., Eisen,J.A., Fraser,C.M., et
al. (1999) Evidene for lateral gene transfer between
Arhaea and bateria from genome sequene of Thermotoga maritima. Nature
323-9.
399
Saunders,N.J., Hood,D.W. and Moxon,E.R. (1999)
Baterial evolution: bateria play pass the gene.
Curr. Biol R180-3.
9
Tettelin,H., Saunders,N.J., Heidelberg,J., Jeffries,A.C., Nelson,K.E., Eisen,J.A., Kethum,K.A.,
Hood,D.W., Peden,J.F., Dodson,R.J., Nelson,W.C.,
Gwinn,M.L., DeBoy,R., Peterson,J.D., Hikey,E.K.,
Haft,D.H.,
Salzberg,S.L.,
White,O.,
Fleishmann,R.D., Dougherty,B.A., Mason,T., Cieko,A.,
Parksey,D.S., Blair,E., Cittone,H., Clark,E.B., Cotton,M.D., Utterbak,T.R., Khouri,H., Qin,H.,
Vamathevan,J., Gill,J., Sarlato,V., Masignani,V., Pizza,M., Grandi,G., Sun,L., Smith,H.O.,
Fraser,C.M., Moxon,E.R., Rappuoli,R. and Venter,J.C. (2000) The omplete genome sequene of
Neisseria meningitidis serogroup B strain MC58.
Siene
1809-1815.
278
Vasquez,J.A., Berron,S., O'Rourke,M., Carpenter,G., Feil,E., Smith,N.H. and Spratt,B.G. (1995)
Interspeies reombination in nature: a meningoo11
95
Referenes
[Forterre and Philippe, 1999℄ Forterre P, Philippe H.
Where is the root of the universal tree of life?
[Karlin and Burge, 1995℄ Karlin,S. and Burge,C.
Bioessays. 1999 Ot;21(10):871-9.
(1995) Dinuleotide relative abundane extremes:
a genomi signature. Tren. Genet.,
283-290.
[Nelson et al., 1999℄ Nelson KE, Clayton RA, Gill
SR, Gwinn ML, Dodson RJ, Haft DH, Hikey EK,
[Doolittle, 2000℄ Doolittle,W.F. (2000) Uprooting
Peterson JD, Nelson WC, Kethum KA, MDonald
the tree of life. Si. Am.
90-5.
L, Utterbak TR, Malek JA, Linher KD, Garrett
MM, Stewart AM, Cotton MD, Pratt MS, Phillips
[Arber, 1993℄ Arber,W. (1993) Evolution of prokaryCA, Rihardson D, Heidelberg J, Sutton GG, Fleisoti genomes. Gene
49-56.
hmann RD, Eisen JA, Fraser CM, et al. Evidene
[Lawrene et al., 1998℄ Lawrene,J.G., Ohman,H.
for lateral gene transfer between Arhaea and ba(1998) Moleular arhaeology of the Esherihia
teria from genome sequene of Thermotoga maroli genome. Pro. Natl. Aad. Si. USA 9413-7.
itima. Nature. 1999 May 27;399(6734):323-9.
[Maddox, 1998℄ Maddox J: What Remains to Be Disovered. TheFree Press, Simon & Shuster, New [Hou, 1999℄ Hou YM: Transfer RNAs and
pathogeniity islands. Trends Biohem Si.
York. 1998.
1999 Aug;24(8):295-8.
[Martin, 1999℄ Martin W: Mosai baterial hromosomes: a hallenge enroute to a tree of genomes. [Haker et al., 1997℄ Haker J, Blum-Oehler G,
Muhldorfer I, Tshape H: Pathogeniity islands
BioEssays 1999 21:99-104.
of virulent bateria: struture, funtion and
[Faguy and Doolittle, 2000℄ Faguy DM, Doolittle
impat on mirobial evolution. Mol Mirobiol 1997
WF: Horizontal transfer of atalase-peroxidase
23(6):1089-1097.
genes between arhaea and pathogeni bateria.
Trends Genet. 2000 May;16(5):196-7.
[Alfano et al., 2000℄ Alfano JR, Charkowski AO,
Deng WL, Badel JL, Petniki-Owieja T, van Dijk
[Doolittle, 1999℄ Doolittle WF. Lateral genomis.
K, Collmer A: The pseudomonas syringae hrp
Trends Cell Biol. 1999 De;9(12):M5-8.
pathogeniity island has a tripartite mosai struture omposed of a luster of type III seretion
[Brown and Doolittle, 1999℄ Brown JR, Doolittle
genes bounded by exhangeable eetor and onWF. Gene desent, dupliation, and horizonserved eetor loi that ontribute to parasiti ttal transfer in the evolution of glutamyl- and
ness and pathogeniity in plants. Pro Natl Aad
glutaminyl-tRNA synthetases. J Mol Evol. 1999
Si U S A. 2000 Apr 25;97(9):4856-61.
Ot;49(4):485-95.
11
282
135
95
[Woese, 1998℄ Woese C. The universal anestor. Pro [Groisman and Ohman, 1996℄ Groisman
EA,
Natl Aad Si U S A. 1998 Jun 9;95(12):6854-9.
Ohman H: Pathogeniity islands: baterial
evolution in quantum leaps. Cell. 1996 Nov
[Jain et al., 1999℄ Jain et al. Horizontal gene trans29;87(5):791-4.
fer among genomes: The omplexity hypothesis.
PNAS 1999 96:3801.
[Kroll et al., 1998℄ Kroll JS, Wilks KE, Farrant JL,
[Moreire, 2000℄ Moreire: Multiple independent horiLangford PR: Natural geneti exhange between
zontal transferes of informational genes from baHaemophilus and Neisseria: intergeneri transteria to plasmids and phages: impliations for the
fer of hromosomal genes between major huorigin of baterial repliation mahinery. Mol Miman pathogens. Pro Natl Aad Si USA 1998
robiol 2000 35:1.
95(21):12381-5.
12
[Saunders et al., 1999℄ Saunders NJ, Hood DW, [Burge et al., 1992℄ Burge C, Campbell AM, Karlin
S: Over- and under-representation of short oligonuMoxon ER: Baterial evolution: bateria play pass
leotides in DNA sequenes. Pro Natl Aad Si
the gene. Curr Biol 1999 9(5):R180-3.
USA (1992) 89(4): 1358- 1362.
[Vasquez et al., 1995℄ Vasquez JA, Berron S,
O'Rourke M, Carpenter G, Feil E, Smith NH, [Tettelin et al., 2000℄ Herv Tettelin, Nigel J. Saunders, John Heidelberg, Alex C. Jeries, Karen E.
Spratt BG: Interspeies reombination in nature:
Nelson, Jonathan A. Eisen, Karen A. Kethum,
a meningoous that has aquired a gonooal
Derek W. Hood, John F. Peden, Robert J. Dodson,
PIB porin. Mol Mirobiol 1995 15:1001-1007.
William C. Nelson, Mihelle L. Gwinn, Robert DeBoy, Jeremy D. Peterson, Erin K. Hikey, Daniel H.
[Lawrene and Ohman, 1997℄ Lawrene
JG,
Haft, Steven L. Salzberg, Owen White, Robert D.
Ohman H: Amelioration of baterial genomes:
Fleishmann, Brian A. Dougherty, Tanya Mason,
rates of hange and exhange. J Mol Evol 1997
Anne Cieko, Debbie S. Parksey, Eri Blair, Henry
44(4): 383-397.
Cittone, Emily B. Clark, Matthew D. Cotton,
Terry R. Utterbak, Hoda Khouri, Haiying Qin,
[Karlin et al., 1998℄ Karlin S, Campbell AM, Mrazek
Jessia Vamathevan, John Gill, Vinenzo Sarlato,
J: Comparative DNA analysis aross diverse
Vega Masignani, Mariagrazia Pizza, Guido Grandi,
genomes. Annual Rev Genet 1998 32:185-225.
Li Sun, Hamilton O. Smith, Claire M. Fraser,
[Josse et al., 1961℄ Josse K, Kaiser AD, Kornberg A:
E. Rihard Moxon, Rino Rappuoli, and J. Craig
Enzymati synthesis of deoxyribonulei aid VIII.
Venter. The omplete genome sequene of NeisseFrequenies of neaarest neighbor base sequenes in
ria meningitidis serogroup B strain MC58. Siene
deoxyribonulei aid. J. Biol Chem 1961 263: 864(2000) 278: 1809-1815
875.
[Karlin et al., 1997℄ Karlin S, Mrazek J, Campbell
AM: Compositional Biases of Baterial Genomes
[Karlin et al., 1994.a℄ Karlin S, Ladunga I, Blaisdell
and Evolutionary Impliations. Journal of BateBE: Heterogeneity of genomes: measures and valriology 1997 179(12): 3899-3913.
ues. Pro Natl Aad Si USA 1994 91: 1283712841.
[Karlin et al., 1994.b℄ Karlin S, Moarski ES,
Shahtel GA: Moleular evolution of herpesviruses:
genomi and protein sequene
omparisons. J Virol 1994 68: 1886-1902.
[Karlin et al., 1994.℄ Karlin S, Ladunga I: Comparisons of eukaryoti genomi sequenes. Pro Natl
Aad Si USA 1994 20 91: 12832-12836.
[Kaldewaij, 1990℄ Kaldewaij A: Programming: The
Derivation of Algorithms. Prentie Hall Int Series
in Comp. Siene, Prentie Hall, New York. 1990.
[Mirsky, 1999℄ Mirsky J: Genome Analysis Methodologies for the Identiation of Horizontally Aquired DNA. Oxford University Computing Laboratory M.S. thesis. 1999.
13