Calculus for Biologists Lab Math 1180-002 Spring 2012
advertisement

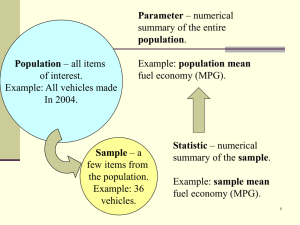
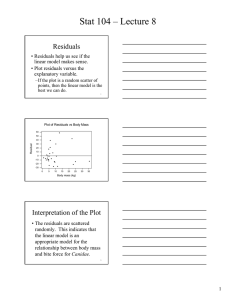
Calculus for Biologists Lab Math 1180-002 Spring 2012 Lab #14 - Regression in R Report due date: Friday, April 27, 2012 at 11:59 p.m. Goal: To perform linear regression in R. You will also analyze results based on r2 and p-values. ? Create a new script, either in R (laptop) or with a text editor (Linux computers). Linear regression is a useful tool for determining characteristic behaviors of data. In this lab, you will perform regressions on simulated data. To start, you will need to import the four data files into R. To do this, first check your current directory in R by executing this command: getwd() You may see something that looks similar to this: /home/1020/ma/yourname You should have a folder yourname somewhere on your computer into which you need to download the data files. Do not rename the files when you download them. If you have a mac or pc, you will also need to download the files to the appropriate folder. Once you’ve done that, you can import the files, which should be called data1, data2, data3 and data4. We do this using read.table and construct a data table with as.data.frame. data1 data2 data3 data4 = = = = as.data.frame(read.table("data1",header=TRUE)) as.data.frame(read.table("data2",header=TRUE)) as.data.frame(read.table("data3",header=TRUE)) as.data.frame(read.table("data4",header=TRUE)) You will notice that each table imported contains column headers. You can check these by executing, for example, names(data1) which should result in [1] "glucose" "insulin" You can access the individual columns, as we have done previously, by executing data1$insulin, as an example. Regression: the easiest thing to do in R As you probably guessed, data1 contains simulated glucose and insulin data, as a nice follow-up to our discussion of diabetes last week. For each glucose value, there is a corresponding insulin measurement, which we can view with a plot. par(mfrow=c(1,2)) plot(insulin~glucose,data1,pch="+",main="Regression") Notice how I’ve told R to plot the data. For consistency with the remainder of the lab, we will use the y~x notation, which tells R, “plot y as a function of x.” Note that this is exactly equivalent to executing plot(glucose,insulin,...). You may see a trend in the plot of these data, and it is our goal is to determine if it’s a significant one. So, we run a linear regression to see how closely the data lie on a line. In R, this amounts to using the lm(y~x, data=data.set.name) command, which calculates the slope a and intercept b for the best-fit line y = ax + b to the data. Typing y~x tells R to assume that the output y is a linear function of x. We will name the results of our hard work regr. regr = lm(insulin~glucose, data=data1) What information does regr give us? summary will, you guessed it, give you a summary of the results of the regression, including r2 and the statistical significance of the fit, among other things. regr.summ = summary(regr) should give you the following mass of information: 1 of 3 L14 Call: lm(formula = insulin ~ glucose, data = data1) Residuals: Min 1Q -8.5332 -2.7982 Median 0.2384 3Q 2.7895 Max 8.1475 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 0.1886 2.4983 0.075 0.94 glucose 10.0032 0.5199 19.241 <2e-16 *** --Signif. codes: 0 *** 0.001 ** 0.01 * 0.05 . 0.1 1 Residual standard error: 3.79 on 98 degrees of freedom Multiple R-squared: 0.7907,Adjusted R-squared: 0.7886 F-statistic: 370.2 on 1 and 98 DF, p-value: < 2.2e-16 But, all we care about are R-squared and Pr(>|t|) for glucose. So what do we do? r.sq = regr.summ$r.squared pval = regr.summ$coefficients[2,4] We also want to see how the regression line matches up against our data. Thankfully, there’s abline to save us from doing any real work. In previous labs, we’ve used this command to add horizontal and vertical lines to plots. As it happens, abline will also magically draw regression lines using the information contained in regr. abline(regr,col="dodgerblue",lwd=2) What do you think of the fit? We can add some more things to the figure window, like residuals, to get a better idea of how things fare. In the rest of the plot commands, you’ll see regr$residuals. Embedded in regr is also a list of the residual values at each point. mtext(side=3,bquote(r^2 == .(r), list(r = format(r.sq,digits=4))),adj=0) ## show me r^2 mtext(side=3,bquote(p == .(p), list(p = format(pval,digits=4))),adj=1) ## show me p ## plot residuals plot(regr$residuals~data1$glucose, pch="+", xlab = "glucose", ylab="Residuals", main="Residuals") abline(h=0,col="dodgerblue",lwd=2) Plot 14.1: Save this to include in your assignment. More regression, because it’s easy We have a few more experiments to run. Execute the following lines of code and to generate the necessary plots, saving them where applicable. Between each figure, there is a command to clear all regression variables, as they will be repeatedly redefined. rm(list=c("regr","regr.summ","pval","r.sq")) regr = lm(insulin~glucose, data=data2) regr.summ = summary(regr) pval = regr.summ$coefficients[2,4] r.sq = regr.summ$r.squared par(mfrow=c(1,2)) plot(insulin~glucose,data2,pch="+",main="Regression") abline(regr,col="dodgerblue",lwd=2) mtext(side=3,bquote(r^2 == .(r), list(r = format(r.sq,digits=4))),adj=0) mtext(side=3,bquote(p == .(p), list(p = format(pval,digits=4))),adj=1) plot(regr$residuals~data2$glucose, pch="+", xlab = "glucose", ylab="Residuals", main="Residuals") abline(h=0,col="dodgerblue",lwd=2) 2 of 3 L14 Plot 14.2: Save this to include in your assignment. rm(list=c("regr","regr.summ","pval","r.sq")) regr = lm(productivity~procrastination, data=data3) regr.summ = summary(regr) pval = regr.summ$coefficients[2,4] r.sq = regr.summ$r.squared par(mfrow=c(1,2)) plot(productivity~procrastination,data3, pch="+", main="Regression") abline(regr,col="limegreen",lwd=2) mtext(side=3,bquote(r^2 == .(r), list(r = format(r.sq,digits=4))),adj=0) mtext(side=3,bquote(p == .(p), list(p = format(pval,digits=4))),adj=1) plot(regr$residuals~data3$procrastination, pch="+", xlab="procrastination", ylab="Residuals", main="Residuals") abline(h=0,col="limegreen",lwd=2) Plot 14.3: Save this to include in your assignment. rm(list=c("regr","regr.summ","pval","r.sq")) regr = lm(effect~concentration,data=data4) regr.summ = summary(regr) pval = regr.summ$coefficients[2,4] r.sq = regr.summ$r.squared par(mfrow=c(1,2)) plot(effect~concentration,data4,pch="+",main="Regression") abline(regr,col="magenta",lwd=2) mtext(side=3,bquote(r^2 == .(r), list(r = format(r.sq,digits=4))),adj=0) mtext(side=3,bquote(p == .(p), list(p = format(pval,digits=4))),adj=1) plot(regr$residuals~data4$concentration, pch="+", xlab="concentration", ylab="Residuals", main="Residuals") abline(h=0,col="magenta",lwd=2) Plot 14.4: Save this to include in your assignment. ? Save your script so that you can use it for your assignment. 3 of 3 L14