Document 11559665

AN ABSTRACT OF THE THESIS OF
Jennifer L. Rosenberger for the degree of Master of Science in Chemical Engineering presented on July 2, 2010.
Title: Two-Stage Photobioreactor Cultivation for Enhancing Lipid Production from
Diatom Cells by Controlled Silicon Limitation
Abstract approved:
.
.
Gregory L. Rorrer
There is significant interest in utilizing microalgae as a source for biofuels. Diatoms are a class of single-celled microalgae which make silica cell walls and require dissolved silicon as a substrate for cell division. Manipulation of soluble silicon delivery to the culture offers a route to control cell cycle and lipid production. A twostage, semi-continuous photobioreactor cultivation process was developed to induce the production of lipid-rich algal biomass from the centric diatom Cyclotella sp. In
Stage I of the process, algal cells were grown up to high density until all of the dissolved silicon in the feed medium was consumed. The cells were in the siliconstarved state when the cell density was constant for 24 hours and the dissolved silicon concentration was near-zero and ceased to change. In Stage II, fresh medium containing dissolved silicon was perfused into the reactor for a 48, 72 or 96 hours. The silicon was rapidly consumed by the silicon-starved cells under a surge uptake mechanism, maintaining the same near-zero silicon concentrations that were present at the end of the first stage, so that silicon starvation was maintained within the system.
Cells grown with 48 and 72 hour perfusion yielded high lipid concentrations (>45% of dry cell weight) and high productivity in the algal biomass while maintaining biomass
generation. The lipid composition was analyzed by GC/MS and seven fatty acids were identified including the three main fatty acids: palmitoleic acid, palmitic acid and eicosapentanoic acid. It was determined by GC analysis that the lipid composition remains constant once silicon starvation is achieved, regardless of perfusion addition of additional silicon. This study illustrates that a bioreactor cultivation strategy enhances lipid production by algal cultures for the purpose of algal biodiesel production.
©Copyright by Jennifer L. Rosenberger
July 2, 2010
All Rights Reserved
Two-Stage Photobioreactor Cultivation for Enhancing Lipid Production from Diatom Cells by Controlled Silicon Limitation by
Jennifer L. Rosenberger
A THESIS submitted to
Oregon State University in partial fulfillment of the requirements for the degree of
Master of Science
Presented July 2, 2010
Commencement June 2011
Master of Science thesis of Jennifer L. Rosenberger presented on July 2, 2010
APPROVED:
.
Major Professor, representing Chemical Engineering
.
Head of the School of Chemical, Biological and Environmental Engineering
.
Dean of the Graduate School
.
.
.
.
I understand that my thesis will become part of the permanent collection of Oregon
State University libraries. My signature below authorizes release of my thesis to any reader upon request.
.
Jennifer L. Rosenberger, Author
ACKNOWLEDGEMENTS
The author graciously acknowledges the support for this research from BEST, Oregon
Built Environmental and Sustainable Technologies Center.
The author expresses sincere gratitude to advisor Gregory L. Rorrer for his generosity and mentoring, to my fellow graduate students Debra Gale, Aaron Goodwin and
Jeremy Campbell for helping me in my research, and most importantly to my husband,
Jason Rosenberger, who has made this all possible through his support and love.
TABLE OF CONTENTS
Page
Two-Stage Photobioreactor Cultivation without Controlled Silicon Delivery ........ 10
Two-Stage Photobioreactor Cultivation with Controlled Silicon Delivery ............. 11
Two-Stage Photobioreactor Cultivation without Controlled Silicon Delivery ........ 16
Two-Stage Photobioreactor Cultivation With Controlled Silicon Delivery ............. 25
Silicon Concentration and Total Lipid Weight Percent ........................................... 48
TABLE OF CONTENTS, CONTINUED
Page
Appendix F – Correlation of cell number to dry cell mass .................................... 182
LIST OF FIGURES
Figure Page
1. Diatom cell structure .................................................................................................. 2
2. Silicon uptake by transport proteins ........................................................................... 3
3. Fatty acid de novo synthesis pathway. ....................................................................... 5
4. Triacylglyceride synthesis pathway ........................................................................... 6
5. Cyclotella sp. cell suspension in culture flask ........................................................... 8
6. Cyclotella sp. cell under 100x magnification ............................................................. 8
7. Schematic of photobioreactor .................................................................................. 10
8. Charge addition (a) Total number of cells and Si vs.
cultivation time (b) Total number of cells and pH vs.
cultivation time .............................................................................................. 21
9. Charge addition (a) Total Lipids and Si vs. cultivation
time (b) Total DCW and Total Lipid productivity vs.
cultivation time .............................................................................................. 23
10. 48 h perfusion (a) Total number of cells and Si vs.
time, (b) Total number of cells and pH vs. time ......................................... 31
11. 48 h perfusion (a) Total lipids (wt %/DCW) and Si
concentration vs. time, (b) Total DCW productivity
and total lipids productivity vs. time ............................................................ 33
LIST OF FIGURES, CONTINUED
Figure Page
12. 72 h perfusion (a) Total number of cells and Si
vs. time, (b) Total number of cells and pH vs. time .................................... 37
13. 72 h perfusion (a) Total lipids (wt%/DCW) and Si
vs. time, (b) Total DCW productivity and Total
Lipid productivity vs. time .......................................................................... 39
14. 96 h perfusion (a) Total number of cells and Si vs.
time, (b) Total number of cells and pH vs. time ......................................... 43
15. 96 h perfusion (a) Total lipids (wt%/DCW) and Si
vs. time, (b) Total DCW productivity and Total
Lipid productivity vs. time .......................................................................... 46
16. Representative cultivation with 48 h perfusion ...................................................... 51
17. (a) – (j) Nile red lipid stain images ........................................................................ 52
18. Fatty Acid composition during cultivation for
a) 48 h perfusion b) 72 h perfusion ............................................................. 55
19. Representative GC chromatogram ........................................................................ 56
LIST OF TABLES
Table Page
1. Two-stage photobioreactor cultivation with charge addition ................................... 18
2. Two-stage photobioreactor cultivation with 48 h perfusion ................................... 26
3. Two-stage photobioreactor cultivation with 72 h perfusion .................................... 27
4. Two-stage photobioreactor cultivation with 96 h perfusion .................................... 28
6. Fatty acid composition, 48 and 72 h perfusion experiments .................................... 54
LIST OF APPENDIX FIGURES
Figure Page
C 1. Calibration data for Silicon dioxide detection at 360 nm on 11/15/2008 ........... 148
C 4. Calibration data for Silicon detection at 360 nm on 6/11/2009.......................... 150
C 7. Calibration data for Silicon detection at 360 nm on 12/15/2009........................ 152
C 10. Calibration data for Silicon detection at 360 nm on 7/13/2009 ....................... 154
LIST OF APPENDIX FIGURES, CONTINUED
Figure Page
C 19. Calibration data for lipid detection at 350 nm on 8/15/2009 ........................... 160
C 21. Statistical analysis of lipid calibration data from 8/15/2009 ............................ 161
LIST OF APPENDIX FIGURES, CONTINUED
Figure Page
E 1. k’, apparent light attenuation constant ………………………………………. 181
E 2. k c
, specific light attenuation constant (mL/cells-cm) ………………………... 181
F 1. Correlated data ……………………………………………………………….. 183
LIST OF APPENDIX TABLES
Table Page
B 11. Cy-JA-10, Si concentration by spectrophotometer .......................................... 102
LIST OF APPENDIX TABLES, CONTINUED
Table Page
B 16. Cy-JA-11, Si concentration by spectrophotometer .......................................... 111
B 21. Cy-JA-12, Si concentration by spectrophotometer .......................................... 121
LIST OF APPENDIX TABLES, CONTINUED
Table Page
B 26. Cy-JA-13, Si concentration by spectrophotometer .......................................... 129
B 33. Cy-JA-14, Si concentration by spectrophotometer .......................................... 138
B 40. Cy-JA-15, Si concentration by spectrophotometer .......................................... 146
D 1.72 h perfusion GC data ……………………………………………………...…168
LIST OF APPENDIX TABLES, CONTINUED
Table Page
D 2. 48 h perfusion GC data ………………………………………………………. 171
Table E 1. Light attenuation data ………………………………………………….. 180
Table F 1. Tabulated Data …………………………………………………………. 182
Two-Stage Photobioreactor Cultivation for Enhancing Lipid Production from Diatom Cells by Controlled Silicon Limitation
Introduction
As concern for global warming and dependence on fossil fuels grows, interest in clean and renewable energy sources has increased dramatically. Alternative energy sources such as wind, solar, geothermal and biofuels have risen to the forefront as possible solutions to this energy crisis. Biofuels are attractive because they utilize existing technologies and are compatible with current infrastructures. Because biofuels are derived from plants, they can be considered carbon neutral energy sources. Additionally, microalgae have simple cellular structures, short cultivation cycles (1-10 days), the ability to accumulate large quantities of lipids (40-80%) per dry cell weight biomass
(Chisti 2007; Metting 1996; Spolaore 2006), potential for growth in harsh conditions and on land that is inadequate for food crops (Searchinger 2008), less nutrient and fertilizer requirements (Cantrell 2008; Rodolfi 2008), and the capability to grow year round in controlled conditions using specially designed bioreactors (Verma 2010). While there are many advantages to algal biodiesel there are some challenges as well. Improving the cost effectiveness of algal biodiesel is necessary before large scale commercialization is feasible. One important area of improvement is increasing algal lipid productivity. Nutrient deprivation can be used to increase lipid concentrations within cells by altering the lipid biosynthetic pathways. Typically nitrogen is limited but other limiting other nutrients can have a similar effect (Basova 2005). However, nutritionally stressed algae have much lower growth rates then when grown in a healthy environment, which yields high lipid concentrations within the cells but low lipid productivity overall.
Diatoms are by far the most prolific class of microalgae which make lipids that are readily converted to biodiesel (Sheehan 1998). Diatoms are single-celled
photosynthetic algae of the class Bacillariophyceae that possess silica shells called frustules, which they make by a process called biosilicification.
Biosilicification is the process by which an organism uptakes soluble silicon from the environment, precipitates the silicon into amorphous silicon oxides and creates a frustule. Each diatom frustule is composed of two overlapping valves (Figure 1).
2
Figure 1. Diatom cell structure
As a diatom prepares to divide, silicon in the form of Si(OH)
4
is transported across the cell wall and into the cytoplasm (Figure 2) by silicon transport proteins (Azam 1974). Soluble silicon in the cytoplasm is transported to the silicon deposition vesicle, or SDV (Hildebrand 1997). The SDV is a specialized cell compartment located along the central axis of a diatom.
Within the SDV soluble silicon is polymerized to form solid silica (SiO
2
) and this SiO
2
is used to construct the structures that become new diatom frustules.
When the new frustule is completed, the diatom divides. A complete review of silicon metabolism in diatoms (Martin-Jezequel et al., 2000) and diatom frustule formation (Round et al., 1990; van den Hoek, et al., 1995) can be found elsewhere.
3
Figure 2. Silicon uptake by transport proteins
Silicon metabolism in the diatom is linked to the four phases of its cell cycle:
G1 (photosynthetic growth), S (DNA replication), G2 (Si uptake and new valve formation), and M (mitosis) as reviewed by Martin-Jézéquel et al.
(2000). Studies suggest three different modes of silicon uptake in diatoms: surge uptake, internally controlled uptake, and externally controlled uptake
(Conway et al., 1976; Conway et al., 1977). Surge uptake occurs when siliconstarved cells are exposed to high silicon concentrations and there is a large concentration gradient between the extracellular and intracellular environments causing rapid uptake. Internally controlled uptake is regulated by the intracellular rate at which silicon deposition into the cell wall occurs.
Externally controlled uptake occurs when extracellular concentrations are at very low levels and is a function of substrate concentration.
4
Silicon biochemistry influences and controls not only development of silica structures but other parts of the metabolism of diatoms. Of particular interest to this study are the effects of silicon on lipid metabolism which produces lipids of various classes, including triglycerides, the major raw lipid precursor for biodiesel production. Triglyceride synthesis can be divided into three steps
(Verma 2010): Step one is the formation of acetyl-CoA, which occurs with photosynthesis. Step two is fatty acid chain elongation. Step three is the formation of triacylglycerides.
Under optimal conditions of growth diatoms synthesize fatty acids for esterification into membrane lipids by a de novo synthesis pathway. Fatty acids perform a structural role in the cell and are necessary for cell division.
De novo synthesis of fatty acids in algae occurs mainly in the chloroplast (Hu
2008). Figure 3 depicts the generalized scheme for de novo fatty acid biosynthesis. In this synthesis scheme the committed step is Reaction 1, in which acetyl CoA is converted to malonyl CoA. This reaction is catalyzed by the enzyme acetyl CoA carboxylase (ACCase) (Verma 2010).
5
Figure 3. Fatty acid de novo synthesis pathway.
In silicon-deficient cells a substantial increase in the activity of acetyl-CoA carboxylase has been observed. It was determined that silicon deficiency promotes transcription of nuclear genes responsible for the formation of
ACCase (Roessler 1988), thus increasing fatty acid content within the diatom.
This increased fatty acid production drives the formation of triacylglycerides
(TAG) (Figure 4) by the glycerol-3-phosphate pathway (Roessler 1994).
Unlike membrane lipids, TAGs do not perform a structural role but instead are transferred to the cytoplasm as lipid vesicles and serve primarily to store carbon and energy.
6
Figure 4. Triacylglyceride synthesis pathway
Investigations into the extent of enhanced lipid production as a consequence of silicon-starvation in batch culture systems demonstrated lipid content doubling during exponential growth and increasing by as much as 20% when maintained at silicon starvation (Shifrin 1981; Roessler 1988; Lombardi 1995) but with biomass limited by the amount of initial silicon provided to the system.
Further studies considered two-stage systems in which a second charge addition of silicon was added to the reactor once silicon-starvation had been achieved. These experiments saw additional biomass generation but lipid concentrations decreased as cells began to uptake silicon and divide (Coombs
1967; Taguchi 1987; Lynn 2000). One experiment untilized a turbidostat to deliver a steady flow of silicon at low concentration (Lombardi 1991) and saw increased lipid concentrations but low lipid production overall due to a decrease in cell growth rate. These studies made no attempt to control silicon delivery, but simply studied systems of silicon depletion or added instant charges of additional silicon for cell growth.
This study designs and tests a unique bioprocess strategy which hopes to take advantage of increased lipid formation during times of silicon-starvation but also enable cell growth and allow for truly enhanced lipid production. A twostage photobioreactor cultivation scheme will mimic a fed-batch system and control silicon delivery. The silicon will be added at low concentrations over extended time periods, such that silicon-starved cells with increased lipid concentrations will receive silicon by perfusion at rates low enough to maintain
7 a silicon-depleted state and yet receive sufficient silicon for continued biomass production while maintaining high lipid levels.
The first objective of this study was to measure the lipid concentration over cultivation time in a fed-batch system. The second objective of this study was to determine if the lipid profile is affected by the rate of fed-batch perfusion.
The third objective was to characterize the fatty acid composition over the course of cultivation. Based upon these results, a method for enhanced lipid production using a two-stage photobioreactor with perfusion was developed.
Materials and Methods
Cell Culture
Pure stock cultures of the marine diatom Cyclotella sp. obtained from UTEX
Culture Collection of Algae (UTEX# 1269) were used in the following experiments. Cyclotella sp. is a centric diatom comprised of two adjoining disc-shaped valves, with a 10 µm diameter, resting on top of a ring-shaped girdle band. Stock cultures were maintained on a modified Harrison’s artificial seawater medium supplemented with f/2 nutrients. The modified artificial seawater base medium contained 347 mM NaCl, 23.8 mM Na
2
SO
4
, 7.68 mM
KCl, 1.97 mM NaHCO
3
, 690 µM KBr, 354 µM H
3
BO
3
, 63.3 µM NaF, 45.1 mM MgCl
2
• 6H
2
O, 8.74 mM CaCl
2
• 2H
2
O, and 78.0 µM SrCl
2
• 6H
2
O. The f/2 enrichment medium components consisted of 35.9 nM Na
2
MnO
4
• 2H
2
O,
0.954 nM Na
2
SeO
3
, 6.0 nM NiCl
2
• 6H
2
O, 165 nM ZnSO
4
• 7H
2
O, 45.3 nM
CuSO
4
• 5H
2
O, 24.7 nM CoSO
4
• 7H
2
O, 2.31 µM MnSO
4
• 4H
2
O, 19.7 µM
FeCl
3
• 6H
2
O, and 21.9 uM ethylenedinitrol tetraacedi acid disodium salt
(C
10
H
1
4O
8
N
2
Na
2
• 2H
2
O). The cultures were grown in a 22°C incubator without agitation in 500 mL foam-stoppered flasks under cool white fluorescent light at 100 µE/m
2
-s incident light flux with a photoperiod of 14 hour light/10 hour dark. The cell suspension was subcultured every 14 days.
8
Images of the culture are presented in Figures 5 and 6. Figure 5 shows the cell culture as it appears in suspension in the culture flask. Figure 6 is a microscopic image at 100x magnification.
Figure 5. Cyclotella sp. cell suspension in culture flask
Figure 6. Cyclotella sp. cell under 100x magnification
9
Photobioreactor Design
The bubble-column photobioreactor shown in Figure 7 was used to cultivate the Cyclotella sp. cell suspension under controlled conditions. The bioreactor vessel was composed of a glass column with a 10.5 cm inner diameter, 4.8 mm wall thickness, and 70.5 cm height to provide a total volume of 6 L and a working volume of 5 L. The glass column was mounted onto two stainless steel support plates at the base and top. The baseplate assembly contained a stainless steel sparge plate consisting of four 1.0 mm diameter holes on a 3.6 cm square pitch. Pressurized house air was particulate filtered, metered through a flowmeter, sterile filtered at 0.2 µm, and introduce to the baseplate.
The headplate assembly contained 8 ports, including a fresh medium delivery port, thermocouple port, a 4.6 mm sampling tube, 11 mm. D.O. electrode port, two air outlet ports, and 9.5 mm outer diameter by 1.09 m long stainless steel internal U-tube heat exchanger. Controlled sampling of the liquid suspension within the vessel was accomplished by pressurizing the vessel headspace and collecting the liquid in a sterile culture bottle. Water from a temperaturecontrolled chilling circulator was pumped through the internal heat exchanger to provide constant temperature within the bioreactor vessel. The bioreactor was externally illuminated by six 20 W cool fluorescent lamps 57 cm in length.
The lamps were vertically positioned in a hexagonal array approximately 1-2 cm from the vessel surface. The lamps were connected to a timer to control the photoperiod. The incident light flux was measured with a LI-COR SA 190
PAR quantum sensor.
10
Figure 7. Schematic of photobioreactor
Two-Stage Photobioreactor Cultivation without Controlled Silicon Delivery
A control experiment was performed in which silicon delivery was not controlled but was instead added as a single charge. Medium composition for cultivation of Cyclotella sp. in the photobioreactor was identical to the flask culture with the exception of the dissolved silicon concentration. Flask
11 cultured diatom cells harvested 14-21 days after subculture were used for bioreactor inoculation.
In Stage I of the two-stage photobioreactor cultivation process, culture growth was designed for silicon depletion. Sterile transfer of a defined inoculum volume targeted a cell density of 2 - 3 x10
5
cells/mL and the initial dissolved silicon concentration (as soluble Na
2
SiO
3
) was targeted to provide 1.6 x 10
6 cells/mL, or three cell doublings. After the soluble silicon was consumed and achieved an unchanging near-zero concentration, the culture was kept in a silicon-starved state at constant stationary phase cell density for at least 24 hours. At the point of silicon starvation, and after at least 24 hours at constant cell density, Stage I was complete. The reactor volume was then drawn down to approximately 2 L. During Stage I, cell suspension was sampled at 24 hour intervals for determination of cell number density, pH, and dissolved silicon concentration. Cell suspension was sampled at 72 hour intervals for total lipid content.
Stage II began with a rapid charge of 2 L medium with composition identical to Stage I with the exception of the dissolved silicon concentration. In Stage II the dissolved silicon concentration was calculated to provide sufficient silicon for one cell division based on the cell density at the end of Stage I. During
Stage II, the culture was sampled twice during the lighted portion of the photoperiod throughout the perfusion and at 24 hour intervals after the completion of perfusion for cell number density, pH, dissolved silicon concentration and total lipid content.
Two-Stage Photobioreactor Cultivation with Controlled Silicon Delivery
Medium composition for cultivation of Cyclotella sp. in the photobioreactor was identical to the flask culture with the exception of the dissolved silicon
12 concentration. Flask cultured diatom cells harvested 14-21 days after subculture were used for bioreactor inoculation.
In Stage I of the two-stage photobioreactor cultivation process, culture growth was designed for silicon depletion. Sterile transfer of a defined inoculum volume targeted a cell density of 2 - 3 x10
5
cells/mL and the initial dissolved silicon concentration (as soluble Na
2
SiO
3
) was targeted to provide 1.6-2.4 x
10
6
cells/mL, or 3 or 4 cell doublings. After the soluble silicon was consumed and achieved an unchanging near-zero concentration, the culture was kept in a silicon-starved state at constant stationary phase cell density for at least 24 hours. At the point of silicon starvation, and after at least 24 hours at constant cell density, Stage I was complete. The reactor volume was then drawn down to approximately 2 L. During Stage I, cell suspension was sampled at 24 hour intervals for determination of cell number density, pH, and dissolved silicon concentration. Cell suspension was sampled at 72 hour intervals for total lipid content.
Stage II began with a perfusion of 2 L medium with composition identical to
Stage I with the exception of the dissolved silicon concentration. The dissolved silicon concentration was calculated to provide sufficient silicon for one cell division based on the cell density at the end of Stage I. The medium was perfused for 48, 72, and 96 hour (depending on the experimental design) using a peristaltic pump. During Stage II, the culture was sampled twice during the lighted portion of the photoperiod throughout the perfusion and at
24 hour intervals after the completion of perfusion for cell number density, pH, dissolved silicon concentration and total lipid content.
Culture Suspension Assays
To determine cell number density a 0.1 mL sample of the cell suspension was diluted in 10.0 mL of Isoton II electrolyte solution (Beckman-Coulter). Cells
13 in the cell suspension were counted on the Beckman Z2 Coulter Counter at a threshold of 6 µm (duplicate assay). Immediately following cell count, the cell suspension was centrifuged at 1000 rpm for 15 minutes to separate the cell mass from the filtrate. During Stage I and Stage II, at time points designated for measurement of total lipid content, the cell mass was lyophilized and stored at -20°C. During Stage I, in-process monitoring of dissolved silicon concentration in the filtrate was determined by spectrophotometeric assay at
360 nm following derivatization of 5.0 mL with 0.2 mL 13.3% w/v ammonium molybdate reagent in water and 0.1 mL 13.7% v/v HCl in water (Fanning and
Pilson 1973). Dissolved silicon concentration in filtrate samples from Stage I and Stage II was determined by inductively coupled plasma (ICP) analysis using a Varian (Liberty 150) ICP atomic emission spectrometer. The analysis wavelength for Si is 251.611 nm and the limit of detection for Si in the assay solution was 0.07 mg/L. The nitrate concentration in the medium was assayed using LaMotte nitrate test kit (model NCR 3110). The nitrate concentration was measured spectrophotometrically at 530 nm.
Lipid Extraction from Dry Cell Mass
To isolate the lipids within the cells the cell mass isolated from cell suspension was washed three times with 100 mM sodium chloride and then lyophilized for
12 hours. Once lyophilized, 25.0 mg dry cell mass was homogenized and rehydrated for 12 hours in 5.0 mL 2:1 (v:v) CHCl
3
:MeOH and the lipid extract was separated from the cell mass. Twice more 5.0 mL 2:1 (v:v) CHCl
3
:MeOH was added to the cell mass and vortexed for 60 seconds before allowing the cell mass solids to settle. Once the solids had settled the solvent was separated and all of the extractions were pooled. The pooled lipid extract was combined with 5.0 mL 0.88% (w/v) KCl in H
2
O, vortexed for 60 seconds and then separated again by centrifugation at 1000 rpm for 10 minutes. The lipid extract was then combined with 5.0 mL 1:1 MeOH:H
2
O, vortexed for 60 seconds and then separated by centrifugation at 1000 rpm for 10 minutes.
14
Finally, the lipid extract was dried using anhydrous Na
2
SO
4
, the final volume of extract was recorded and then stored at -20°C. This process is described as a modified Folch method (Ways 1964).
Lipid Extraction from Medium
The medium was extracted to test for the presence of lipids by combining 10.0 mL medium with 1.0 mL methanol. 2.0 mL chloroform was added and the mixture was vortexed for 60 sec then centrifuged at 2000 rpm for 2 minutes to separate. The bottom layer was kept and dried with Na
2
SO
4
. The volume was recorded and then the extract was stored at -20°C.
Total Lipid Weight Percent Analysis
The total lipid content as a weight percent of the dry cell weight in the lipid extract was determined after a reaction with K
2
Cr
2
O
7
. A 0.2 mL aliquot of lipid extract was evaporated under nitrogen and combined with 1.0 mL (2.5 g/L) K
2
Cr
2
O
7
in H
2
SO
4
. The mixture was heated at 100°C for 45 minutes then cooled to room temperature. After cooling, 0.5 mL of the reaction mixture was diluted in 4.5 mL H
2
O. The diluted reaction solution was allowed to cool to room temperature and then assayed spectrophotometrically for total lipids at
350 nm.
Nile Red Fluorescence Determination of Lipids
In addition to direct measurement by isolation of the lipids, the lipid content within the cells themselves can be qualitatively analyzed by staining. A 0.4 mL aliquot of cell suspension of known cell density, usually between 3 x 10
6 and 4 x 10
6
cells/mL, was combined with 0.03 mL 50 µg Nile Red/mL
Acetone. 2.57 mL 25% (v/v) DMSO/H2O was added and the mixture was heated at 40°C for 10 minutes. After the algal cell suspension was stained with Nile red fluorescence was observed using a Leica fluorescent microscope with a 535 nm excitation wavelength and a 610 nm emission wavelength.
15
Images of both the unstained and stained cells were recorded during a twostage cultivation experiment with a 48 hour perfusion of fresh media for a qualitative assessment of changing lipid content within the cells. Images were taken during the early exponential growth phase, late exponential growth phase, stationary phase, mid-perfusion and post-perfusion. The lipids were extracted from samples taken at the same time points and the total lipid content was determined as previously described.
Transesterification of Fatty Acids in Lipid Extracts
Before the fatty acid components of lipids can be analysed by GC, it is necessary to convert them to low molecular weight non-polar derivatives, in this case transesterification to fatty acid methyl esters. A 1.0 mL aliquot of lipid extract was evaporated to dryness with nitrogen. To the dried extract was added 0.5 mL 1% (v/v) H
2
SO
4
/MeOH. The mixture was heated at 100°C for
60 minutes then cooled to room temperature. Once cooled, 0.5 mL H
2
O was added followed by 0.5 mL hexane. The sample was vortexed for 60 seconds then separated by centrifugation at 2000 rpm for 3 minutes. The upper solvent phase was removed by pipette and dried with anhydrous Na
2
SO
4
.
Fatty Acid Identification by GC-MS
The major fatty acid constituents present in the lipid extract were identified by
GC-MS. GC-MS analyses were performed using a Hewlitt Packard HP- 6890
GC-MS instrument equipped with an HP-5 capillary column (30 m × 0.25 mm i.d.; film thickness 0.25 µm). The carrier gas was helium with a flow rate of 1 mL/min and the inlet temperature was 280ºC. The column temperature was initially 40ºC (held for 3 minutes) and then increased to 200ºC at 20ºC min
-1
, held for 10 minutes, and then increased to 210ºC at 10ºC min
-1
. The mass spectrometer was operated in positive EI mode with an ionization energy of 70 eV. Spectra were monitored from 45 to 400 m/z and the fatty acid components were identified by comparison of their mass spectra with reference mass
16 spectra from the American Oil Chemists Society database. Three representative transesterified time point samples were chosen for GC-MS analysis: one sample from Stage I mid-exponential growth phase, one sample from Stage I stationary phase and one sample from Stage II post perfusion. A
1.0 mg/mL methyl nonadecanoate solution (Sigma) in hexane was used as an internal standard.
Fatty Acid Quantification by GC
The major fatty acid constituents present in the lipid extract were quantified by
GC. GC analyses were performed using an HP5890 GC instrument, equipped with an HP-5 capillary column (30m × 0.32 mm i.d.; film thickness 0.25 µm).
The carrier gas was helium and the inlet temperature was 280ºC. The column temperature was initially 40ºC (held for 3 minutes) and then increased to
200ºC at 20ºC min
-1
, held for 10 minutes, and then increased to 210ºC at 10ºC min
-1
. Three representative transesterified time point samples from 48 hour and 72 hour perfusion experiments were chosen for GC-MS analysis: one sample from Stage I mid-exponential growth phase, one sample from Stage I stationary phase and one sample from Stage II post perfusion. 1 mg/mL methyl nonadecanoate (Sigma) in hexane was used as an internal standard.
The area counts were converted to concentrations using external calibration curves.
Results
Two-Stage Photobioreactor Cultivation without Controlled Silicon Delivery
A two-stage photobioreactor cultivation process without controlled silicon limitation was used as a control experiment. In Stage I, soluble silicon was added to the cell suspension culture and the culture was grown up to silicon starvation. In Stage II soluble silicon was added in one charge. The cultivation
17 conditions for Stages I and II are provided in Table 1. Refer to Appendix B for tabulated data.
Table 1. Two-stage photobioreactor cultivation with charge addition
Run ID #:
Date Started:
Time Started:
Date Ended:
Experiment Duration:
Run Description
Experimental Parameters
Cy-JA-05
12/5/08
16:00
12/22/08
404 hr
Temperature Setpoint: 22.0 °C
Incident Light Intensity: 150 µE
Light Distance From Vessel Surface: 10 mm
Light Period : 14 h
Dark Period:
Reactor Type:
Working Liquid Volume
Air sparger:
Air flow rate:
10 h
5 L Bubble Column
4 L
Stainless Perforated Plate (4 1mm holes)
1000 mL/min
Culture Loading
Culture:
Age of Inoculum:
Base Medium:
Sterilization Method:
Medium Volume:
Inoculum Volume:
Initial Cell Density:
Initial Si Concentration:
Cyclotella sp.
12 days
Harrison’s Artificial Seawater
Autoclave
4000 mL
240 mL
1.56 ± 0.01 x 10
5
cells/mL
0.35 ± 0.01 mmol/L
Stage II Summary
Experiment Day Perfusion Started:
Date Charge Added:
Volume of Charged Media:
Initial Cell Density:
Si Concentration in Charged Media:
Day 12
12/17/08
1900 mL
1.60 ± 0.02 x 10
6
cells/mL
0.27 ± 0.01 mmol/L
18
19
Total number of cells and dissolved silicon concentration in the culture medium vs. cultivation time for Stages I and II for the control experiment are presented in Figure 8 (a). In Stage I, the amount of silicon added to the culture was calculated to provide enough silicon for three cell doublings, so that at an initial cell number density of 2.0 × 10
5
cells/mL yielded a nominal final cell number density of 1.6 × 10
6
cells/mL. In Stage I of cultivation, silicon consumption was correlated to growth, and the cell number density increase over time was proportional to the decrease in dissolved silicon concentration. When the cells achieved silicon starvation and the dissolved silicon concentration in culture medium was depleted, the cell number density leveled off and then became constant. Silicon starvation was defined as the cultivation state in which cell number density and the dissolved silicon concentration were constant for at least two photoperiods (48 hours). Figure 8
(b) presents total number of cells and pH vs. cultivation time. The start of each photoperiod is denoted on the graph by a faint dotted line. In Stage I of cultivation, the pH rose from 8.4 at inoculation to 9.6 at the end of stationary phase, as seen in Figure 8 (b), with pH values cyclically decreasing during dark periods and rising over the course of light periods. Rising levels of pH indicate greater consumption of CO
2
by the cells. The variation in pH with photoperiod was due to a shift in the carbon equilibrium. The carbon equilibrium can be described by (Lobban 1985):
CO
2
H
2
O H
2
CO
3
HCO
3
H CO
3
2
H HCO
3
CO
2
OH
During the light period CO
2
was consumed by photosynthesis. Consumption of CO
2
caused a shift in the carbon equilibrium resulting in the dissociation of carbonate and the release of hydroxyl groups. During the dark period, CO
2 was released due to respiration processes such as the oxidation of sugars in the energy pathways of glycolysis and the Krebs cycle. The released CO
2
reacted
20 with free hydroxyl group to form carbonate, reducing the pH. Rising levels of pH with cultivation time indicates greater consumption of CO
2
by the cells.
In Stage II of cultivation, a charge of fresh media with soluble silicon was added to the silicon-starved culture 5 hours into the light period. The cultivation was allowed to proceed for an additional 114 hours. The amount of silicon added to the Stage II culture provided enough silicon for one cell number doubling. In Stage II, the silicon concentration spiked to an initial concentration almost equal to the concentration within the perfusion media itself. There was no controlled addition in this experiment but instead one large addition of media.
5.0E+09
4.0E+09
3.0E+09
2.0E+09
1.0E+09
0.0E+00
0
Cells
Si no Si uptake
0.4
0.2
100 200 300
Time (h)
400 500
0.0
21
Figure 8. Charge addition Cy-JA-05 (a) Total number of cells and Si vs. cultivation time (b) Total number of cells and pH vs. cultivation time
Note: Light vertical lines on Figures 8 (b) and (d) represent the photoperiod while heavy vertical lines represent the charge addition of medium.
0.6
1,400
1,200
1,000
800
600
400
200
0
0
Cells
Si
0.4
0.2
100 200
Time (h)
300 400 500
0.0
22
Figure 9. Charge addition - Exp Cy-JA-06 (c) Total number of cells and Si vs. cultivation time (d) Total number of cells and pH vs. cultivation time
Note: Light vertical lines on Figures 8 (b) and (d) represent the photoperiod while heavy vertical lines represent the charge addition of medium.
100 0.4
80
60
Lipids
Si no Si uptake 0.3
0.2
40
0.1
20
0
0 500
0.0
100 200 300
Time (hr)
400
1.2
1.0
0.8
0.6
DCW
Lipids
0.4
0.2
0.0
0 100 200
Time (h)
300 400 500
Figure 10. Charge addition – Cy-JA-05 (a) Total Lipids and Si vs. cultivation time (b) Total DCW and Total Lipid productivity vs. cultivation time
23
24
100
80
60
40
20
0
0
4.0
3.0
2.0
125
DCW
Lipids
Lipids
250
Time (hr)
375
0.6
0.4
0.2
500
0.0
1.0
0.0
0 125 250
Time (h)
375 500
Figure 11. Charge addition – Exp Cy-JA-06 (a) Total Lipids and Si vs. cultivation time (b) Total DCW and Total Lipid productivity vs. cultivation time
Two-Stage Photobioreactor Cultivation With Controlled Silicon Delivery
A two-stage photobioreactor cultivation process with controlled silicon limitation was used to increase lipid content within the cells of Cyclotella sp. while simultaneously generating biomass. In Stage I, soluble silicon was added to the cell suspension culture and the culture was grown up to silicon starvation. In Stage II, soluble silicon was added by perfusion to the siliconstarved culture. The cultivation conditions for Stages I and II are provided in
Tables 2, 3 and 4 for 48, 72 and 96 hour perfusions, respectively. Refer to
Appendix B for raw data.
25
Table 2. Two-stage photobioreactor cultivation with 48 h perfusion
Run ID #:
Date Started:
Time Started:
Date Ended:
Experiment Duration:
Run Description
Experimental Parameters
Cy-JA-10
06/06/09
17:00
06/26/09
474 hr
Temperature Setpoint: 22.0 °C
Incident Light Intensity: 150 µE
Light Distance From Vessel Surface: 10 mm
Light Period :
Dark Period:
Reactor Type:
Working Liquid Volume
Air sparger:
Air flow rate:
14 h
10 h
5 L Bubble Column
4 L
Stainless Perforated Plate (4 1mm holes)
Culture Loading
1000 mL/min
Culture:
Age of Inoculum:
Base Medium:
Sterilization Method:
Medium Volume:
Inoculum Volume:
Initial Cell Density:
Cyclotella sp.
12 days
Harrison’s Artificial Seawater
Autoclave
4400 mL
550 mL
3.81 ± 0.31 x 10
5
cells/mL
Initial Si Concentration:
Initial Nitrate Concentration:
0.42 ± 0.01 mmol/L
9.71 ± 0.80 mmol/L
Stage II Summary
Experiment Day Perfusion Started:
Date Perfusion Started:
Time Perfusion Started
Date Perfusion Stopped:
Time Perfusion Stopped:
Length of Perfusion:
Volume of Perfusion Media:
Perfusion Flow Rate:
Initial Cell Density:
Si Concentration in Perfusion Media:
Initial Nitrate Concentration:
Nitrate Concentration at t = 379 h:
Day 14
06/20/09
11:00
6/22/09
11:00
48 h
1900 mL
39.6 mL/h
2.12 ± 0.50 x 10
6
cells/mL
0.65 ± 0.01 mmol/L
3.22 ± 0.23 mmol/L
3.80 ± 0.20 mmol/L
26
Table 3. Two-stage photobioreactor cultivation with 72 h perfusion
Run ID #:
Date Started:
Time Started:
Date Ended:
Experiment Duration:
Run Description
Experimental Parameters
Cy-JA-12
7/28/09
11:40
8/25/09
624 h
Temperature Setpoint: 22.0 °C
Incident Light Intensity: 150 µE
Light Distance From Vessel Surface: 10 mm
Light Period : 14 h
Dark Period:
Reactor Type:
Working Liquid Volume
Air sparger:
Air flow rate:
10 h
5 L Bubble Column
4 L
Stainless Perforated Plate (4 1mm holes)
1000 mL/min
Culture Loading
Culture:
Age of Inoculum:
Base Medium:
Sterilization Method:
Medium Volume:
Inoculum Volume:
Initial Cell Density:
Initial Si Concentration:
Initial Nitrate Concentration:
Stage II Summary
Cyclotella sp.
14 days
Harrison’s Artificial Seawater
Autoclave
4400 mL
375 mL
2.06 ± 0.18 x 10
5
cells/mL
0.46 ± 0.04 mmol/L
9.51 ± 1.08 mmol/L
Experiment Day Perfusion Started:
Date Perfusion Started:
Time Perfusion Started
Date Perfusion Stopped:
Time Perfusion Stopped:
Length of Perfusion:
Volume of Perfusion Media:
Perfusion Flow Rate:
Initial Cell Density:
Si Concentration in Perfusion Media:
Initial Nitrate Concentration:
Nitrate Concentration at t = 504 h:
21
8/19/09
11:00
8/22/09
11:00
72 h
1900 mL
26.4 mL/h
1.61 ± 0.15 x 10
6
cells/mL
0.76 ± 0.01 mmol/L
3.18 ± 0.29 mmol/L
3.82 ± 0.25 mmol/L
27
Table 4. Two-stage photobioreactor cultivation with 96 h perfusion
Run ID #:
Date Started:
Time Started:
Date Ended:
Experiment Duration:
Run Description
Cy-JA-14
01/05/2010
2:00 pm
02/10/2010
676 h
Experimental Parameters
Temperature Setpoint: 22.0 °C
Incident Light Intensity: 150 µE
Light Distance From Vessel Surface: 10 mm
Light Period : 14 h
Dark Period:
Reactor Type:
Working Liquid Volume
Air sparger:
Air flow rate:
10 h
5 L Bubble Column
4 L
Stainless Perforated Plate (4 1mm holes)
1000 mL/min
Culture Loading
Culture:
Age of Inoculum:
Base Medium:
Sterilization Method:
Medium Volume:
Inoculum Volume:
Initial Cell Density:
Initial Si Concentration:
Initial Nitrate Concentration:
Stage II Summary
Cyclotella sp.
14 days
Harrison’s Artificial Seawater
Autoclave
4400 mL
425 mL
2.75 ± 0.18 x 10
5
cells/mL
0.45 ± 0.05 mmol/L
10.00 ± 0.18 mmol/L
Experiment Day Perfusion Started:
Date Perfusion Started:
Time Perfusion Started
Date Perfusion Stopped:
Time Perfusion Stopped:
Length of Perfusion:
Volume of Perfusion Media:
Perfusion Flow Rate:
Initial Cell Density:
Si Concentration in Perfusion Media:
Initial Nitrate Concentration:
Nitrate Concentration at t = 504 h:
21
8/19/09
11:00
8/22/09
11:00
96 h
2000 mL
26.4 mL/h
1.61 ± 0.15 x 10
6
cells/mL
0.76 ± 0.01 mmol/L
3.18 ± 0.29 mmol/L
3.82 ± 0.25 mmol/L
28
29
48 hour perfusion
Total number of cells and dissolved silicon concentration in the culture medium vs. cultivation time for Stages I and II for the 48 hour perfusion experiment are presented in Figure 10 (a). In Stage I, the amount of silicon added to the culture was calculated to provide enough silicon for three cell doublings, so that at an initial cell number density of 3.0 × 10
5
cells/mL yielded a nominal final cell number density of 2.4 × 10
6
cells/mL. In Stage I of cultivation, silicon consumption was correlated to growth, and the cell number density increase over time was proportional to the decrease in dissolved silicon concentration. When the cells achieved silicon starvation and the dissolved silicon concentration in culture medium was depleted, the cell number density leveled off and then became constant. Silicon starvation was defined as the cultivation state in which cell number density and the dissolved silicon concentration were constant for at least two photoperiods (48 hours).
Figure 10 (b) presents the total number of cells and pH vs. cultivation time.
The start of each photoperiod is denoted on the graph by a faint dotted line. In
Stage I of cultivation, the pH rose from 8.6 at inoculation to 9.6 at the end of stationary phase, as seen in Figure 6 (b), with pH values cyclically decreasing during dark periods and rising over the course of light periods.
Rising levels of pH with cultivation time indicates greater consumption of CO
2
by the cells.
In Stage II of cultivation, perfusion of fresh media with soluble silicon by peristaltic pump was added to the silicon-starved culture 3 hours into the light period and continued perfusing for 48 hours. The cultivation was allowed to proceed for an additional 95 hours. The amount of silicon added to the Stage II culture provided enough silicon for one cell number doubling. However, due to sampling and reactor volume constraints, the experiment was terminated prior to achieving a full cell doubling. In Stage II, the silicon concentration
remained at the same silicon-depleted concentrations as were achieved at the end of Stage I and maintained within the cell culture a silicon-starved state.
30
31
1.4E+10
1.2E+10
1.0E+10
8.0E+09
6.0E+09
4.0E+09
2.0E+09
0.0E+00
0
Cells
Si
100 200 300
Time (hr)
400
1.0
0.8
0.6
0.4
0.2
500
0.0
1.4E+10
1.2E+10
Cells pH
11.0
10.5
1.0E+10
10.0
8.0E+09
6.0E+09
9.5
9.0
4.0E+09
2.0E+09
0.0E+00
500
8.5
8.0
0 100 200 300
Time (hr)
400
Figure 12. 48 h perfusion Cy-JA-10 (a) Total number of cells and Si vs. time,
(b) Total number of cells and pH vs. time
Note: Light vertical lines on Figures 10 (b) and (d) represent the photoperiod while heavy vertical lines represent the charge addition of medium.
32
1.4E+10
1.2E+10
1.0E+10
8.0E+09
6.0E+09
4.0E+09
2.0E+09
0.0E+00
0
Cells
Si
600
1.0
0.9
0.8
0.7
0.6
0.5
0.4
0.3
0.2
0.1
0.0
200
Time (hr)
400
1.4E+10
1.2E+10
1.0E+10
8.0E+09
6.0E+09
4.0E+09
2.0E+09
0.0E+00
Cell s
600
10
9.8
9.6
9.4
9.2
9
8.8
8.6
8.4
8.2
8
0 200
Time (hr)
400
Figure 13. 48 h perfusion – Cy-JA-09 (c) Total number of cells and Si vs. time,
(d) Total number of cells and pH vs. time
Note: Light vertical lines on Figures 10 (b) and (d) represent the photoperiod while heavy vertical lines represent the charge addition of medium.
100
80
Lipids
Si
No Si uptake
1.0
0.8
60
40
0.6
0.4
20 0.2
0
0 500
0.0
100 200 300
Time (hr)
400
4.0
3.5
3.0
2.5
2.0
1.5
1.0
0.5
0.0
0
Lipids
DCW
100 200 300
Time (hr)
400 500
Figure 14. 48 h perfusion Cy-JA-10 (a) Total lipids (wt %/DCW) and Si concentration vs. time, (b) Total DCW productivity and total lipids productivity vs. time
33
34
100
75
50
25
0
0
Lipids
Si
125 250
Time (hr)
375
6.0
5.0
4.0
3.0
2.0
1.0
0.0
0
DCW
Lipids
125 250
Time (hr)
375 500
Figure 15. 48 h perfusion Cy-JA-09 (c) Total lipids (wt %/DCW) and Si concentration vs. time, (d) Total DCW productivity and total lipids productivity vs. time
500
0.6
0.5
0.4
0.3
0.2
0.1
0.0
1.0
0.9
0.8
0.7
35
72 hour perfusion
Total number of cells and dissolved silicon concentration in the culture medium vs. cultivation time for Stages I and II for the 72 hour perfusion experiment are presented in Figure 12 (a). In Stage I, the amount of silicon added to the culture was calculated to provide enough silicon for three cell doublings, so that at an initial cell number density of 2.0 × 10
5
cells/mL yielded a nominal final cell number density of 1.6 × 10
6
cells/mL. As with the
48 hour experiment, in Stage I of cultivation, silicon consumption was correlated to growth, and the cell number density increase over time was proportional to the decrease in dissolved silicon concentration. When the cells achieved silicon starvation and the dissolved silicon concentration in culture medium was depleted, the cell number density leveled off and then became constant. Silicon starvation was defined as the cultivation state in which cell number density and the dissolved silicon concentration were constant for at least two photoperiods (48 hours).
Figure 12 (b) presents total number of cells and pH vs. cultivation time. In
Stage I of cultivation, the pH rose from 8.3 at inoculation to 9.42 at the end of stationary phase, as seen in Figure 12 (b). The start of each photoperiod is denoted on the graph by a faint dotted line.
Rising levels of pH with cultivation time indicate greater consumption of CO
2
by the cells.
In Stage II of cultivation, perfusion of fresh media with soluble silicon by peristaltic pump was added to the silicon-starved culture 3 hours into the light period and continued perfusing for 72 hours. The cultivation was allowed to proceed for an additional 94 hours. The amount of silicon added to the Stage II culture provided for one more cell number doubling. In Stage II, the silicon concentration remained at the same silicon-depleted concentrations as were
achieved at the end of Stage I and maintained within the cell culture a siliconstarved state.
36
37
1.2E+10 1.0
1.0E+10
Cells
Si 0.8
8.0E+09
6.0E+09
0.6
0.4
4.0E+09
2.0E+09
0.2
0.0E+00
0 200 400
Time (hr)
600
0.0
1.6E+10
1.4E+10
1.2E+10
1.0E+10
8.0E+09
6.0E+09
4.0E+09
2.0E+09
0.0E+00
0
Cells pH
600
10.0
9.5
9.0
8.5
8.0
200 400
Time (hr)
Figure 16. 72 h perfusion Cy-JA-12 (a) Total number of cells and Si vs. time,
(b) Total number of cells and pH vs. time
Note: Light vertical lines on Figures 12 (b) and (d) represent the photoperiod while heavy vertical lines represent the charge addition of medium.
38
1.2E+10
1.0E+10
8.0E+09
6.0E+09
4.0E+09
2.0E+09
0.0E+00
0
Cells
200 400
Time (hr)
600
1.2E+10
1.0E+10
8.0E+09
6.0E+09
4.0E+09
2.0E+09
0.0E+00
0
Cells pH
200 600
Figure 17. 72 h perfusion Cy-JA-11 (c) Total number of cells and Si vs. time,
(d) Total number of cells and pH vs. time
Note: Light vertical lines on Figures 12 (b) and (d) represent the photoperiod while heavy vertical lines represent the charge addition of medium.
10
9.5
9
8.5
11.5
11
10.5
8
1.0
0.8
0.6
0.4
0.2
0.0
100
80
60
40
20
0
0
Lipids
Si no Si uptake
200 400
Time (hr)
600
6
5
4
3
2
1
0
0
DCW
Lipids
200 400
Time (hr)
600 800
Figure 18. 72 h perfusion Cy-JA-12 (a) Total lipids (wt%/DCW) and Si vs. time, (b) Total DCW productivity and Total Lipid productivity vs. time
0.2
800
0.0
1.0
0.8
0.6
0.4
39
40
100
80
60
Lipids
1.0
0.8
0.6
40
20
0.4
0.2
0
0 800
0.0
200 400
Time (hr)
600
6.0
5.0
4.0
3.0
2.0
DCW
Lipids
1.0
0.0
0 200 400
Time (hr)
600 800
Figure 19. 72 h perfusion Cy-JA-11 (c) Total lipids (wt%/DCW) and Si vs. time, (d) Total DCW productivity and Total Lipid productivity vs. time
41
96 hour perfusion
Total number of cells and dissolved silicon concentration in the culture medium vs. cultivation time for Stages I and II for the 96 hour perfusion experiment are presented in Figure 14 (a). In Stage I, the amount of silicon added to the culture was calculated to provide enough silicon for three cell doublings, so that at an initial cell number density of ~3.0 × 10
5
cells/mL yielded a nominal final cell number density of 2.4 × 10
6
cells/mL. As with the
48 & 72 hour experiments, in Stage I of cultivation, silicon consumption was correlated to growth, and the cell number density increase over time was proportional to the decrease in dissolved silicon concentration. When the cells achieved silicon starvation and the dissolved silicon concentration in culture medium was depleted, the cell number density leveled off and then became constant. Silicon starvation was defined as the cultivation state in which cell number density and the dissolved silicon concentration were constant for at least two photoperiods (48 h).
Figure 14 (b) presents total number of cells and pH vs. cultivation time. In
Stage I of cultivation, the pH rose from 8.3 at inoculation to 9.05 at the end of stationary phase, as seen in Figure 14 (b). The start of each photoperiod is denoted on the graph by a faint dotted line. Rising levels of pH with cultivation time indicate greater consumption of CO
2
by the cells.
In Stage II of cultivation, perfusion of fresh media with soluble silicon by peristaltic pump was added to the silicon-starved culture 3 hours into the light period and continued perfusing for 96 hours. The cultivation was allowed to proceed for an additional 240 hours. The amount of silicon added to the Stage
II culture provided for one more cell number doubling. In Stage II, the silicon concentration did not remain at the same silicon-depleted concentrations as
were achieved at the end of Stage I but instead rose steadily throughout the perfusion and then began to decrease once the perfusion had ended.
42
43
2.5E+10
2.0E+10
1.5E+10
1.0E+10
5.0E+09
0.0E+00
0
Cells
Si
200 400
Time (hr)
600
1.00
0.80
0.60
0.40
0.20
800
0.00
2.5E+10
2.0E+10
1.5E+10
Cells pH
12.0
11.0
10.0
1.0E+10
5.0E+09
9.0
0.0E+00 8.0
0 200 600
Figure 20. 96 h perfusion – Cy-JA-14 (a) Total number of cells and Si vs. time,
(b) Total number of cells and pH vs. time
Note: Light vertical lines on Figures 12 (b) and (d) represent the photoperiod while heavy vertical lines represent the charge addition of medium.
44
1.8E+10
1.6E+10
1.4E+10
1.2E+10
1.0E+10
8.0E+09
6.0E+09
4.0E+09
2.0E+09
0.0E+00
0
Cells
Si
200 400
Time (hr)
600
0.40
0.20
800
0.00
1.00
0.80
0.60
2.5E+10
2.0E+10
1.5E+10
1.0E+10
5.0E+09
Cells pH
12.5
12.0
11.5
11.0
10.5
10.0
9.5
9.0
8.5
8.0
0.0E+00
0 200 400
Time (hr)
600
Figure 21. 96 h perfusion Cy-JA-13 (c) Total number of cells and Si vs. time,
(d) Total number of cells and pH vs. time
45
Note: Light vertical lines on Figures 12 (b) and (d) represent the photoperiod while heavy vertical lines represent the charge addition of medium.
46
100
80
60
40
20
0
0
Lipids
Si no Si uptake
200 400
Time (hr)
600
6
5
4
DCW
Lipids
3
2
1
0
0 200 400
Time (hr)
600 800
Figure 22. 96 h perfusion Cy-JA-14 (a) Total lipids (wt%/DCW) and Si vs. time, (b) Total DCW productivity and Total Lipid productivity vs. time
1.0
0.8
0.6
0.4
0.2
800
0.0
47
100
80
60
40
20
0
0
Lipids
Si
200 400
Time (hr)
600
1.00
0.80
0.60
0.40
0.20
800
0.00
6.0
5.0
4.0
3.0
2.0
1.0
0.0
0
DCW
Lipids
200 400
Time (hr)
600 800
Figure 23. 96 h perfusion Cy-JA-13 (c) Total lipids (wt%/DCW) and Si vs. time, (d) Total DCW productivity and Total Lipid productivity vs. time
48
Silicon Concentration and Total Lipid Weight Percent
Charge addition
The total lipid concentration, as a percentage of the biomass dry cell weight, and dissolved silicon concentration for Stages I and II are presented in Figure 9
(a). At inoculation, when silicon concentration measured 0.35 mmol Si/L, total lipid concentration was 9% of the dry cell weight. As the cells consumed the silicon and divided, silicon was depleted and the cells became silicon-starved.
Once silicon was depleted and cells became silicon-starved the total lipid concentration within the cells began to increase. By the end of Stage I, total lipid concentration had reached 47% of the dry cell weight. Stage II began with a charge of fresh media into the system and total lipid concentrations decreased to a low of 34% by the time Stage II was ended. The dotted line on
Figure 9 (a) represents the silicon concentration profile if there was no cellular uptake.
48 hour perfusion experiment
The total lipid concentration, as a percentage of the biomass dry cell weight, and dissolved silicon concentration in the culture medium vs. cultivation time for Stages I and II are presented in Figure 11 (a). At inoculation, when silicon concentration measured 0.42 mmol Si/L, total lipid concentration was 12% of the dry cell weight. As the cells consumed the silicon and divided, silicon was depleted and the cells became silicon-starved. Once silicon was depleted and cells became silicon-starved the total lipid concentration within the cells began to increase. By the end of Stage I, total lipid concentration had reached 43% of the dry cell weight. Stage II began with perfusion of fresh media into the system and total lipid concentrations decreased to a low of 32%. The addition of silicon by perfusion was controlled such that the silicon concentration remained in a state of depletion. The dotted line on Figure 11 (a) represents the silicon concentration profile if there was no cellular uptake. The total lipid
49 concentration began to increase ~71 hours after the start of Stage II and ~23 hours after perfusion ceased. The total lipid concentration reached 40% by the time Stage II was ended.
72 hour perfusion experiment
The total lipid concentration, as a percentage of the biomass dry cell weight, and dissolved silicon concentration in the culture medium vs. cultivation time for Stages I and II are presented in Figure 13 (a). At inoculation, when silicon concentration measured 0.48 mmol Si/L, total lipid concentration was 10% of the dry cell weight. As the cells consumed the silicon and divided, silicon was depleted and the cells became silicon-starved. Once silicon was depleted and cells became silicon-starved the total lipid concentration within the cells began to increase. By the end of Stage I, total lipid concentration had reached 46% of the dry cell weight. Stage II began with perfusion of fresh media into the system and total lipid concentrations decreased to a low of 35%. The addition of silicon by perfusion was controlled such that the silicon concentration remained in a state of depletion. The dotted line on Figure 13 (a) represents the silicon concentration profile if there was no cellular uptake. The total lipid concentration began to increase ~71 hours after the start of Stage II and ~23 hours after perfusion ceased. The total lipid concentration reached 48% by the time Stage II was ended.
96 hour perfusion experiment
The total lipid concentration, as a percentage of the biomass dry cell weight, and dissolved silicon concentration in the culture medium vs. cultivation time for Stages I and II are presented in Figure 15 (a). At inoculation, when silicon concentration measured 0.45 mmol Si/L, total lipid concentration was 20% of the dry cell weight. As the cells consumed the silicon and divided, silicon was depleted and the cells became silicon-starved. Once silicon was depleted and
50 cells became silicon-starved the total lipid concentration within the cells began to increase. By the end of Stage I, total lipid concentration had reached 34% of the dry cell weight. Stage II began with perfusion of fresh media into the system and total lipid concentrations decreased to a low of 19% by the end of the perfusion. In this experiment the lipids did not increase as dramatically as with shorter lengths of perfusion. Also, the silicon concentration did not remain at silicon-depleted concentrations but instead began to increase ~45 hours into the perfusion. The dotted line on Figure 15 (a) represents the silicon concentration profile if there was no cellular uptake.
Biomass and Lipid Productivity
As cell growth proceeds during two-stage cultivation, total biomass productivity and total lipid productivity can be determined. Total biomass productivity (as dry cell weight) and total lipid productivity vs. cultivation time for a 4 L bioreactor for each experiment are presented in Table
Table 5. Dry cell weight and Lipid productivity
Experiment
Charge addn
DCW, initial
(g)
0.16 ± 0.01
DCW, final
(g)
1.07 ± 0.01
Lipid, initial
(g)
0.01 ± 0.07
Lipid, final
(g)
0.40 ± 0.07
48 h perfusion 0.47 ± 0.01 3.17 ± 0.01 0.03 ± 0.07 1.50 ± 0.01
72 h perfusion 0.25 ± 0.02 3.47 ± 0.09 0.02 ± 0.01 1.70 ± 0.05
96 h perfusion 0.33 ± 0.12 5.47 ± 0.06 0.08 ± 0.01 0.89 ± 0.01
Due to varying initial cell densities a direct comparison of the dry cell weight and lipid productivities is not possible. However, it is possible to compare the relative changes for each experiment. The increase in biomass for each experiment was comparable, based on the scales of the individual experiments.
The charge addition and 96 hour perfusion show only minor increases in lipid
51 productivity compared the increases in productivity for 48 and 72 hour perfusion.
Nile Red Fluorescence Determination of Lipids
Figure 12 presents a representative two-stage cultivation scheme, with 48 hour perfusion, used to generate cells for observation of intracellular lipids over the course of cultivation. Images were captured at the points represented by large triangles on the cells data line.
4.5E+09
4.0E+09
3.5E+09
3.0E+09
2.5E+09
2.0E+09
1.5E+09
1.0E+09
5.0E+08
0.0E+00
0
Cells
Si
100 200
Time (hr)
300
Figure 24. Representative cultivation with 48 h perfusion
Figures 17 (a) – (j) demonstrate the qualitative analysis of lipid content that is possible by staining living cells with Nile red and then observing intracellular lipids with fluorescent microscopy. The samples include three time points in
Stage I (early exponential growth phase, mid-exponential growth phase,
1.0
0.8
0.6
0.4
0.2
0.0
52 stationary phase, mid-perfusion) and two time points in stage II (mid-perfusion and shutdown).
(a) t = 26.5 h (b) 15.1 ± 2.0 wt% lipid
(c) t = 168 h (d) 32.6 ± 8.8 wt% lipid
(e) t = 217 h (f) 42 ± 1.5 wt% lipid
Figure 25. (a) – (j) Nile red lipid stain images
(g) t = 266 h (h) 37.3 ± 6.4 wt% lipid
53
(i) t = 340 h (j) 42.5 ± 2.1 wt% lipid
Figure 17. (a) – (j) Continued
It is possible to qualitatively assess the lipid content within the cell by comparing the presence and relative size of lipid vesicles within the cells.
Figure 17.b (t = 26.5 h) image clearly shows a lack of distinct lipid vesicles within the cell. Instead, the cell walls and a few internal membranes, which are comprised of lipids, are stained. Figures 17.d, f, h and j all show significant lipid increases when compared with Figure 17.b and clearly exhibit lipid vesicles within the cells.
54
Fatty Acid Composition and Profile
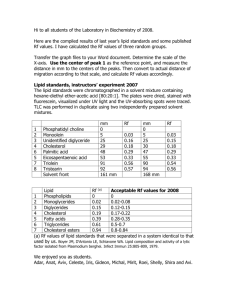
Cyclotella sp. cells harvested at three distinct points in cultivation for both 48 and 72 hour perfusion experiments were analyzed for fatty acid composition.
Those time points are as follows: Stage I mid-exponential growth, Stage I stationary phase, and late Stage II. Seven fatty acid constituents were identified by GC/MS including myristic acid (14:0), hexadecatrienoic acid
(16:3), palmitoleic acid (16:1), palmitic acid (16:0), stearic acid (18:0), octadecanoic acid (18:1), and eicosapentanoic acid (20:5). The fatty acid composition as percentages of total fatty acid are reported in Table 6. Refer to
Appendix D for GC and GC/MS data.
Table 6. Fatty acid composition, 48 and 72 h perfusion experiments
Fatty
Acid
48 h Perfusion
(% of Total Fatty Acids)
Mid-Exp.
Phase
Stationary
Phase
Late
Stage II
72 h Perfusion
(% of Total Fatty Acids)
Mid-Exp.
Phase
Stationary
Phase
Late
Stage II
14:0 5.5 ± 0.1 5.7 ± 0.4 5.7 ± 0.1 5.6 ± 0.1 4.9 ± 0.1 5.3 ± 0.1
16:3
16:1
16:0
18:1
10.7 ± 0.1
36.3 ± 0.4
23.2 ± 0.1
1.2 ± 0.2
8.0 ± 0.1 8.0 ± 0.1 8.0 ± 0.8 7.9 ± 0.3 9.6 ± 0.1
34.4 ± 0.7
24.7 ± 0.6
2.3 ±0.2
34.4 ±
0.5
24.7 ±
0.1
46.8 ±
0.1
24.4 ±
0.6
49.8 ± 0.4
22.9 ± 0.2
2.3 ± 0.1 0.8 ± 0.1 0.8 ± 0.1
39.8 ±
0.2
27.4 ±
0.2 n.d.
18:0
1. 2 ± 0.2 3.3 ±1.0
20:5
12.6 ± 0.3 13.4 ±0.6
3.3 ± 0.1 1.0 ± 0.1 1.6 ± 0.1 4.7 ± 0.1
13.4 ±
0.4
8.3 ± 0.1 8.0 ± 0.1 7.1 ± 0.3 n.d.
not detected, 1 S.D. n = 2
55
Figures 18 and 19 demonstrate the fatty acid components at the previously described time points.
60
Stage I: Mid-Exponential Phase, t = 164 h
Stage I: Stationary phase, t = 308 h
Stage II: End, t = 402 h
40
20
0
14:0 16:3 16:1 16:0
Fatty Acid
18:1 18:0 20:5
60
40
Stage I: Mid-exponential phase, t = 263 h
Stage I: Stationary phase, t = 480 h
Stage II: End, t = 624 h
20
0
14:0 16:3 16:1 16:0
Fatty Acid
18:1 18:0 20:5
Figure 26. Fatty Acid composition during cultivation for a) 48 h perfusion b)
72 h perfusion
56
The predominant fatty acids present in both experiments are palmitoleic acid, palmitic acid and eicosapentanoic acid. The relative amounts of each component remained unchanged over the course of the experiment and thus, remained unchanged regardledss of silicon concentration.
A representative GC chromatogram is presented in Figure 19, with the seven fatty acid peaks indicated. Several minor peaks in Figure 19 not labeled were determined to be impurities in the sample (mass spectra didn’t match with fatty acids).
120000
100000
80000
60000
40000
20000
0
0 5 10 15 20
Time (min)
25 30 35
Figure 27. Representative GC chromatogram
57
Discussion
This study demonstrated that lipid production by Cyclotella sp. can be enhanced using a bioprocess strategy of controlled silicon addition. This was achieved by utilizing a two-stage photobioreactor cultivation scheme in which silicon-starved cells with increased lipid concentrations receive silicon by perfusion at rates low enough to maintain a silicon-depleted state and yet provided sufficient silicon for continued biomass production while maintaining high lipid levels. While it is known that silicon-starvation induces high lipid concentrations within diatoms by driving fatty acid synthesis with increased formation of ACCase (Roessler 1988), this is the first study to actively design and implement a system by which lipid production is enhanced along with lipid concentrations. Prior to this study, investigations into the extent of enhanced lipid production as a consequence of silicon-starvation in batch systems demonstrated lipid content doubling during exponential growth and increasing even further when maintained at silicon starvation by as much as
20% (Shifrin 1981; Roessler 1988; Lombardi 1995) but with biomass limited by the amount of initial silicon provided to the system. Further studies considered two-stage systems in which a second charge addition of silicon was added to the reactor once silicon-starvation had been achieved. These experiments saw additional biomass generation but lipid concentrations decreased as cells began to uptake silicon and divide (Coombs 1967; Taguchi
1987; Lynn 2000). One experiment untilized a turbidostat to deliver a steady flow of silicon at low concentration (Lombardi 1991) and saw increased lipid concentrations but low lipid production overall due to a decrease in cell growth rate.
58
These experiments were similar to the charge-addition control experiment performed in this study, in which a surge uptake mechanism was utilized for silicon replenishment. Lipid concentration decreased as the cellular uptake of silicon resumed and cells doubled, allowing for low lipid productivity and high biomass productivity. By considering an alternative delivery system for silicon more control over the cellular lipid metabolic pathways can be exerted. Using slow perfusion addition of a large-volume, low-silicon-concentration medium the system was maintained at silicon starvation concentrations of silicon.
Since the intracellular concentrations of silicon would have been quite low this new silicon supply was taken up quickly by the starved cells. In both the 48 and 72 hour perfusion experiments the silicon delivery profile without cellular uptake can be seen in Figures 7-a and 9-a. Had the cellular uptake not been rapid, the silicon in the system would have accumulated to significant levels.
The lipid concentrations of both the 48 and 72 hour experiments decreased somewhat during the silicon-uptake and cell division but the overall lipid productivity continued to increase (see Figures 7-b and 9-b) because the biomass continued to increase.
However, the 96 hour perfusion showed a different trend. After the start of the perfusion the cells grew much more slowly and the silicon uptake was slower than previous experiments, allowing for a small increase in system silicon concentration, but not one that approached the no-silicon uptake profile (see
Figure 15-a,b). As a consequence of the slow silicon uptake, lipid concentrations decreased and had not increased by the end of the experiment.
This demonstrates a limit to the length of perfusion, with the rate of silicon addition being too low for the cells in the system. This slow rate of addition would have affected the metabolism of the cells. The cells were unable to respond rapidly, unlike in the 48 and 72 hour experiments, but instead continued slowly removing silicon from the medium until the experiment was shutdown. As a consequence, silicon starvation was not maintained and lipid
59 concentrations decreased from their previously enhanced levels. The increased presence of silicon would have slowed the formation of ACCase and thus fatty acid synthesis. Additionally, fatty acids would be used in cellular components when generating new cells.
While it has been shown that lipid production is affected by silicon concentration, GC analysis of samples taken at various points throughout 48 and 72 hour perfusion experiments showed no changes in lipid composition with respect to silicon availability. The distribution of fatty acids (see Figures
18 and 19) showed no significant change either during perfusion or after when compared with the fatty acid distribution prior to perfusion.
By utilizing a perfusion of silicon and taking advantage of the externally controlled silicon uptake mechanism, a silicon-depleted state was conserved with enhanced lipid productivity while producing biomass. To date, enhanced productivity of lipids in combination with biomass production has not been achieved. As a consequence of this study a new method has been proposed by which to cultivate microalgae for enhanced lipid production along with biomass production. In a comprehensive review of biofuels from microalgae,
Brennan and Owende (2010) concluded that continued development of technologies to optimize microalgae lipid production are needed in order to provide a sustainable biofuel source that can meet large-scale demands costeffectively. This study develops a unique method by which to accomplish this goal.
60
References
Azam, F., Hemmingsen, B.B. & Volcani, B.E. "Role of silicon in diatom metabolism. V. Silicic acid transport and metabolism in the heterotrophic diatom Nitzschia alba." Archives of Microbiology , 1974: 103-14.
Basova, M.M. "Fatty acid composition of lipids in microalgae." International
Journal of Algae , 2005: 33-57.
Cantrell, K.B., Ducey, T., Ro, K.S., & Hunt, P.G. "Livestock waste-tobioenergy generation opportunities." Bioresource Technology , 2008: 7941-
7953.
Chisti, Y. "Biodiesel from microalgae." Biotechnology Advances , 2007: 294-
306.
Christie, William W. Gas Chromatography and Lipids.
1989.
Conway, H.L. & Harrison, P.J. "Marine diatoms grwon in chemostats under silicate or ammonium limitation IV. Transient response of Chaetoceros debilis,
Skeletonema costatum, and Thalassiosira gravida to a single addition of the limiting nutrient." Marine Biology , 1977: 33-43.
Conway, H.L., Harrison, P.J., & Davis, C.O. "Marine diatoms grown in chemostats under silicate or ammonium limitation II. Transient response of
Skeletonema coastatum to a single addition of the limiting nutrient." Marine
Biology , 1976: 187-199.
Coombs, J., Darley, W.M., Holm-Hansen, O., & Volcani, B.E. "Studies on the biochemistry and fine structure of silica shel formation in diatoms. Chemical composition of Navicula pelliculosa during silicon-starvation synchrony."
Plant Physiology , 1967: 1601-1606.
Fanning, K.A., and M.E.Q. Pilson. Anal. Chem.
, no. 45 (1973): 136.
Hildebrand, M., Volcani, B.E., Gassman, W. & Schroeder, J.I. "A gene familyl of silicon transporters." Nature , 1997: 688-689.
61
Hu, Q., Sommerfeld, M., Jarvis, E., Ghirardi, M., Posewitz, M., Seibert, M. &
Darzins, A. "Microalgal triacylglycerols as feestocks for biofuel production: perspectcive and advances." The Plant Journal , 2008: 621-639.
Lobban, C.S., Harrison, P.L., & Duncan, M.J. The Physiological Ecology of
Seaweeds.
Cambridge: Cambridge University Press, 1985.
Lombardi, A.T. & Wangersky, P.J. "Influence of phosphorous and silicon on lipid class production by the marine diatom Chaetoceros gracilis grown in turbidostat cage cultures." Marine Ecology Progress Series , 1991: 39-47.
Lombardi, A.T. & Wangersky, P.J. "Particulate lipid class composition of three marine phytoplankters Chaetoceros gracilis, Isochrysis galbana (Tahiti) and
Dunaliella tertiolecta grown in batch culture." Hydrobiologia , 1995: 1-6.
Lynn, S.G., Kilham, S.S., Kreeger, D.A. & Interlandi, S.J. "Effect on nutrient availability on the biochemical and elemental stoichiometry in the freshwater diatom Stephanodiscus minutuls (Bacillariophyceae)." Journal of Phycology ,
2000: 510-522.
Martin-Jezequel, V., Hildebrand, M. & Brzezinski, M. A. "Silicon metabolism in diatoms: Implications for growth." Journal of Phycology , 2000: 821-840.
Metting, F.B. "Biodiversity and application of microalgae." Journal of
Industrial Microbiology , 1996: 477-489.
Rodolfi, L., Zittelli, G.C., Bassi, N., Padovani, G., Biondi, N., Bonini, G. et al.
"Microaglae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor." Biotechnology and
Bioengineering , 2008: 100-112.
Roessler. "Changes in the activities of various lipid and carbohydrate biosynthetic enzymes in the diatom Cyclotella cryptica in response to silicon deficiency." Archives of Biochemistry and Biophysics , 1988: 521-528.
Roessler, P., Bleibaum, J.L., Thompson,G.A. & Ohlrogge, J.B.
"Characteristics of the gene that encodes acetyl-CoA carboxylase in the diatom
Cyclotella cryptica." Annals of The New York Academy of Sciences , 1994: 250-
256.
Round, F. E., Crawford, R. M.,& Mann, D. G. The Diatoms: Biology and
Morphology of the Genera.
Cambridge: Cambridge University Press, 1990.
62
Searchinger, T., Heimlich, R., Houghton, R.A., Dong, F., Elobeid, A., Fabios,
J. et al. "Use of U.S. croplands for biofels increases greenhouse gases through emissions from land-use change." Science , 2008: 1238-1240.
Sheehan, J., Dunahay, T., Benemann, J. & Roessler, P. A look back at the U.S.
Department of Energy's Aquatic Species Program - Biodiesel from algae.
Report NREL/TP-580-24190, Golden, CO: National Renewable Energy
Laboratory, 1998.
Shifrin, N.S. & Chisholm, S.W. "Phytoplankton lipids: Interspecific differences and effects of nitrate, silicate and light-dark cylces." Journal of
Phycology , 1981: 374-384.
Spolaore, P., Joannis-Cassan, C., Duran, E. & Isambert. A. "Commercial applications of microbiology." Journal of Bioscience and Bioengineering ,
2006: 87-96.
Taguchi, S., Hirata, J.A. & Laws, E.A. "Silicate deficiency and lipid synthesis of marine diatoms." Journal of Phycology , 1987: 260-267. van den Hoek, C., Mann, D. G., & Jahns, H. M. Algae: An Introduction to
Phycology.
Cambridge: Cambridge University Press, 1995.
Verma, N.M., Mehrotra, S., Shukla, A. & Mishra, B.N. "Prospective of biodiesel production utilizing microalgae as the cell factories: A comprehensive discussion." African Journal of Biotechnology , 2010: 1402-
1411.
Ways, P. & Hanahan, D.J. "Characterization and quantification of red cell lipids in normal man." Journal of Lipid Research , 1964: 318-328.
Appendices
63
64
Appendix A- Procedures
Bioreactor Sterilization
Equipment:
Autoclave
Photobioreactor assembly
Photobioreactor cradle
Procedures:
Sparge gas hydrator:
1.
Disconnect the 1 L sparge gas hydrator bottle from the photobioreactor and air lines and empty the water.
2.
Fill the sparge gas hydrator bottle with 800 mL of reverse osmosis water.
3.
Put the sparge gas hydrator top back on the bottle but do not thread the cap.
4.
Confirm that appropriate filters are in place on sparge gas hydrator top.
5.
Cover all openings with aluminum foil and secure the foil with autoclave tape.
Photobioreactor:
1.
Disconnect the photobioreactor from the cooling bath.
2.
Disassemble the photobioreactor headplate and base plates from the photobioreactor glass.
3.
Thoroughly wash all parts with soap and water.
4.
Rinse thoroughly with reverse osmosis water to remove any soap residues.
5.
Reassemble the photobioreactor and tighten the headplate and base plate into the silicone gasket, forming an air tight seal.
6.
Attach the desired silicone tubes and filters to the headplate and cover the ends with aluminum foil and secure the foil with autoclave tape.
65
7.
Cover the sparge gas inlet tube with foil and secure the foil with autoclave tape.
8.
Place the photobioreactor on the photobioreactor cradle and slide the cradle into the autoclave.
9.
Autoclave the photobioreactor and the sparge gas hydrator for 30 minutesat
121 °C and 23 psig.
10.
Remove the photobioreactor cradle holding the photobioreactor and sparge gas hydrator from the autoclave and allow to cool overnight.
11.
Connect the inlet gas line to the sparge gas hydrator and connect the sparge gas hydrator to the photobioreactor.
12.
Turn on the inlet gas and allow it to run for 12 hours before adding medium.
13.
Attach the photobioreactor to the cooling bath.
14.
Turn on the cooling bath and the photobioreactor lights.
15.
Check the assembly for leaks and nonfunctioning light bulbs and fix any problems.
66
Bioreactor Loading
Materials:
Sterile Photobioreactor
4.0 L Harrison’s ASM + Guillards f/2 enrichment medium
Two 5 L Pyrex bottles with caps
“loading cap” and 3/8” O.D. ¼” I.D. silicone tubing.
Equipment:
Autoclave
Laminar flow hood (Edge Gard Hood US pat # 3,318,076)
Procedures:
1.
Distribute 4.0 L of Harrison’s ASM + Guillards f/2 enrichment medium equally into two 5 L Pyrex bottles.
2.
Loosely place a cap on each of the 5 L Pyrex bottles.
3.
Prepare a loading cap with enough silicone tubing attached to the inside of the cap to reach the bottom of the bottle and the outside of the cap to reach the loading port on the top of the photobioreactor. Place aluminum foil secured with autoclave tape over the tube connected to the top side of the cap. Completely wrap the loading cap and tube assembly in aluminum foil and secure the foil with autoclave tape.
4.
Autoclave the bottles and loading cap assembly for 40 minutesat 123 °C,
23 psig.
5.
Place the autoclaved bottles and loading cap assembly in the laminar flow hood to cool.
6.
Once cooled, combine the two bottles of medium into one of the 5 L Pyrex bottles. Set the other aside, as it is no longer needed.
67
7.
Open the foil containing the loading cap assembly and tighten the loading cap assembly onto the neck of the 5 L Pyrex bottle.
8.
Attach the top tube of the loading cap to the loading port of the photobioreactor.
9.
Attach the loading cap air inlet port to the air outlet needle valve of the photobioreactor fluidics system.
10.
Open the air outlet needle valve ¼ turn and allow the bottle headspace to pressurize, forcing the medium from the bottle in the photobioreactor.
11.
When the medium bottle is empty, close the air outlet needle valve and aseptically clamp the silicone loading tube.
68
Bioreactor Inoculation
Materials:
Cyclotella sp .
cell culture
500 mL Pyrex bottle with “loading cap”
3/8” O.D. x ¼” I.D. silicone tubing
Sterile 25 mL pipette
Sterile 1 mL pipette tips and 1 mL pipettor
Z2 Beckman Coulter Counter cuvettes
Sterile 500 mL graduated cylinder
Equipment:
Autoclave
Z2 Beckman Coulter Counter
Pipet-Aid pipetting device
Procedures:
1.
Loosely attach a “loading cap” to a 500 mL Pyrex bottle making sure the silicone tubing on the inside of the loading cap reaches the bottom of the bottle and the silicone tubing on the top of the loading cap reaches the loading port of the photobioreactor.
2.
Cover all openings with aluminum foil and secure the foil with autoclave tape.
3.
Autoclave the bottle/loading cap assembly for 30 min. at 121 °C, 23 psig.
4.
Determine the volume of inoculum cell culture required to give the desired initial cell number density, e.g. 4 x 10
4
cells/mL using the formula:
69 where V inoculum
is the volume of inoculum to be added, C desired is the desired final cell number density, V medium is the medium volume before inoculation and C inoculum
is the measured inoculum cell density.
5.
Sanitize gloves and the laminar flow hood, remove four culture flasks from the incubator and place in the laminar flow hood according to the culture maintenance subculturing protocol.
6.
Combine the cell culture from four flasks into one flask and mix thoroughly by swirling.
7.
Remove 100 µL of culture with a sterile 1 mL pipette and place the aliquot in a Coulter Counter cuvette. Add 10 mL of diluent. Determine the cell number density according to the cell number density determination protocol. Repeat for a duplicate measurement.
8.
Measure out V inoculum with the sterile 500 mL graduated cylinder and transfer the culture to the inoculation bottle.
9.
Use the photobioreactor fluidics air outlet needle valve to load the cell culture according to the bioreactor loading protocol to transfer the culture into the medium.
70
Bioreactor Sampling
Materials:
125 mL sampling bottle with sampling cap
50 mL centrifuge tube
Equipment:
Photobioreactor
Procedures:
1.
Increase the airflow to the photobioreactors to the maximum level for 15 minutesbefore sampling to reduce flocculation and dislodge cell sediment from the reactor bottom.
2.
Loosen the cap on the sampling bottle.
3.
Release the clamp on the sampling port. Pressure in the headspace of the photobioreactor will cause cell culture to flow into the sampling bottle.
4.
Fill the sampling bottle with 20 mL of cell culture and then immediately remove the cap and fill 20 mL of cell culture into each of two 50 mL centrifuge tubes.
5.
Analyze the cell culture in the centrifuge tubes according to the cell number density and liquid phase determination protocols.
71
Cell Density Measurement with Z2 Beckman Coulter Counter, Cyclotella sp.
Materials:
Z2 Beckman Coulter Counter cuvette
Cell culture sample
Coulter Counter diluents (10 g NaCl/L, sterile filtered)
Equipment:
Z2 Beckman Coulter Counter
Procedure:
1.
Vortex the centrifuge tube containing the cell culture sample to be analyzed at high speed for three repetitions of five seconds.
2.
Immediately transfer 0.100 mL of the sample to a Z2 Beckman Coulter
Counter Cuvette.
3.
Immediately add 10 mL of diluents to the sample, cover with the lid and invert 10 times.
4.
Confirm the dilution setting on Coulter Counter is “101”.
5.
Lower the elevated sample holder and remove the cuvette holding electrolyte diluent.
6.
Remove the cuvette cap and place the cuvette with the sample in the sample holder.
7.
Raise the sample platform until the aperture is submerged in the sample.
8.
Push “Start” on the keypad.
9.
Record the output.
10.
Repeat steps 8 and 9 twice more for a total of three measurements.
11.
Repeat steps 1-10 for the replicate cell culture sample.
12.
Spray the aperture with distilled water from a polyethylene distilled water bottle and pat dry with a Kimwipe.
72
13.
Return the cuvette containing only diluent to the sample platform.
14.
Repeat steps 5-9 with fresh diluent until the cell number density is less than
1000/mL.
73
Aqueous Phase Silicon Concentration Determination
Reagents:
18.7% HCl in water by volume
Ammonium molybdate reagent: Dissolve 10 g (NH
4
)
6
Mo
7
O
24
· 4H
2
O in 75 mL distilled water (warmed/stirring). Dilute to 100 mL with distilled water. Adjust and record pH to 7-8 by adding 1.2M NaOH
(aq)
dropwise.
Store in a polyethylene bottle.
Equipment:
Photobioreactor
Procedure:
1.
Remove a 20 mL sample from the reactor according to the sampling protocol.
2.
Prepare sample vial labeled with sample number.
3.
Pipette 5.00 mL from sample into sample vial.
4.
To 5.00 mL sample add in rapid succession 0.100 mL 18.7 % HCl and
0.200 mL ammonium molybdate reagent.
5.
Mix by 10 inversions and let stand 10 min.
6.
Place liquid sample into spectrophotometer cuvette.
7.
Have a calibration curve prepared by measuring the absorbance at 360 nm of 0.00, 0.010, 0.050, 0.100, 0.200, and 0.300 mM soluble silicon solutions prepared from sodium metasilicate (Na
2
O
3
Si) dissolved in DI H
2
O.
8.
Plot the absorbance (in absorbance units) at 360 nm versus the concentration of the solutions.
9.
Fit the absorbance of the stock solutions to the following linear relationship where a
Si,1
is the slope of the calibration curve and a
Si,Δ
is the error (one standard error) of the slope of the calibration curve.
74
Si a A
Si ,1
360 a A 360
Si ,
10.
Measure the absorbance of the reactor sample and determine the concentration of silicon in the sample by substation into the calibration equation.
Note: The measured absorbance should be in the linear range, 0.4-0.9. If the absorbance is above 0.9 units then dilutions to the sample should be made to give the sample an absorbance in the linear range.
11.
Repeat steps 1-13 for a duplicate measurement.
12.
Collect the assay waste and dispose of down the drain.
13.
Wipe down the spectrophotometer cleaning away all liquids and salts.
75
Preparation of Bioreactor Liquid Phase Samples for ICP analysis
Equipment: pH meter stir plate and stir bar
Reagents:
Sodium Hydroxide solid
Nitric acid, 15.8N
Procedure:
1.
Dissolve 10.0 g NaOH pellets in 2.0 L distilled IH
2
O using a stir plate and stir bar, to achieve a solution concentration = 5.0 g NaOH/L.
2.
While continuing to stir, submerge a pH probe in the solution.
3.
Add HNO
3
dropwise until a pH of 7.0 ± 1.0 is achieved.
4.
If the pH drops below 6.0 add a solution of 1.2 M NaOH drop wise until a pH 7.0 ± 1.0 is achieved.
5.
Place 2.0 mL of the bioreactor liquid phase sample in a 20 mL plastic vial.
6.
Dilute the sample to 20.0 mL with the 5.0 g NaOH/L solution.
76
Analysis of Bioreactor Liquid Phase for Si with Varian 150 ICP
Equipment:
Varian 150 ICP Operation
Reagents:
Silicon Standard Solution, 1 mg/mL Si in 2% NaOH, Acros Organics, catalog no. 196291000
ICP Standard solutions
Distilled H
2
O
Procedure:
Stock Solutions:
1.
Prepare a 5 ppm Si standard for ICP using Silicon Standard Solution, 1 mg/mL.
2.
Acquire 200 mL of DDH
2
O for baseline measurements.
3.
Have sample solutions properly diluted according to the procedure entitled
“Preparation
of Bioreactor Liquid Phase Samples for ICP analysis”.
4.
Verify that the samples are clear and free of any particulates.
Varian 150 ICP Operation:
1.
Start the torch chiller which sits behind the instrument.
2.
Place the sample inlet lines in the peristaltic pumping mechanism.
3.
Turn on the RF power.
4.
Turn on the exhaust fan.
5.
Click the “shortcut to Liberty.exe” icon on the desktop
77
6.
Select “Worksheet” → “New” and enter the name of the new worksheet protocol.
7.
Select “Develop” → “Edit Method” to select the elements and their analysis wavelengths from the software generated periodic table according to Table 1 .
8.
Select the elements as “analyte” and click “ok”.
9.
Select the “Conditions” tab to set the integration time and uptake delay according to Table 2 .
10.
Select the “Standards” tab and enter the concentration of Si in the standard solution, 5 ppm Si.
11.
Select the “Calibrations” tab and set the calibration to “linear”
12.
Select the “Labels” tab and set to “manual sample source”. Failure to follow this step will cause the Varian 150 ICP to use the non-existent auto sampler.
13.
Open the argon gas cylinder and adjust the argon gas pressure regulator to
80 psi.
14.
Submerge the sample inlet tube in the blank.
15.
Press the torch icon on the “Worksheet” tab to start the torch.
16.
Allow the torch to stabilize for 10 minutes before beginning an analysis.
17.
Select “Analyze” → “Start Analysis” to initiate the analysis.
18.
Follow the prompts for standards, blanks, and samples.
19.
After the completion of the analysis insert a 3 ½” floppy disk into the A: drive and select the “Export Data” function. The data is now copied to the disk.
20.
Open Excel.
21.
Open the A: drive and look for all file types.
22.
Change the file extension of the data file to .xls.
23.
Open the file and select the data spacing type to be “comma”.
24.
Save the new file name to the disk.
78
25.
Make sure the sample inlet tube is in the blank solution and allow the blank to rinse the instrument for 10 minutes.
26.
Turn off the torch by selecting the “torch off” icon on the worksheet page
27.
Turn off the RF power, exhaust fan and argon gas.
28.
Remove the sample lines from the peristaltic pump.
29.
Turn off the torch chiller five minutes after the rest of the instrument has been shut down.
30.
Close the Varian software.
31.
To analyze using a previously developed method select “Open” after step 5 and proceed directly to step 13.
Table 1.
Element ICP analysis parameters
Element
Silicon wavelength
(nm)
251.611 intensity k-factor
850
Table 2.
Operating parameters for elemental analysis
Experiment
Silicon
Integration time
(sec)
10.0
Uptake Delay
(sec)
15
79
Extraction of Lipids from Freeze Dried Diatoms
Reagents:
2:1 (v:v) Chloroform:Methanol
1:1 (v:v) Methanol:H
2
O
0.88 wt% potassium chloride in H
2
O
Materials:
16 mL glass vials with PTFE-lined cap
50 mL centrifuge tubes
Equipment:
Centrifuge
Vortex mixer
Procedures:
14.
Place 25 mg free-dried diatom cells in a 16 mL glass vial with a PTFElined cap.
15.
Add 10 mL 2:1 Chloroform:Methanol.
16.
Vortex the mixture for 60 seconds then store in the dark at 4ºC for 12 hours.
17.
Recover the organic extract and place in a 16 mL glass vial with a PTFElined cap. Set the organic extract aside.
18.
Add 5 mL 2:1 Chloroform:MeOH to the solids, vortex for 60 seconds then let the solids settle.
19.
Recover the organic extract and combine with the previous extract in the
16 mL glass vial.
20.
Add 5 mL 2:1 Chloroform:MeOH to the solids, vortex for 60 seconds then let the solids settle.
80
21.
Recover the organic extract and combine with the previous extracts in the
16 mL glass vial. Dispose of solids as waste.
22.
To the combined extracts add 5 mL 0.88wt% KCl in H
2
O.
23.
Vortex for 60 seconds then transfer the solution to a 50 mL centrifuge tube and centrifuge at 1000 rpm for 10 minutes.
24.
Remove lower extract layer by pipette and dispose of the upper layer and any precipitates at the interface. Transfer the extract layer to a clean centrifuge tube.
25.
Add 5 mL 1:1 Methanol:H
2
O to the extract layer.
26.
Vortex for 60 seconds then centrifuge at 1000 rpm for 10 minutes.
27.
Remove and dispose of the upper layer and any precipitates at the interface, keeping only the lower extract layer.
28.
Recover the extract volume, transfer to a glass vial and store at 0ºC or less until needed for analysis.
81
Analysis of Lipid Content of Freeze-Dried Diatoms
Equipment:
Spectrophotometer
Vortex mixer
Reacti-Therm Heating Module
Reagents:
Dichromate solution in conc. H
2
SO
4
(2.5 g/L), prepared by dissolving 2.5 g
K
2
Cr
2
O
7
in 1.0 L conc. H
2
SO
4
Procedure:
1.
Place 0.2 mL lipid extract into a 1 mL glass Reacti-Therm vial.
2.
Evaporate to dryness with N
2
.
3.
Heat Reacti-Therm Heating Module to 100ºC.
4.
Add 1 mL dichromate solution and cap with PTFE-lined cap
5.
Place in Reacti-Therm Heating Module for 45 minutes.
6.
After 45 minutes remove from Reacti-Therm and cool to room temperature.
7.
Combine 0.5 mL sample with 4.5 mL DDIH
2
O. Caution: This addition is exothermic.
8.
Let the diluted sample cool to room temperature then measure the absorbance at 350 nm using a spectrophotometer. Note: Use water as the blank to set the baseline on the spectrophotometer.
9.
Repeat steps 1-7 with a blank sample of chloroform in place of the lipid extract.
Nile Red Fluorescent Lipid Determination
Equipment:
Fluorescent microscope
Reagents:
Nile red in acetone, 50 µg/mL
25% (v/v) DMSO/H
2
O
Procedure:
1.
Place 0.4 mL aliquot of cell suspension of known cell density into a 4 mL vial.
2.
Combine with 0.03 mL 50 µg Nile Red/mL Acetone.
3.
Add 2.57 mL 25% (v/v) DMSO/H
2
O.
4.
Heat at 40°C for 10 minutes.
5.
Observe using a fluorescent microscope with a 535 nm excitation wavelength and a 610 nm emission wavelength.
82
Transesterification of Fatty Acids in Lipid Extracts
Equipment:
Reacti-Therm Heating Module
Vortex mixer
Centrifuge
Reagents:
1% (v/v) H
2
SO
4
/MeOH
Hexane
Procedure:
1.
Evaporate 1.0 mL of lipid extract to dryness with nitrogen.
2.
Add 0.5 mL 1% (v/v) H
2
SO
4
/MeOH.
3.
Heat at 100°C for 60 minutes then cool to room temperature.
4.
Once cooled, add 0.5 mL H
2
O followed by 0.5 mL hexane.
5.
Vortex for 60 seconds.
6.
Separated the aqueous from solvent layers by centrifugation at 2000 rpm for 3 minutes.
7.
Remove the upper solvent phase by pipette and dry with anhydrous
Na
2
SO
4
.
83
Appendix B – Tabulated Data
Table B 0. Cy-JA-05, Sample Information
Sample# Date
Sample time medium
Inoculum
Cy-JA-05-01-1 12/5/2008 4:00 PM
Cy-JA-05-02-1 12/6/2008 5:00 PM
Cy-JA-05-03-1 12/7/2008 4:45 PM
Cy-JA-05-04-1 12/9/2008 4:15 PM
Cy-JA-05-05-1 12/11/2008 1:30 PM
Cy-JA-05-06-1 12/12/2008 1:45 PM
Cy-JA-05-07-1 12/13/2008 3:00 PM
Cy-JA-05-08-1 12/14/2008 3:00 PM
Cy-JA-05-09-1 12/15/2008 2:30 PM
Cy-JA-05-10-1 12/16/2008 4:30 PM
Cy-JA-05-11-1 12/17/2008 4:00 PM
Media Charge 12/17/2008 4:30 PM
Cy-JA-05-12-1 12/18/2008 4:00 PM
Cy-JA-05-13-1 12/20/2008 1:15 PM
Cy-JA-05-14-1 12/22/2008 12:00 PM
Cultivation
Time (hr)
Culture pH
0.0
25.0
48.8
96.3
143.0
167.3
192.5
216.5
240.0
266.0
289.5
290.0
313.5
360.25
404.25
8.60
8.80
9.20
9.50
9.50
9.40
8.40
8.40
8.40
9.60
9.5
9.6
9.5
9
220
120
120
120
120
180
220
200
Sample volume
(mL)
40
220
220
220
220
220
220
2660
2540
2420
2300
2180
3980
3200
2980
2780
Medium volume
(mL)
4000
240
3980
3760
3540
3320
3100
2880
Table B 1.Cy-JA-05, Cell density and growth
Sample# Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Sample average
(cells/mL)
Sample +/-
1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/- 1
S.D.
Inoculum
Inoculum
Cy-JA-05-01-1
Cy-JA-05-01-2
Cy-JA-05-02-1
Cy-JA-05-02-2
Cy-JA-05-03-1
Cy-JA-05-03-2
Cy-JA-05-04-1
Cy-JA-05-04-2
Cy-JA-05-05-1
Cy-JA-05-05-2
Cy-JA-05-06-1
Cy-JA-05-06-2
Cy-JA-05-07-1
Cy-JA-05-07-2
Cy-JA-05-08-1
Cy-JA-05-08-2
Cy-JA-05-0-9-
Cy-JA-05-10-1
3.501E+06
3.540E+06
1.586E+05
1.721E+05
2.870E+05
3.040E+05
3.763E+05
3.575E+05
5.838E+05
5.713E+05
8.959E+05
8.761E+05
1.080E+06
1.051E+06
1.220E+06
1.143E+06
1.227E+06
1.294E+06
1.458E+06
1.417E+06
1.468E+06
3.699E+06
3.462E+06
1.553E+05
1.561E+05
3.357E+05
2.929E+05
3.836E+05
3.618E+05
6.014E+05
5.557E+05
9.035E+05
8.975E+05
1.075E+06
1.015E+06
1.261E+06
1.144E+06
1.201E+06
1.292E+06
1.440E+06
1.434E+06
1.526E+06
3.320E+06
3.456E+06
1.416E+05
1.555E+05
2.897E+05
2.975E+05
3.618E+05
3.535E+05
5.727E+05
5.446E+05
9.104E+05
8.773E+05
1.055E+06
1.029E+06
1.252E+06
1.153E+06
1.188E+06
1.277E+06
1.446E+06
1.404E+06
1.543E+06
3.507E+06
3.486E+06
1.518E+05
1.613E+05
3.041E+05
2.982E+05
3.739E+05
3.576E+05
5.859E+05
5.572E+05
9.033E+05
8.836E+05
1.070E+06
1.032E+06
1.244E+06
1.147E+06
1.205E+06
1.288E+06
1.448E+06
1.418E+06
1.512E+06
1.896E+05
4.686E+04
9.010E+03
9.393E+03
2.738E+04
5.580E+03
1.111E+04
4.141E+03
1.446E+04
1.339E+04
7.290E+03
1.203E+04
1.323E+04
1.815E+04
2.155E+04
5.508E+03
1.986E+04
9.292E+03
9.165E+03
1.504E+04
3.932E+04
3.496E+06
1.566E+05
3.011E+05
3.658E+05
5.716E+05
8.935E+05
1.051E+06
1.196E+06
1.247E+06
1.433E+06
1.509E+06
1.240E+05
9.717E+03
1.797E+04
1.166E+04
2.009E+04
1.397E+04
2.535E+04
5.531E+04
4.718E+04
1.970E+04
2.640E+04
6.231E+08
1.132E+09
1.295E+09
1.898E+09
2.770E+09
3.026E+09
3.180E+09
3.166E+09
3.468E+09
3.470E+09
3.867E+07
6.759E+07
4.126E+07
6.668E+07
4.331E+07
7.300E+07
1.471E+08
1.198E+08
4.768E+07
6.072E+07
86
Table B 1. Continued
Sample#
Cy-JA-05-10-2
Cy-JA-05-11-1
Cy-JA-05-11-2
Media Charge
Cy-JA-05-12-1
Cy-JA-05-12-2
Cy-JA-05-13-1
Cy-JA-05-13-2
Cy-JA-05-14-1
Cy-JA-05-14-2
Trial #1
(cells/ mL)
1.495E+06
1.569E+06
1.604E+06
1.177E+06
1.203E+06
1.295E+06
1.274E+06
1.432E+06
1.378E+06
1.517E+06
1.513E+06
Trial #2
(cells/ mL)
1.501E+06
1.587E+06
1.633E+06
1.190E+06
1.225E+06
1.272E+06
1.255E+06
1.411E+06
1.333E+06
1.485E+06
1.549E+06
Trial #3
(cells/ mL)
1.519E+06
1.600E+06
1.609E+06
1.200E+06
1.207E+06
1.292E+06
1.237E+06
1.341E+06
1.341E+06
1.536E+06
1.545E+06
Trial average
(cells/mL)
1.505E+06
1.585E+06
1.615E+06
1.189E+06
1.212E+06
1.286E+06
1.255E+06
1.395E+06
1.351E+06
1.513E+06
1.536E+06
Trial +/- 1
S.D.
(cells/mL)
Sample average
(cells/mL)
Sample
+/- 1 S.D.
(cells/mL)
1.249E+04
1.557E+04
1.550E+04
1.168E+04
1.163E+04
1.250E+04
1.850E+04
4.765E+04
2.401E+04
2.577E+04
1.973E+04
1.600E+06
1.200E+06
1.271E+06
1.419E+06
1.543E+06
2.152E+04
1.614E+04
2.209E+04
7.836E+04
6.842E+03
Total No. cells in reactor
3.489E+09
3.489E+09
4.067E+09
4.230E+09
4.290E+09
Total No. cells in reactor +/- 1
S.D.
4.691E+07
6.423E+07
7.067E+07
2.335E+08
1.902E+07
Table B 2. Silicon concentration by spectrophotometer
Sample# medium medium
Cy-JA-05-01-1
Cy-JA-05-01-2
Cy-JA-05-02-1
Cy-JA-05-02-2
Cy-JA-05-03-1
Cy-JA-05-03-2
Cy-JA-05-04-1
Cy-JA-05-04-2
Cy-JA-05-05-1
Cy-JA-05-05-2
Cy-JA-05-06-1
Cy-JA-05-06-2
Cy-JA-05-07-1
Cy-JA-05-07-2
Cy-JA-05-08-1
Cy-JA-05-08-2
Cy-JA-05-0-9-1
Cy-JA-05-09-2
Cy-JA-05-10-1
Cy-JA-05-10-2
Cy-JA-05-11-1
Cy-JA-05-11-2
A at 360 nm (mAU)
0.265
0.279
0.265
0.279
0.265
0.279
0.265
0.279
0.265
0.279
0.484
0.526
0.814
0.829
0.764
0.750
0.691
0.647
0.413
0.432
0.132
0.441
0.224
0.220
Dilution
1.0
1.0
1.0
1.0
1.0
1.0
1.0
1.0
1.0
1.0
2.5
2.5
2.5
1.0
1.0
1.0
2.5
2.5
2.5
2.5
2.5
2.5
2.5
2.5
Csi (mmol/L)
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.18
0.18
0.06
0.07
0.04
0.04
0.21
0.45
0.35
0.35
0.32
0.32
0.29
0.27
0.05
0.05
0.05
0.05
0.05
Average Csi
(mmol/L)
0.33
0.35
0.32
0.28
0.18
0.07
0.04
0.00
0.00
0.00
0.00
0.00
Csi +/- 1S.D.
(mmol/L)
0.17
0.00
0.00
0.01
0.01
0.01
0.00
87
Table B 2. Continued
Sample#
Cy-JA-05-12-1
Cy-JA-05-12-2
Cy-JA-05-13-1
Cy-JA-05-13-2
Cy-JA-05-14-1
Cy-JA-05-14-2
A at 360 nm (mAU)
0.647
0.705
0.55
0.501
0.38
0.333
Dilution
1.0
1.0
1.0
1.0
1.0
1.0
Csi (mmol/L)
0.11
0.12
0.09
0.09
0.06
0.06
Average Csi
(mmol/L)
0.11
0.09
0.06
Csi +/- 1S.D.
(mmol/L)
0.01
0.01
0.01
88
89
Table B 3. Cy-JA-05, Lipid data
Sample#
Cy-JA-05-01-1
Cy-JA-05-01-2
Cy-JA-05-04-1
Cy-JA-05-04-2
Cy-JA-05-08-1
Cy-JA-05-08-2
Cy-JA-05-10-1
Cy-JA-05-10-2
Cy-JA-05-12-1
Cy-JA-05-12-2
Cy-JA-05-14-1
Cy-JA-05-14-2
Dry cell mass
(mg)
19.5
19.1
23.6
21.6
7.1
7.8
8.2
9.2
10.5
10.7
24.9
26
Total vol lipid extract
(mL)
Extract assay vol
(mL)
13
13
13
13
13
13
13
13
13
13
13
13
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
Dilution factor
(mL/mL)
Abs at
350 nm
0.05 1.427
0.05 1.536
0.05
0.05
1.41
1.39
0.05 1.272
0.05 1.308
0.05 1.246
0.05 1.249
0.05 1.374
0.05 1.288
0.05 0.793
0.05 0.768 calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at
350 nm
1.80 8.44 1.42
0.68 1.05 1.414
1.97 7.93 1.39
2.17 9.89 1.445
3.38 52.14 1.333
3.01 41.34
3.64 49.35
1.35
1.29
3.61 43.55 1.187
2.34 22.37 1.239
3.21 32.61 1.266
8.27 40.39 1.049
8.52 39.95 0.93 calc. palmitic acid in cuvette
(mg/L)
1.87
1.93 wt% lipid
8.92
9.52
2.17
1.61
9.05
6.51
2.76 40.74 average wt%
6.98
8.35
42.10 st dev
3.98
1.46
7.43
2.58 34.19
3.19 42.23
4.24 52.49
3.71 39.42
3.44 35.34
5.65 26.75
6.87 31.69
46.91
32.44
34.69
4.84
7.27
6.64
Table B 4. Cy-JA-09, Cultivation information
Sample# Date medium
Inoculum
Cy-JA-09-01-1 6/6/2009
Cy-JA-09-02-1 6/7/2009
Cy-JA-09-03-1 6/8/2009
5:00 PM
1:00 PM
2:30 PM
Cy-JA-09-04-1 6/9/2009 4:45 PM
Cy-JA-09-05-1 6/10/2009 12:45 PM
Cy-JA-09-06-1 6/11/2009 3:00 PM
Cy-JA-09-07-1 6/12/2009 10:45 AM
Cy-JA-09-08-1 6/14/2009 1:15 PM
Cy-JA-09-09-1 6/15/2009 2:15 PM
Cy-JA-09-10-1 6/16/2009
Cy-JA-09-11-1 6/17/2009
2:50 PM
3:15 PM
Cy-JA-09-12-1 6/18/2009 12:15 PM
Cy-JA-09-13-1 6/18/2009 4:30 PM
Cy-JA-09-14-1 6/19/2009 12:15 PM
Cy-JA-09-15-1 6/19/2009 3:30 PM
Cy-JA-09-16-1 6/19/2009
Cy-JA-09-17-1 6/20/2009
5:30 PM
6:15 PM
Cy-JA-09-18-1 6/21/2009 11:15 AM
Cy-JA-09-19-1 6/21/2009 3:15 PM
Cy-JA-09-20-1 6/22/2009 11:00 AM
Cy-JA-09-21-1 6/22/2009 3:20 PM
Cy-JA-09-22-1 6/23/2009 11:00 AM
Cy-JA-09-23-1 6/24/2009 3:00 PM
Cy-JA-09-24-1 6/25/2009 11:00 AM
Cy-JA-09-25-1 6/26/2009 11:00 AM
Sample time
287.5
308.25
311.50
313.50
338.25
355.25
359.25
379.00
382.33
402.00
430.00
454.00
478.00
0.0
20.0
45.5
71.8
91.8
118.0
137.8
164.3
189.3
213.8
238.3
259.3
Cultivation Time
(hr)
Culture pH
180
180
180
180
180
180
180
180
220
220
180
180
180
Sample volume, includes 20 mL to clear lines (mL)
320
320
320
60
320
220
220
220
60
60
60
220
8.95
9.53
8.73
9.17
8.74
9.2
8.55
8.45
8.96
9.18
9.42
9.6
8.81
9.37
9.45
9.63
9.56
9.26
8.61
8.38
8.50
8.72
8.83
9.23
Medium volume
(mL)
2650
2430
2250
2070
2177
2850
3008
3790
3922
3742
3562
3382
3202
3490
3270
3210
3150
3090
2870
4400
550
4950
4630
4310
4250
3930
3710
Table B 5. Cy-JA-09, Cell density
Sample#
Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Sample average
(cells/mL)
Sample
+/- 1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
Cy-JA-09-01-1
334108
Cy-JA-09-01-2
319564
Cy-JA-09-02-1 567014
Cy-JA-09-02-2 556914
341582
341968
583174
327644
304414
566610
3.34E+05
3.22E+05
5.72E+05
6.98E+03
1.89E+04
9.45E+03
3.28E+05
5.74E+05
1.45E+04
1.20E+04
1.625E+09
2.839E+09
7.153E+07
5.935E+07
579134 588628 5.75E+05 1.63E+04
Cy-JA-09-03-1
719928
Cy-JA-09-03-2 701142
692254
298920
704172
686598
7.05E+05 1.39E+04
5.62E+05 2.28E+05
6.34E+05 1.64E+05 3.137E+09 8.141E+08
Cy-JA-09-04-1
1.170E+06 1.265E+06 1.032E+06 1.16E+06 1.17E+05 1.03E+06 1.52E+05 5.122E+09 7.540E+08
Cy-JA-09-04-2
9.391E+05 9.011E+05 9.017E+05 9.14E+05 2.18E+04
Cy-JA-09-05-1
1.124E+06 1.162E+06 1.118E+06 1.13E+06 2.39E+04 1.09E+06 5.49E+04 5.381E+09 2.717E+08
Cy-JA-09-05-2
1.036E+06 1.055E+06 1.028E+06 1.04E+06 1.39E+04
Cy-JA-09-06-1 1.355E+06 1.342E+06 1.376E+06 1.36E+06 1.72E+04 1.35E+06 2.45E+04 6.694E+09 1.212E+08
Cy-JA-09-06-2 1.334E+06 1.385E+06 1.322E+06 1.35E+06 3.35E+04
Cy-JA-09-07-1 1.510E+06 1.497E+06 1.490E+06 1.50E+06 1.01E+04 1.53E+06 3.09E+04 7.550E+09 1.528E+08
Cy-JA-09-07-2 1.560E+06 1.560E+06 1.534E+06 1.55E+06 1.50E+04
Cy-JA-09-08-1 1.854E+06 1.779E+06 1.858E+06 1.83E+06 4.45E+04 1.84E+06 6.47E+04 9.087E+09 3.204E+08
Cy-JA-09-08-2 1.834E+06 1.936E+06 1.753E+06 1.84E+06 9.17E+04
Cy-JA-09-10-1 1.943E+06 1.951E+06 1.901E+06 1.93E+06 2.69E+04 2.01E+06 9.45E+04 9.959E+09 4.677E+08
Cy-JA-09-10-2 2.146E+06 2.074E+06 2.057E+06 2.09E+06 4.72E+04
Cy-JA-09-11-1 2.230E+06 2.150E+06 2.243E+06 2.21E+06 5.04E+04 2.23E+06 4.07E+04 1.104E+10 2.016E+08
Cy-JA-09-11-2 2.253E+06 2.262E+06 2.243E+06 2.25E+06 9.50E+03
Cy-JA-09-12-1 2.164E+06 2.139E+06 2.116E+06 2.14E+06 2.40E+04 2.17E+06 6.06E+04 1.073E+10 2.999E+08
Cy-JA-09-12-2 2.135E+06 2.163E+06 2.285E+06 2.19E+06 7.98E+04
Cy-JA-09-13-1 2.113E+06 2.100E+06 2.049E+06 2.09E+06 3.38E+04 2.11E+06 3.62E+04 1.045E+10 1.793E+08
Table B 5. Continued
Sample#
Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Sample average
(cells/mL)
Sample +/-
1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
Cy-JA-09-13-2 2.152E+06 2.141E+06 2.115E+06 2.14E+06 1.90E+04
Cy-JA-09-14-1 2.049E+06 2.089E+06 2.046E+06 2.06E+06 2.40E+04 2.04E+06 3.24E+04 1.008E+10 1.604E+08
Cy-JA-09-14-2 2.021E+06 2.012E+06 1.999E+06 2.01E+06 1.11E+04
Cy-JA-09-15-1 2.113E+06 2.100E+06 2.049E+06 2.09E+06 3.38E+04 2.07E+06 2.99E+04 1.027E+10 1.478E+08
Cy-JA-09-15-2 2.049E+06 2.089E+06 2.046E+06 2.06E+06 2.40E+04
Cy-JA-09-16-1 2.200E+06 2.122E+06 2.139E+06 2.15E+06 4.10E+04 2.12E+06 4.95E+04 1.050E+10 2.450E+08
Cy-JA-09-16-2 2.113E+06 2.100E+06 2.049E+06 2.09E+06 3.38E+04
Cy-JA-09-17-1 1.820E+06 1.878E+06 1.878E+06 1.86E+06 3.35E+04 1.91E+06 5.96E+04 9.432E+09 2.952E+08
Cy-JA-09-17-2 1.981E+06 1.962E+06 1.914E+06 1.95E+06 3.45E+04
Cy-JA-09-18-1 1.684E+06 1.692E+06 1.724E+06 1.70E+06 2.12E+04 1.74E+06 1.11E+05 1.028E+10 5.485E+08
Cy-JA-09-18-2 1.669E+06 1.703E+06 1.962E+06 1.78E+06 1.60E+05
Cy-JA-09-19-1 1.679E+06 1.688E+06 1.961E+06 1.78E+06 1.60E+05 1.73E+06 1.15E+05 1.047E+10 5.711E+08
Cy-JA-09-19-2 1.685E+06 1.666E+06 1.677E+06 1.68E+06 9.54E+03
Cy-JA-09-20-1 1.352E+06 1.362E+06 1.358E+06 1.36E+06 5.03E+03 1.40E+06 3.82E+04 9.590E+09 1.890E+08
Cy-JA-09-20-2 1.331E+06 1.320E+06 1.260E+06 1.30E+06 3.82E+04
Cy-JA-09-21-1 1.478E+06 1.439E+06 1.405E+06 1.44E+06 3.65E+04 1.43E+06 1.19E+05 9.984E+09 5.870E+08
Cy-JA-09-21-2 1.251E+06 1.257E+06 1.190E+06 1.23E+06 3.71E+04
Cy-JA-09-22-1 1.409E+06 1.419E+06 1.447E+06 1.43E+06 1.97E+04 1.46E+06 4.29E+04 1.014E+10 2.122E+08
Cy-JA-09-22-2 1.513E+06 1.461E+06 1.504E+06 1.49E+06 2.78E+04
Cy-JA-09-23-1 1.484E+06 1.475E+06 1.430E+06 1.46E+06 2.89E+04 1.45E+06 2.75E+04 1.010E+10 1.362E+08
Cy-JA-09-23-2 1.438E+06 1.418E+06 1.472E+06 1.44E+06 2.73E+04
Cy-JA-09-24-1 1.660E+06 1.606E+06 1.564E+06 1.61E+06 4.81E+04 1.62E+06 3.45E+04 1.124E+10 1.707E+08
Cy-JA-09-24-2 1.649E+06 1.617E+06 1.605E+06 1.62E+06 2.27E+04
92
Cy-JA-09-01-1
Cy-JA-09-01-2
Cy-JA-09-02-1
Cy-JA-09-02-2
Cy-JA-09-03-1
Cy-JA-09-03-2
Cy-JA-09-04-1
Cy-JA-09-04-2
Cy-JA-09-05-1
Cy-JA-09-05-2
Cy-JA-09-06-1
Cy-JA-09-06-2
Cy-JA-09-07-1
Cy-JA-09-07-2
Cy-JA-09-08-1
Cy-JA-09-08-2
Cy-JA-09-10-1
Cy-JA-09-10-2
Cy-JA-09-11-1
Cy-JA-09-11-2
Cy-JA-09-12-1
Cy-JA-09-12-2
Cy-JA-09-13-1
Cy-JA-09-13-2
Cy-JA-09-14-1
Cy-JA-09-14-2
Cy-JA-09-15-1
Cy-JA-09-15-2
Cy-JA-09-16-1
Cy-JA-09-16-2
Cy-JA-09-17-1
Cy-JA-09-17-2
Cy-JA-09-18-1
Cy-JA-09-18-2
Cy-JA-09-19-1
Cy-JA-09-19-2
Cy-JA-09-20-1
Cy-JA-09-20-2
Cy-JA-09-21-1
Cy-JA-09-21-2
Cy-JA-09-22-1
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
Table B 6. Cy-JA-09, Si concentration by Spectrophotometer
Sample# Dilution Csi (mmolSi/L)
Csi +/-
1S.D.
(mmolSi/L)
0.045
0.036
0.045
0.071
0.023
0.016
0.012
0.008
0.069
0.068
0.061
0.089
0.011
0.027
0.001
0.004
0.014
0.005
0.009
0.023
0.006
0.144
0.202
0.144
0.144
0.071
0.089
0.064
0.129
0.237
0.188
0.269
0.250
0.186
0.248
0.164
0.075
0.135
0.072
Averag e Csi
(mmolS i/L)
0.441
0.341
0.298
0.055
0.176
0.112
0.177
0.227
0.176
0.112
0.068
0.093
0.194
0.087
0.054
0.101
0.077
0.072
0.135
0.124
0.188
0.285
0.236
0.141
0.226
0.313
0.313
0.187
0.178
0.460
0.422
0.342
0.341
0.295
0.302
0.258
0.238
0.168
0.161
0.081
0.068
0.151
0.119
0.076
A at
360 nm
(mAU)
0.055
0.175
0.111
0.176
0.226
0.175
0.111
0.068
0.093
0.193
0.087
0.054
0.100
0.077
0.072
0.134
0.123
0.187
0.284
0.235
0.140
0.225
0.311
0.311
0.186
0.177
0.458
0.420
0.340
0.339
0.294
0.300
0.257
0.237
0.167
0.160
0.081
0.068
0.150
0.118
0.076
Table B 6. Continued
Sample#
Cy-JA-09-22-2
Cy-JA-09-23-1
Cy-JA-09-23-2
Cy-JA-09-24-1
Cy-JA-09-24-2
Cy-JA-09-25-1
Cy-JA-09-25-2
A at
360 nm
(mAU)
0.193
0.472
0.478
0.327
0.332
0.342
0.315
Dilution
5
5
5
5
5
5
5
Csi (mmolSi/L)
0.194
0.474
0.480
0.329
0.334
0.344
0.317
Averag e Csi
(mmolS i/L)
0.477
0.331
0.330
Csi +/-
1S.D.
(mmolSi/L)
0.004
0.004
0.019
94
Table B 7. Cy-JA-09, Si concentration by ICP
Sample#
Cy-JA-09-01-1
Cy-JA-09-01-2
Cy-JA-09-02-1
Cy-JA-09-02-2
Cy-JA-09-03-1
Cy-JA-09-03-2
Cy-JA-09-04-1
Cy-JA-09-04-2
Cy-JA-09-05-1
Cy-JA-09-05-2
Cy-JA-09-06-1
Cy-JA-09-06-2
Cy-JA-09-07-1
Cy-JA-09-07-2
Cy-JA-09-08-1
Cy-JA-09-08-2
Cy-JA-09-10-1
Cy-JA-09-10-2
Cy-JA-09-11-1
Cy-JA-09-11-2
Cy-JA-09-12-1
Cy-JA-09-12-2
Cy-JA-09-13-1
Emission at
251.611 nm
Trial #1
Emission at
251.611 nm
Trial #2
Emission at
251.611 nm
Trial #3
Trial
Avg.
365
381
395
349
375
360
345
1287
1336
1026
1058
918
899
687
705
596
541
408
364
423
402
462
363
382
373
393
337
357
376
335
1289
1322
1035
1057
918
887
699
693
588
534
405
372
413
413
461
383
359
366
394
354
356
357
332
1289
1309
1043
1075
939
892
713
701
592
536
412
380
411
396
464
362
Assay
Conc.
(ppm)
1288 1.1595
1322 1.1901
1035 0.9312
1063 0.957
925 0.8325
893 0.8034
700 0.6297
700 0.6297
592 0.5328
537 0.4833
408 0.3675
372 0.3348
416 0.3741
404 0.3633
462 0.4161
369 0.3324
369 0.3318
373 0.336
394 0.3546
347 0.312
363 0.3264
364 0.3279
337 0.3036 dilution
Sample conc.
(ppm)
Si conc.
(mmol
Si/L)
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
11.595 0.413
11.901 0.424
9.312
9.57
8.325
8.034
6.297
6.297
5.328
4.833
3.675
3.348
3.741
3.633
4.161
3.324
3.318
3.36
3.546
3.12
3.264
3.279
3.036
0.332
0.341
0.296
0.286
0.224
0.224
0.190
0.172
0.131
0.119
0.133
0.129
0.148
0.118
0.118
0.120
0.126
0.111
0.116
0.117
0.108
0.119
0.119
0.116
0.108
0.418
0.336
0.291
0.224
0.181
0.125
0.131
0.133
Average
Si conc
(mmol
Si/L)
Average Si conc +/- 1
S.D.
(mmol
Si/L)
0.001
0.011
0.000
0.000
0.008
0.006
0.007
0.000
0.012
0.008
0.003
0.021
Table B 7. Continued
Sample#
Cy-JA-09-13-2
Cy-JA-09-14-1
Cy-JA-09-14-2
Cy-JA-09-15-1
Cy-JA-09-15-2
Cy-JA-09-16-1
Cy-JA-09-16-2
Cy-JA-09-17-1
Cy-JA-09-17-2
Cy-JA-09-18-1
Cy-JA-09-18-2
Cy-JA-09-19-1
Cy-JA-09-19-2
Cy-JA-09-20-1
Cy-JA-09-20-2
Cy-JA-09-21-1
Cy-JA-09-21-2
Cy-JA-09-22-1
Cy-JA-09-22-2
Cy-JA-09-24-2
Cy-JA-09-25-1
Cy-JA-09-25-2
Emission at
251.611 nm
Trial #1
Emission at
251.611 nm
Trial #2
414
446
323
402
403
440
392
417
329
372
355
444
418
394
410
408
397
319
334
352
327
384
411
439
318
409
390
424
389
419
331
366
340
431
417
400
417
402
392
316
325
351
321
382
Emission at
251.611 nm
Trial #3
Trial
Avg.
412
435
327
409
399
434
385
424
348
369
358
426
427
395
419
410
389
324
328
348
320
390
Assay
Conc.
(ppm)
336 0.3024
369 0.3321
351 0.3159
434 0.3903
421 0.3786
396 0.3567
415 0.3738
407 0.366
412 0.3711
440 0.396
323 0.2904
407 0.366
397 0.3576
433 0.3894
389 0.3498
420 0.378
393 0.3534
320 0.2877
329 0.2961
350 0.3153
323 0.2904
385 0.3468 dilution
Sample conc.
(ppm)
Si conc.
(mmol
Si/L)
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
3.711
3.96
2.904
3.66
3.576
3.894
3.498
3.78
3.024
3.321
3.159
3.903
3.786
3.567
3.738
3.66
3.534
2.877
2.961
3.153
2.904
3.468
0.108
0.118
0.112
0.139
0.135
0.127
0.133
0.130
0.132
0.141
0.103
0.130
0.127
0.139
0.125
0.135
0.126
0.102
0.105
0.112
0.103
0.123
0.122
0.129
0.132
0.130
0.115
0.137
0.130
0.131
0.104
0.112
0.113
Average
Si conc
(mmol
Si/L)
Average Si conc +/- 1
S.D.
(mmol
Si/L)
0.027
0.002
0.010
0.006
0.004
0.003
0.004
0.001
0.002
0.014
96
97
Table B 8. Cy-JA-09, Lipid data
Cy-JA-09-01-1 15
Cy-JA-09-01-2 15.6
Cy-JA-09-02-1 17.1
Cy-JA-09-02-2 19
Cy-JA-09-05-1 21
Cy-JA-09-05-2 21
Cy-JA-09-08-1 15.2
Cy-JA-09-08-2 16.5
Cy-JA-09-12-1 22.7
Cy-JA-09-12-2 26.5
Cy-JA-09-14-1 27.3
Cy-JA-09-14-2 28.7
Cy-JA-09-17-1 27
Cy-JA-09-17-2 21
Cy-JA-09-18-1 20
Cy-JA-09-18-2 17.6
Cy-JA-09-19-1 18.9
Cy-JA-09-19-2 20.3
Cy-JA-09-20-1 14.1
Dry cell mass
(mg)
Total vol lipid extract
(mL)
Extract assay vol
(mL)
Trial 1 Trial 2
Dilution
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Trial 3
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid average wt% st dev adjusted wt% adj stdev
11.4
11.4
11.4
13
13
13
13
11.4
8.5
8.5
8.5
8.5
12.7
11.4
11
11
11
11
11
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.05
0.05
0.05
0.05
0.05
0.050
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
1.489
1.314
1.228
1.274
1.449
1.116
0.908
0.778
0.016
0.448
0.374
0.641
0.645
0.982
1.094
1.035
1.064
1.26
3.00
3.85
3.39
1.66
4.96
7.01
8.30
15.85
11.57
9.62
6.28
5.17
5.76
5.47
3.02 1.230
12.34 1.293
15.49 1.196
11.90 1.253
5.61 1.379
22.94 1.173
45.48 0.938
50.48 0.828
62.75
43.67
48.25
36.09
30.36
0.596
0.527
12.30 45.97 0.450
9.66 37.70 0.561
0.680
1.054
32.82 1.054
34.58 1.064
1.017
3.83
3.20
4.16
3.60
2.35
4.39
6.72
7.81
10.10
10.79
11.55
10.45
9.27
5.57
5.57
5.47
5.94
17.55
13.47
17.07
12.84
9.81
19.87
43.33
47.18
38.91
40.52
42.98
41.04
46.37
31.46
35.74
32.61
33.34
1.484
0.040
1.212
1.319
1.320
1.221
0.955
0.829
0.587
0.429
0.481
0.604
0.692
1.044
1.083
1.084
1.045
1.31
15.61
4.00
2.95
2.94
3.92
6.55
7.80
10.19
11.76
11.24
10.02
9.15
5.67
5.28
5.27
5.66
3.30
81.07
16.28
9.91
13.34
17.29
42.11
47.11
39.28
44.43
41.76
39.24
45.73
32.10
33.62
31.24
31.56
9.94
13.91
14.81
45.95
6.48
2.80
6.48
3.01
46.98 13.66
43.06
33.64
32.28
4.30
1.91
1.54
12.91
14.13
15.08
45.78
39.10
43.22 1.93 43.21
43.10
33.12
32.19
32.08
0.80
2.09
4.42
1.81
0.26
1.14
3.50
2.06
0.96
3.71
98
Table B 8. Continued
Dry cell mass
(mg)
Total vol lipid extract
(mL)
Extract assay vol
(mL)
Trial 1 Trial 2
Dilution
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L)
Trial 3 wt% lipid
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid average wt% st dev adjusted wt% adj stdev
Cy-JA-09-20-2 10.7
Cy-JA-09-22-1 14.8
Cy-JA-09-22-2 14.2
Cy-JA-09-25-1 15
Cy-JA-09-25-2 18.7
11.8
11.8
11.2
11.4
11.2
0.2
0.2
0.2
0.2
0.2
0.05
0.05
0.05
0.05
0.05
1.273
1.106
1.195
1.191
0.972
3.40
5.05
4.17
4.21
6.38
29.46 1.225
34.48 1.170
27.16 1.100
26.47 1.080
33.85 0.982
3.88
4.42
5.11
5.31
6.28
34.70
29.43
34.58
34.82
33.25
0.248
1.135
1.041
1.080
0.957
13.55
4.77
5.70
5.31
6.53
141.37 32.08 3.71
32.19 32.84 4.25 32.67
39.18
34.82
34.74
32.99 3.26 34.30
2.43
0.71
Table B 9. Cy-JA-10, Cultivation information
Sample# Date medium
Inoculum
Cy-JA-10-1-1 6/6/2009
Cy-JA-10-2-1 6/7/2009
Cy-JA-10-3-1 6/8/2009
5:00 PM
1:00 PM
2:30 PM
Cy-JA-10-4-1 6/9/2009 4:45 PM
Cy-JA-10-5-1 6/10/2009 12:45 PM
Cy-JA-10-6-1 6/11/2009 3:00 PM
Cy-JA-10-7-1 6/12/2009 10:45 AM
Cy-JA-10-8-1 6/14/2009 1:15 PM
Cy-JA-10-9-1 6/15/2009 2:15 PM
Cy-JA-10-10-1 6/16/2009
Cy-JA-10-11-1 6/17/2009
2:50 PM
3:15 PM
Cy-JA-10-12-1 6/18/2009 12:15 PM
Cy-JA-10-13-1 6/18/2009 4:30 PM
Cy-JA-10-14-1 6/19/2009 12:15 PM
Cy-JA-10-15-1 6/19/2009 3:30 PM
Cy-JA-10-16-1 6/19/2009
Cy-JA-10-17-1 6/20/2009
5:30 PM
6:15 PM
Cy-JA-10-18-1 6/21/2009 11:15 AM
Cy-JA-10-19-1 6/21/2009 3:15 PM
Cy-JA-10-20-1 6/22/2009 11:00 AM
Cy-JA-10-21-1 6/22/2009 3:20 PM
Cy-JA-10-22-1 6/23/2009 11:00 AM
Cy-JA-10-23-1 6/24/2009 11:00 AM
Cy-JA-10-24-1 6/25/2009 11:00 AM
Cy-JA-10-25-1 6/26/2009 11:00 AM
Sample time
Cultivation Time
(hr)
359
379
382
402
426
450
474
0
308
312
314
338
355
138
164
189
214
238
259
0
20
46
72
92
118
Culture pH
9.64
8.95
9.54
8.97
8.95
8.87
8.88
8.64
9.32
9.68
9.78
9.74
9.07
8.92
9.48
9.58
9.67
9.74
9.38
8.64
8.48
8.57
8.82
8.77
9.3
180
180
180
180
180
180
180
320
220
180
180
180
180
Sample volume, includes 20 mL to clear lines (mL)
320
320
320
60
320
220
220
220
60
60
60
220
Medium volume
(mL)
2430
2250
2070
2177
2850
3008
3790
3922
3742
3562
3382
3202
4950
4400
550
4950
4630
4310
4250
3930
3710
3490
3270
3210
3150
3090
2870
Table B 10. Cy-JA-10, Cell density
Sample#
Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Sample average
(cells/mL)
Sample
+/- 1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor,
S.D.
Cy-JA-10-1-1
Cy-JA-10-1-2
Cy-JA-10-2-1
Cy-JA-10-2-2
Cy-JA-10-3-1
Cy-JA-10-3-2
Cy-JA-10-4-1
Cy-JA-10-4-2
407434
342188
537118
504596
659934
658116
875670
889406
419150
368448
526008
538734
654480
614484
866176
882740
393900
352086
510858
508232
656500
636906
862338
883346
4.07E+05
3.54E+05
5.25E+05
5.17E+05
6.57E+05
6.37E+05
8.68E+05
8.85E+05
1.26E+04
1.33E+04
1.32E+04
1.87E+04
2.76E+03
2.18E+04
6.86E+03
3.69E+03
3.81E+05
5.21E+05
6.47E+05
8.77E+05
3.10E+04
1.51E+04
1.79E+04
1.06E+04
1.88E+09
2.58E+09
3.20E+09
4.34E+09
1.71E+07
8.28E+06
9.83E+06
5.82E+06
Cy-JA-10-5-1 9.44E+05 9.75E+05 9.77E+05 9.65E+05 1.85E+04 1.01E+06 4.68E+04 4.98E+09 2.58E+07
Cy-JA-10-5-2 1.05E+06 1.04E+06 1.05E+06 1.05E+06 5.29E+03
Cy-JA-10-6-1 1.19E+06 1.18E+06 1.14E+06 1.17E+06 2.47E+04 1.17E+06 1.87E+04 5.78E+09 1.03E+07
Cy-JA-10-6-2 1.18E+06 1.15E+06 1.16E+06 1.16E+06 1.57E+04
Cy-JA-10-7-1 1.44E+06 1.43E+06 1.44E+06 1.43E+06 3.21E+03 1.47E+06 4.04E+04 7.28E+09 2.22E+07
Cy-JA-10-7-2 1.50E+06 1.51E+06 1.51E+06 1.51E+06 8.50E+03
Cy-JA-10-8-1 1.77E+06 1.71E+06 1.73E+06 1.74E+06 3.11E+04 1.74E+06 2.25E+04 8.59E+09 1.24E+07
Cy-JA-10-8-2 1.75E+06 1.72E+06 1.74E+06 1.73E+06 1.72E+04
Cy-JA-10-10-1 1.95E+06 1.92E+06 1.92E+06 1.93E+06 1.59E+04 2.07E+06 1.55E+05 1.02E+10 8.52E+07
Cy-JA-10-10-2 2.27E+06 2.19E+06 2.15E+06 2.20E+06 5.65E+04
Cy-JA-10-11-1 2.17E+06 2.14E+06 2.14E+06 2.15E+06 1.73E+04 2.17E+06 1.96E+04 1.07E+10 1.08E+07
Cy-JA-10-11-2 2.19E+06 2.17E+06 2.17E+06 2.18E+06 1.19E+04
Cy-JA-10-12-1 2.07E+06 2.01E+06 2.03E+06 2.04E+06 3.02E+04 2.12E+06 9.74E+04 1.05E+10 5.36E+07
Cy-JA-10-12-2 2.23E+06 2.19E+06 2.21E+06 2.21E+06 2.08E+04
Cy-JA-10-13-1 2.20E+06 2.17E+06 2.24E+06 2.20E+06 3.45E+04 2.20E+06 2.94E+04 1.09E+10 1.62E+07
Table B 10. Continued
Sample#
Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Sample average
(cells/mL)
Sample
+/- 1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
Cy-JA-10-13-2 2.23E+06 2.17E+06 2.18E+06 2.20E+06 3.05E+04
Cy-JA-10-14-1 2.36E+06 2.34E+06 2.25E+06 2.32E+06 5.81E+04 2.30E+06 5.65E+04 1.14E+10 3.11E+07
Cy-JA-10-14-2 2.36E+06 2.28E+06 2.23E+06 2.29E+06 6.42E+04
Cy-JA-10-15-1 2.23E+06 2.26E+06 2.21E+06 2.23E+06 2.37E+04 2.25E+06 2.87E+04 1.12E+10 1.58E+07
Cy-JA-10-15-2 2.26E+06 2.30E+06 2.26E+06 2.27E+06 2.12E+04
Cy-JA-10-16-1 2.36E+06 2.18E+06 2.32E+06 2.29E+06 9.53E+04 2.30E+06 6.31E+04 1.14E+10 3.47E+07
Cy-JA-10-16-2 2.32E+06 2.28E+06 2.32E+06 2.31E+06 2.45E+04
Cy-JA-10-17-1 2.12E+06 2.00E+06 2.02E+06 2.05E+06 6.56E+04 2.02E+06 6.56E+04 1.00E+10 3.61E+07
Cy-JA-10-17-2 2.01E+06 1.99E+06 2.01E+06 2.00E+06 1.31E+04
Cy-JA-10-18-1 1.82E+06 1.73E+06 1.72E+06 1.76E+06 5.21E+04 1.70E+06 1.61E+05 1.01E+10 8.85E+07
Cy-JA-10-18-2 1.63E+06 1.68E+06 1.63E+06 1.65E+06 2.77E+04
Cy-JA-10-19-1 1.86E+06 1.87E+06 1.76E+06 1.83E+06 5.99E+04 1.85E+06 4.77E+04 1.12E+10 2.62E+07
Cy-JA-10-19-2 1.90E+06 1.87E+06 1.83E+06 1.87E+06 3.50E+04
Cy-JA-10-20-1 1.46E+06 1.44E+06 1.45E+06 1.45E+06 9.54E+03 1.46E+06 1.92E+04 9.99E+09 1.06E+07
Cy-JA-10-20-2 1.46E+06 1.49E+06 1.45E+06 1.47E+06 2.23E+04
Cy-JA-10-21-1 1.42E+06 1.44E+06 1.47E+06 1.44E+06 2.21E+04 1.52E+06 8.63E+04 1.06E+10 4.75E+07
Cy-JA-10-21-2 1.62E+06 1.60E+06 1.58E+06 1.60E+06 2.06E+04
Cy-JA-10-22-1 1.67E+06 1.67E+06 1.68E+06 1.67E+06 5.77E+03 1.60E+06 7.87E+04 1.11E+10 4.33E+07
Cy-JA-10-22-2 1.52E+06 1.55E+06 1.52E+06 1.53E+06 1.79E+04
Cy-JA-10-23-1 1.68E+06 1.71E+06 1.70E+06 1.70E+06 1.15E+04 1.60E+06 1.03E+05 1.11E+10 5.65E+07
Cy-JA-10-23-2 1.52E+06 1.49E+06 1.53E+06 1.51E+06 1.89E+04
Cy-JA-10-24-1 1.70E+06 1.76E+06 1.71E+06 1.72E+06 3.50E+04 1.81E+06 9.57E+04 1.26E+10 5.26E+07
Cy-JA-10-24-2 1.92E+06 1.90E+06 1.85E+06 1.89E+06 3.56E+04
Cy-JA-10-25-1 1.78E+06 1.76E+06 1.80E+06 1.78E+06 1.70E+04 1.82E+06 5.21E+04 1.27E+10 2.87E+07
101
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
Sample#
A at
360 nm
(mAU)
Cy-JA-10-1-1
Cy-JA-10-1-2
Cy-JA-10-2-1
Cy-JA-10-2-2
Cy-JA-10-3-1
Cy-JA-10-3-2
Cy-JA-10-4-1
Cy-JA-10-4-2
Cy-JA-10-5-1
Cy-JA-10-5-2
Cy-JA-10-6-1
Cy-JA-10-6-2
0.161
0.188
0.065
Cy-JA-10-7-1
Cy-JA-10-7-2
Cy-JA-10-8-1
Cy-JA-10-8-2
0.085
0.097
0.070
0.120
Cy-JA-10-10-1 0.087
0.436
0.418
0.359
0.295
0.267
0.241
0.181
0.186
0.153
Cy-JA-10-10-2 0.061
Cy-JA-10-11-1 0.03
Cy-JA-10-11-2 0.04
Cy-JA-10-12-1 0.103
Cy-JA-10-12-2 0.055
Cy-JA-10-13-1
Cy-JA-10-13-2 0.095
Cy-JA-10-14-1 0.068
Cy-JA-10-14-2 0.122
Cy-JA-10-15-1 0.067
Cy-JA-10-15-2 0.135
Cy-JA-10-16-1 0.061
Cy-JA-10-16-2 0.069
Cy-JA-10-17-1 0.132
Cy-JA-10-17-2 0.145
Cy-JA-10-18-1 0.098
Cy-JA-10-18-2 0.145
Cy-JA-10-19-1 0.072
Cy-JA-10-19-2 0.111
Cy-JA-10-20-1 0.18
Cy-JA-10-20-2 0.093
Cy-JA-10-21-1 0.135
Cy-JA-10-21-2 0.099
Cy-JA-10-22-1 0.297
Table B 11. Cy-JA-10, Si concentration by spectrophotometer
Dilution Csi (mmolSi/L)
0.10
0.10
0.10
0.07
0.14
0.09
0.10
0.07
0.04
0.08
Average
Csi
(mmolSi/
L)
0.43
0.33
0.26
0.18
0.16
0.13
0.12
0.09
0.14
0.12
0.24
0.14
0.06
0.07
0.13
0.15
0.10
0.15
0.06
0.03
0.04
0.10
0.06
0.10
0.07
0.12
0.07
0.07
0.11
0.18
0.09
0.14
0.10
0.30
0.16
0.19
0.07
0.09
0.10
0.07
0.12
0.09
0.44
0.42
0.36
0.30
0.27
0.24
0.18
0.19
0.15
Csi +/-
1S.D.
(mmolSi/L)
0.01
0.01
0.03
0.01
0.03
0.04
0.05
0.03
0.06
0.03
0.08
0.09
0.01
0.04
0.02
0.01
0.05
0.02
0.00
0.01
Table B 11. Continued
Sample#
A at
360 nm
(mAU)
Cy-JA-10-22-2 0.178
Cy-JA-10-23-1 0.126
Cy-JA-10-23-2 0.201
Cy-JA-10-24-1 0.114
Cy-JA-10-24-2 0.0145
Cy-JA-10-25-1 0.157
Cy-JA-10-25-2 0.189
Dilution
5.0
5.0
5.0
5.0
5.0
5.0
5.0
Csi (mmolSi/L)
0.18
0.13
0.20
0.11
0.01
0.16
0.19
Averag e Csi
(mmolS i/L)
0.16
0.06
0.17
Csi +/-
1S.D.
(mmolSi/L)
0.05
0.07
0.02
103
Table B 12. Cy-JA-10, Si concentration by ICP
Sample#
Cy-JA-10-1-1
Cy-JA-10-1-2
Cy-JA-10-2-1
Cy-JA-10-2-2
Cy-JA-10-3-1
Cy-JA-10-3-2
Cy-JA-10-4-1
Cy-JA-10-4-2
Cy-JA-10-5-1
Cy-JA-10-5-2
Cy-JA-10-6-1
Cy-JA-10-6-2
Cy-JA-10-7-1
Cy-JA-10-7-2
Cy-JA-10-8-1
Cy-JA-10-8-2
Cy-JA-10-10-1
Cy-JA-10-10-2
Cy-JA-10-11-1
Cy-JA-10-11-2
Cy-JA-10-12-1
Cy-JA-10-12-2
Cy-JA-10-13-1
Emission at
251.611 nm
Trial #1
Emission at
251.611 nm
Trial #2
365
381
395
349
375
360
345
687
705
596
541
408
364
1287
1336
1026
1058
918
899
423
402
462
363
382
373
393
337
357
376
335
699
693
588
534
405
372
1289
1322
1035
1057
918
887
413
413
461
383
Emission at
251.611 nm
Trial #3
Trial
Avg.
359
366
394
354
356
357
332
713
701
592
536
412
380
1289
1309
1043
1075
939
892
411
396
464
362
Assay
Conc.
(ppm)
369
373
394
347
363
364
337
700
700
592
537
408
372
1288 1.16
1322 1.19
1035 0.93
1063 0.96
925 0.83
893 0.80
416
404
462
369
0.37
0.36
0.42
0.33
0.63
0.63
0.53
0.48
0.37
0.33
0.33
0.34
0.35
0.31
0.33
0.33
0.30 dilution
Sample conc.
(ppm)
Si conc.
(mmol
Si/L)
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
3.32
3.36
3.55
3.12
3.26
3.28
3.04
11.60
11.90
9.31
9.57
8.33
8.03
6.30
6.30
5.33
4.83
3.68
3.35
3.74
3.63
4.16
3.32
0.41
0.42
0.33
0.34
0.29
0.28
0.22
0.22
0.19
0.17
0.13
0.12
0.13
0.13
0.15
0.12
0.12
0.12
0.12
0.11
0.11
0.11
0.11
Average
Si conc
(mmol
Si/L)
Average Si conc +/- 1
S.D.
(mmol
Si/L)
0.001
0.011
0.000
0.000
0.008
0.006
0.007
0.000
0.012
0.008
0.003
0.021
0.12
0.12
0.11
0.11
0.22
0.18
0.12
0.42
0.33
0.29
0.13
0.13
Table B 12. Continued
Sample#
Cy-JA-10-13-2
Cy-JA-10-14-1
Cy-JA-10-14-2
Cy-JA-10-15-1
Cy-JA-10-15-2
Cy-JA-10-16-1
Cy-JA-10-16-2
Cy-JA-10-17-1
Cy-JA-10-17-2
Cy-JA-10-18-1
Cy-JA-10-18-2
Cy-JA-10-19-1
Cy-JA-10-19-2
Cy-JA-10-20-1
Cy-JA-10-20-2
Cy-JA-10-21-1
Cy-JA-10-21-2
Cy-JA-10-22-1
Cy-JA-10-22-2
Cy-JA-10-24-2
Cy-JA-10-25-1
Cy-JA-10-25-2
Emission at
251.611 nm
Trial #1
Emission at
251.611 nm
Trial #2
417
397
319
334
352
327
384
410
408
414
446
323
402
329
372
355
444
418
394
403
440
392
419
392
316
325
351
321
382
417
402
411
439
318
409
331
366
340
431
417
400
390
424
389
Emission at
251.611 nm
Trial #3
Trial
Avg.
424
389
324
328
348
320
390
419
410
412
435
327
409
348
369
358
426
427
395
399
434
385
420
393
320
329
350
323
385
415
407
412
440
323
407
336
369
351
434
421
396
397
433
389
0.38
0.35
0.29
0.30
0.32
0.29
0.35
0.37
0.37
0.37
0.40
0.29
0.37
0.30
0.33
0.32
0.39
0.38
0.36
0.36
0.39
0.35
Assay
Conc.
(ppm) dilution
Sample conc.
(ppm)
Si conc.
(mmol
Si/L)
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
3.78
3.53
2.88
2.96
3.15
2.90
3.47
3.74
3.66
3.71
3.96
2.90
3.66
3.02
3.32
3.16
3.90
3.79
3.57
3.58
3.89
3.50
0.11
0.12
0.11
0.14
0.13
0.12
0.13
0.13
0.13
0.14
0.10
0.13
0.13
0.14
0.12
0.13
0.12
0.10
0.10
0.11
0.10
0.12
0.13
0.10
0.11
0.11
0.13
0.12
0.13
0.11
0.13
0.13
0.13
Average
Si conc
(mmol
Si/L)
Average Si conc +/- 1
S.D.
(mmol
Si/L)
0.006
0.002
0.014
0.004
0.003
0.004
0.001
0.027
0.002
0.010
105
106
Table B 13. Cy-JA-10, Lipid data
Cy-JA-10-1-1
Cy-JA-10-1-2
Cy-JA-10-2-1
Cy-JA-10-2-2
Cy-JA-10-3-1
Cy-JA-10-3-2
Cy-JA-10-5-1
Cy-JA-10-5-2
Cy-JA-10-8-1
Cy-JA-10-8-2
Cy-JA-10-12-1
Cy-JA-10-12-2
Cy-JA-10-14-1
Cy-JA-10-14-2
Cy-JA-10-16-1
Cy-JA-10-16-2
Cy-JA-10-17-1
Cy-JA-10-17-2
Cy-JA-10-20-1
Dry cell mass
(mg)
Total vol lipid extract
(mL)
Extract assay vol
(mL)
Trial 1 Trial 2
Dilution
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Trial 3
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid average wt% st dev adjusted wt% adj stdev
21.4
21
20
23
20.3
15
13.7
23
34.2
17.2
17.1
20.4
20.1
20.4
20.4
25
25
9.7
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
11
11
11
11.5
11.5
11
11.5
11.8
8.5
8.5
8.5
8.5
11.4
11.4
11.8
11.6
10.8
10.5
11.2
0.05 1.401
0.05 1.445
0.05 1.316
0.05 1.024
0.05 1.154
0.05 1.09
0.05 1.111
0.05 1.124
0.05 1.06
0.05 0.903
0.05
0.05
0.05
0.05
0.05
0.05
0.81
0.83
0.05 0.877
0.785
0.05 0.666
0.705
1.263
2.16 6.99
1.72 4.85
3.01 9.44
5.93 21.93
4.63 21.71
5.27 25.29
5.06 25.08
4.93 32.36
5.57 38.04
7.14 29.19
8.07 37.65
7.87 37.32
7.40 36.60
8.32 37.88
9.51 38.57
9.12 38.53
3.54 34.00
1.403
1.408
1.264
1.41
1.185
1.059
1.171
1.342
1.095
0.835
0.786
0.874
1.035
0.788
0.688
0.708
1.305
2.14
2.09
6.89
6.69
3.53 11.60
2.07 5.60
4.32 19.98
5.58 27.02
4.46 21.59
2.75 15.51
5.22 35.28
7.82 32.30
8.31
7.43
5.82
8.29
9.29
9.09
3.12
38.89
35.02
27.91
37.73
37.60
38.39
28.89
1.323
1.323
1.226
1.213
1.159
1.111
1.144
1.147
1.12
0.75
0.767
0.811
0.953
0.744
0.686
0.725
1.274
2.94 10.85
2.94 10.91
3.91 13.19
4.04 13.93
4.58 21.43
5.06 24.11
4.73 23.16
4.70 30.59
4.97 33.31
8.67 36.18
8.50 39.86
8.06 38.32
6.64 32.42
8.73 39.93
9.31
8.92
37.69
37.61
3.43 32.66
7.86
12.62
23.26 2.66 23.14
24.72 6.17
34.05 3.13
38.80
34.60
38.51
38.06
32.00
2.46 6.86
5.46
1.11
3.87
1.23
0.48
2.72
12.04
25.11
34.27 1.78
38.80
35.94
38.51
37.96
31.78
0.16
1.99
1.87
1.11
2.30
1.23
0.46
1.94
107
Table B 13. Continued
Cy-JA-10-22-1
Cy-JA-10-22-2
Cy-JA-10-24-1
Cy-JA-10-24-2
Cy-JA-10-25-1
Dry cell mass
(mg)
Total vol lipid extract
(mL)
Extract assay vol
(mL)
Trial 1 Trial 2
Dilution
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L)
Trial 3 wt% lipid
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid average wt% st dev adjusted wt% adj stdev
19.8
17.4
16.5
17.1
10.1
11.5
11.5
11.8
11.8
11.5
0.2
0.2
0.2
0.2
0.2
0.05 0.985
0.05 1.05
0.05 0.977
0.05 0.899
0.05 1.1
6.32 32.38
5.67 32.55
6.40 40.44
7.18 44.41
5.17 50.38
0.96
0.97
0.993
0.956
1.15
6.57 33.83
6.47 37.84
6.24 39.30
6.61 40.47
4.67 44.69
0.959
0.918
1.026
0.932
1.13
6.58 33.89
6.99 41.27
5.91 36.94
6.85 42.13
4.87 46.97
35.29 3.53
40.61
47.35
2.53
2.31
34.10 2.20
40.58
47.35
1.17
2.31
Table B 14. Cy-JA-11, Cultivation information
Sample# Date medium
Inoculum
Cy-JA-11-1-1 7/28/2009 11:40 AM
Cy-JA-11-2-1 7/29/2009
Cy-JA-11-3-1 7/30/2009
Cy-JA-11-4-1 7/31/2009
Cy-JA-11-5-1 8/3/2009
4:00 PM
2:00 PM
2:15 PM
1:30 PM
Cy-JA-11-6-1 8/4/2009
Cy-JA-11-7-1 8/5/2009
4:45 PM
4:00 PM
Cy-JA-11-8-1 8/6/2009
Cy-JA-11-9-1 8/7/2009
Cy-JA-11-10-1 8/7/2009
Cy-JA-11-11-1 8/8/2009
Cy-JA-11-12-1 8/9/2009
Cy-JA-11-13-1 8/10/2009
4:00 PM
12:00 PM
3:30 PM
1:15 PM
3:00 PM
2:00 PM
Cy-JA-11-14-1 8/12/2009 11:45 AM
Cy-JA-11-15-1 8/12/2009 3:30 PM
Cy-JA-11-16-1 8/13/2009 4:00 PM
Cy-JA-11-17-1 8/15/2009
Cy-JA-11-18-1 8/18/2009
3:45 PM
1:00 PM
Cy-JA-11-19-1 8/19/2009 11:00 AM
Cy-JA-11-20-1 8/19/2009
Cy-JA-11-21-1 8/19/2009
1:30 PM
5:30 PM
Cy-JA-11-22-1 8/20/2009 11:00 AM
Cy-JA-11-23-1 8/20/2009
Cy-JA-11-24-1 8/20/2009
Cy-JA-11-25-1 8/21/2009
2:00 PM
9:30 PM
1:15 PM
Cy-JA-11-26-1 8/22/2009 11:00 AM
Cy-JA-11-27-1 8/24/2009 2:00 PM
Cy-JA-11-28-1 8/25/2009 1:30 PM
Sample time
Cultivation Time
(hr)
334.1
337.8
362.3
410.1
479.6
504.1
508.1
525.6
528.6
536.1
551.8
573.6
600.6
624.1
0.0
0.0
28.3
50.3
72.6
143.8
171.1
194.3
218.3
238.3
241.8
263.6
289.3
312.3
Culture pH
9.38
8.79
9.27
9.76
9.38
9.05
9.03
9.65
9.69
9.45
9.28
9.03
9.56
9.45
8.28
9.71
9.34
9.72
9.44
9.61
9.37
8.28
8.59
9.01
9.42
9.65
9.75
9.75
120
120
120
120
120
120
60
120
120
120
120
120
120
120
320
Sample volume, includes 20 mL to clear lines (mL)
320
60
320
60
320
60
320
180
180
180
180
180
60
Medium volume
(mL)
2027
2488
2568
2765
3181
3755
2395
2275
2155
2035
1915
1921
3635
3515
4555
4500
375
4555
4495
4175
4115
3795
3735
3415
3235
3055
2875
2695
2515
2455
Table B 15. Cy-JA-11, Cell density
Sample#
Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
Cy-JA-11-1-1 2.044E+05 2.085E+05 1.957E+05 2.022E+05 8.334E+03 9.859E+08 4.063E+07
Cy-JA-11-1-2 1.959E+05 2.147E+05 1.941E+05
Cy-JA-11-2-1 3.052E+05 3.159E+05 3.153E+05 3.117E+05 5.929E+03 1.520E+09 2.890E+07
Cy-JA-11-2-2 3.157E+05 3.150E+05 3.030E+05
Cy-JA-11-3-1 4.852E+05 4.767E+05 4.836E+05 4.770E+05 9.603E+03 2.325E+09 4.681E+07
Cy-JA-11-3-2 4.838E+05 4.725E+05 4.602E+05
Cy-JA-11-4-1 7.470E+05 7.383E+05 7.415E+05 7.491E+05 1.145E+04 3.652E+09 5.582E+07
Cy-JA-11-4-2 7.674E+05 7.417E+05 7.585E+05
Cy-JA-11-5-1 1.089E+06 1.099E+06 1.079E+06 1.051E+06 4.250E+04 5.124E+09 2.072E+08
Cy-JA-11-5-2 1.003E+06 1.021E+06 1.015E+06
Cy-JA-11-6-1 1.146E+06 1.160E+06 1.145E+06 1.140E+06 5.274E+04 5.556E+09 2.571E+08
Cy-JA-11-6-2 1.075E+06 1.222E+06 1.090E+06
Cy-JA-11-7-1 1.189E+06 1.191E+06 1.168E+06 1.167E+06 2.476E+04 5.688E+09 1.207E+08
Cy-JA-11-7-2 1.149E+06 1.127E+06 1.176E+06
Cy-JA-11-8-1 1.198E+06 1.176E+06 1.199E+06 1.172E+06 2.749E+04 5.714E+09 1.340E+08
Cy-JA-11-8-2 1.180E+06 1.130E+06 1.149E+06
Cy-JA-11-9-1 1.188E+06 1.173E+06 1.179E+06 1.157E+06 2.882E+04 5.640E+09 1.405E+08
Cy-JA-11-9-2 1.121E+06 1.123E+06 1.157E+06
Cy-JA-11-10-1 1.154E+06 1.150E+06 1.050E+06 1.154E+06 5.737E+04 5.623E+09 2.797E+08
Cy-JA-11-10-2 1.205E+06 1.209E+06 1.153E+06
Cy-JA-11-11-1 1.340E+06 1.328E+06 1.319E+06 1.323E+06 1.876E+04 6.449E+09 9.143E+07
Cy-JA-11-11-2 1.341E+06 1.319E+06 1.290E+06
Cy-JA-11-12-1 1.287E+06 1.320E+06 1.356E+06 1.324E+06 2.719E+04 6.454E+09 1.326E+08
Table B 15. Continued
Sample#
Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
Cy-JA-11-12-2 1.350E+06 1.300E+06 1.330E+06
Cy-JA-11-13-1 1.489E+06 1.466E+06 1.481E+06 1.440E+06 4.668E+04 7.020E+09 2.276E+08
Cy-JA-11-13-2 1.411E+06 1.424E+06 1.369E+06
Cy-JA-11-14-1 1.447E+06 1.423E+06 1.387E+06 1.506E+06 9.676E+04 7.339E+09 4.717E+08
Cy-JA-11-14-2 1.592E+06 1.585E+06 1.599E+06
Cy-JA-11-15-1 1.518E+06 1.513E+06 1.549E+06 1.505E+06 7.212E+04 7.338E+09 3.516E+08
Cy-JA-11-15-2 1.606E+06 1.419E+06 1.426E+06
Cy-JA-11-16-1 1.619E+06 1.551E+06 1.515E+06 1.605E+06 5.886E+04 7.825E+09 2.870E+08
Cy-JA-11-16-2 1.663E+06 1.648E+06 1.635E+06
Cy-JA-11-17-1 1.651E+06 1.686E+06 1.635E+06 1.663E+06 1.821E+04 8.106E+09 8.880E+07
Cy-JA-11-17-2 1.660E+06 1.670E+06 1.675E+06
Cy-JA-11-18-1 1.641E+06 1.639E+06 1.629E+06 1.658E+06 2.791E+04 8.080E+09 1.360E+08
Cy-JA-11-18-2 1.706E+06 1.666E+06 1.664E+06
Cy-JA-11-19-1 1.630E+06 1.601E+06 1.620E+06 1.610E+06 1.492E+04 7.850E+09 7.274E+07
Cy-JA-11-19-2 1.598E+06 1.620E+06 1.593E+06
Cy-JA-11-20-1 1.595E+06 1.491E+06 1.521E+06 1.588E+06 7.364E+04 7.848E+09 3.590E+08
Cy-JA-11-20-2 1.692E+06 1.636E+06 1.595E+06
Cy-JA-11-21-1 1.653E+06 1.623E+06 1.612E+06 1.647E+06 2.920E+04 8.312E+09 1.424E+08
Cy-JA-11-21-2 1.696E+06 1.646E+06 1.652E+06
Cy-JA-11-22-1 1.395E+06 1.408E+06 1.430E+06 1.399E+06 1.715E+04 7.707E+09 8.361E+07
Cy-JA-11-22-2 1.389E+06 1.387E+06 1.386E+06
Cy-JA-11-23-1 1.344E+06 1.357E+06 1.406E+06 1.382E+06 2.556E+04 7.723E+09 1.246E+08
Cy-JA-11-23-2 1.391E+06 1.392E+06 1.403E+06
110
Cy-JA-11-1-1
Cy-JA-11-1-2
Cy-JA-11-2-1
Cy-JA-11-2-2
Cy-JA-11-3-1
Cy-JA-11-3-2
Cy-JA-11-4-1
Cy-JA-11-4-2
Cy-JA-11-5-1
Cy-JA-11-5-2
Cy-JA-11-6-1
Cy-JA-11-6-2
Cy-JA-11-7-1
Cy-JA-11-7-2
Cy-JA-11-8-1
Cy-JA-11-8-2
Cy-JA-11-9-1
Cy-JA-11-9-2
Cy-JA-11-10-1
Cy-JA-11-10-2
Cy-JA-11-11-1
Cy-JA-11-11-2
Cy-JA-11-12-1
Cy-JA-11-12-2
Cy-JA-11-13-1
Cy-JA-11-13-2
Cy-JA-11-14-1
Cy-JA-11-14-2
Cy-JA-11-15-1
Cy-JA-11-15-2
Cy-JA-11-16-1
Cy-JA-11-16-2
Cy-JA-11-17-1
Cy-JA-11-17-2
Cy-JA-11-18-1
Cy-JA-11-18-2
Cy-JA-11-20-1
Cy-JA-11-20-2
Cy-JA-11-21-1
Cy-JA-11-21-2
Cy-JA-11-22-1
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
A at
360 nm
(mAU)
0.115
0.096
0.095
0.101
0.122
0.101
0.079
0.082
0.103
0.100
0.085
0.105
0.102
0.103
0.121
0.109
0.084
0.123
0.116
0.133
0.113
0.125
0.122
0.118
0.127
0.179
0.604
0.595
0.530
0.541
0.329
0.337
0.106
0.089
0.088
0.072
0.132
0.029
0.112
0.080
Table B 16. Cy-JA-11, Si concentration by spectrophotometer
Sample# Dilution Csi (mmolSi/L)
0.08
0.09
0.08
0.09
0.08
0.09
0.10
0.08
0.10
0.10
0.10
0.10
0.15
0.09
0.08
0.09
0.06
0.07
Average
Csi
(mmolSi/
L)
0.51
0.46
0.28
0.10
0.08
0.08
0.09
0.10
0.09
0.07
0.07
0.09
0.09
0.07
0.09
0.09
0.09
0.10
0.09
0.07
0.10
0.10
0.11
0.10
0.11
0.10
0.10
0.11
0.15
0.09
0.08
0.07
0.06
0.11
0.02
0.10
0.07
0.51
0.51
0.45
0.46
0.28
0.29
Csi +/-
1S.D.
(mmolSi/L)
0.00
0.01
0.01
0.00
0.01
0.00
0.00
0.02
0.00
0.01
0.00
0.01
0.00
0.00
0.04
0.05
0.00
0.01
0.01
0.00
Table B 16. Continued
Sample#
Cy-JA-11-22-2
Cy-JA-11-23-1
Cy-JA-11-23-2
Cy-JA-11-24-1
Cy-JA-11-24-2
Cy-JA-11-25-1
Cy-JA-11-25-2
Cy-JA-11-26-1
Cy-JA-11-26-2
Cy-JA-11-27-1
Cy-JA-11-27-2
Cy-JA-11-28-1
Cy-JA-11-28-2
A at
360 nm
(mAU)
0.184
0.163
0.170
0.156
0.143
0.215
0.233
0.240
0.301
0.195
0.174
0.218
0.190
Dilution
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
0.20
0.20
0.26
0.17
0.15
0.19
0.16
0.16
0.14
0.14
0.13
0.12
0.18
Csi (mmolSi/L)
Average
Csi
(mmolSi/
L)
0.14
0.13
0.19
0.23
0.16
0.17
Csi +/-
1S.D.
(mmolSi/L)
0.04
0.01
0.02
0.00
0.01
0.01
112
Table B 17. Cy-JA-11, Si concentration by ICP
Sample#
Cy-JA-11-1-1
Cy-JA-11-1-2
Cy-JA-11-3-1
Cy-JA-11-3-2
Cy-JA-11-5-1
Cy-JA-11-5-2
Cy-JA-11-7-1
Cy-JA-11-7-2
Cy-JA-11-9-1
Cy-JA-11-9-2
Cy-JA-11-11-1
Cy-JA-11-11-2
Cy-JA-11-13-1
Cy-JA-11-13-2
Cy-JA-11-15-1
Cy-JA-11-15-2
Cy-JA-11-17-1
Cy-JA-11-17-2
Cy-JA-11-21-1
Cy-JA-11-21-2
Cy-JA-11-23-1
Cy-JA-11-23-2
Cy-JA-11-25-1
Emission at
251.611 nm
Trial #1
Emission at
251.611 nm
Trial #2
886
1317
955
919
999
942
870
952
961
1060
1101
1134
3342
3425
1912
1913
874
809
913
894
938
917
909
931
1346
948
925
980
948
878
957
988
1058
1085
1068
3373
3382
1850
1926
853
796
864
891
965
971
908
Emission at
251.611 nm
Trial #3
Trial
Avg.
883
1331
973
933
958
974
846
964
960
1058
1128
1079
3402
3438
1908
1956
849
816
848
918
943
932
903
900
1331
959
926
979
955
865
958
970
1059
1105
1094
3372
3415
1890
1932
859
807
875
901
949
940
907
0.38
0.39
0.42
0.44
0.44
0.36
0.53
0.38
0.37
0.39
0.38
0.35
1.35
1.37
0.76
0.77
0.34
0.32
0.35
0.36
0.38
0.38
0.36
Assay
Conc.
(ppm) dilution
Sample conc.
(ppm)
Si conc.
(mmol
Si/L)
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
3.83
3.88
4.23
4.42
4.37
3.60
5.33
3.83
3.70
3.92
3.82
3.46
13.49
13.66
7.56
7.73
3.43
3.23
3.50
3.60
3.79
3.76
3.63
0.476
0.482
0.265
0.271
0.118
0.111
0.121
0.124
0.131
0.130
0.125
0.124
0.185
0.132
0.128
0.135
0.132
0.119
0.132
0.134
0.147
0.153
0.152
0.159
0.132
0.125
0.133
0.150
0.152
0.479
0.268
0.115
0.122
0.130
0.125
Average
Si conc
(mmol
Si/L)
Average Si conc +/- 1
S.D.
(mmol
Si/L)
0.038
0.005
0.009
0.001
0.005
0.001
0.004
0.004
0.005
0.003
0.001
0.001
Table B 17. Continued
Sample#
Cy-JA-11-26-1
Cy-JA-11-26-2
Cy-JA-11-28-1
Cy-JA-11-28-2
Emission at
251.611 nm
Trial #1
319
334
327
384
Emission at
251.611 nm
Trial #2
316
325
321
382
Emission at
251.611 nm
Trial #3
Trial
Avg.
324
328
320
390
320
329
323
385
Assay
Conc.
(ppm)
0.26
0.26
0.26
0.31 dilution
Sample conc.
(ppm)
Si conc.
(mmol
Si/L)
10
10
10
10
2.56
2.63
2.58
3.08
0.087
0.090
0.088
0.106
Average
Si conc
(mmol
Si/L)
0.088
0.097
Average Si conc +/- 1
S.D.
(mmol
Si/L)
0.002
0.013
Table B 18. Cy-JA-11, Lipid data
Cy-JA-11-1-1
Cy-JA-11-1-2
6.0
6.3
Cy-JA-11-5-1 16.5
Cy-JA-11-5-2 15.8
Cy-JA-11-7-1 17.0
Cy-JA-11-7-2 16.6
12.5
12.75
12.75
12.75
11.75
11.75
Cy-JA-11-11-1 17.4
Cy-JA-11-11-2 17.1
11.75
11.75
Cy-JA-11-15-1 15.5 12.5
Cy-JA-11-15-2 13.1 13
Cy-JA-11-19-1 7.8 12.5
Cy-JA-11-19-2
Cy-JA-11-21-1
Cy-JA-11-21-2
Cy-JA-11-22-1
Cy-JA-11-22-2
Cy-JA-11-24-1
Cy-JA-11-24-2
Cy-JA-11-26-1
9.7
20.0
19.5
16.4
17.2
14.7
11.5
10.3
13
12.5
12.5
12.5
12.5
12.5
9
12
Dry cell mass
(mg)
Total vol lipid extract
(mL)
Extract assay vol
(mL)
Trial 1 Trial 2
Dilution
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Trial 3
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid average wt% st dev adjusted wt% adj stdev
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.05
0.05
0.05
0.05
0.05
0.05
1.425
1.42
1.186
1.213
0.992
1.072
0.05
0.05
0.05
0.05
0.859
0.896
0.832
0.815
0.05 0.986
0.05 0.977
0.05 0.559
0.05 0.607
0.05 0.644
0.05 0.657
0.05 0.682
0.05 0.732
0.05
0.375
0.4
7.81
8.10
1.475
1.427
1.57 14.59 1.228
1.435 14.15 1.244
2.54
2.14
19.76 1.002
17.40 1.083
3.205 23.79 0.822
3.02 22.94 0.883
3.34 29.50 0.815
3.425 37.15 0.844
2.57 46.29 0.977
2.615 39.31
4.705 31.40
4.465 30.66
4.28 35.05
4.215 32.95
4.09 37.49
3.84 32.54
7.5 91.09
0.977
0.522
0.502
0.656
0.68
0.638
0.695
0.125
0.365
1.36
1.28
2.49
2.085
9.24
13.83
12.97
12.90
19.41
17.01
1.424
1.425
1.209
1.209
0.992
1.095
3.39
3.085
25.04
23.39
3.425 30.19
3.28 35.71
2.615 47.01
0.791
0.882
0.798
0.793
1.011
2.615 39.31
4.89 32.55
4.99 34.03
4.22 34.59
4.1 32.11
4.31 39.36
4.025 33.99
7.5 91.09
0.961
0.549
0.525
0.653
0.7
0.666
0.691
0.38 14.55
0.375 14.03
1.455 13.70
1.455 14.31
2.54 19.76
2.025 16.59
3.545 26.09
3.09 23.42
3.51 30.87
3.535 38.24
2.445 44.28
2.695 40.38
4.755 31.71
4.875 33.29
4.235 34.71
4 31.38
4.17 38.17
4.045 34.15
7.5 91.09
11.26
13.77
18.32
24.11
33.61
42.77
32.27
33.46
35.95
38.82
3.20
0.71
1.47
1.20
3.86
3.53
1.25
1.53
0.88
4.98
11.66 3.40
14.05 0.31
18.67
23.79
33.48
42.77
32.27
33.46
35.95
38.82
1.35
0.80
3.47
3.53
1.25
1.53
0.88
4.98
Table B 18. Continued
Trial 1 Trial 2 Trial 3
Cy-JA-11-26-2
Dry cell mass
(mg)
Total vol lipid extract
(mL)
Extract assay vol
(mL)
9.7 12 0.2
Dilution
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L)
0.05 0.889 3.055 41.73 0.89 3.05 wt% lipid
41.67
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L)
1.029 wt% lipid
2.355 33.07 average wt% st dev adjusted wt% adj stdev
Cy-JA-11-27-1 7.2
Cy-JA-11-27-2 7.9
Cy-JA-11-28-1 4.5
Cy-JA-11-28-2 4.6
12.5
13
13
13
0.2
0.2
0.2
0.2
0.05
0.05
0.05
0.05
0.975
0.942
1.177
1.204
2.625 51.10
2.79 51.15
1.615 55.85
1.48 50.82
0.975
0.942
1.204
1.164
2.625
2.79
1.48
1.68
51.10
51.15
51.95
56.47
0.958
0.991
1.218
1.164
2.71 52.58
2.545 47.12
1.41 49.93
1.68 56.47
50.70 1.85
53.58 3.02
51.41
54.32
0.65
2.71
Table B 19. Cy-JA-12, Cultivation information
Sample# Date
Sample time medium
Inoculum
Cy-JA-12-1-1 7/28/2009 11:40 AM
Cy-JA-12-2-1 7/29/2009
Cy-JA-12-3-1 7/30/2009
Cy-JA-12-4-1 7/31/2009
Cy-JA-12-5-1 8/3/2009
4:00 PM
2:00 PM
2:15 PM
1:30 PM
Cy-JA-12-6-1
Cy-JA-12-7-1
8/4/2009
8/5/2009
4:45 PM
4:00 PM
Cy-JA-12-8-1
Cy-JA-12-9-1
8/6/2009
8/7/2009
Cy-JA-12-10-1 8/7/2009
4:00 PM
12:00 PM
3:30 PM
Cy-JA-12-11-1 8/8/2009
Cy-JA-12-12-1 8/9/2009
1:15 PM
3:00 PM
Cy-JA-12-13-1 8/10/2009 2:00 PM
Cy-JA-12-14-1 8/12/2009 11:45 AM
Cy-JA-12-15-1 8/12/2009 3:30 PM
Cy-JA-12-16-1 8/13/2009 4:00 PM
Cy-JA-12-17-1 8/15/2009
Cy-JA-12-18-1 8/18/2009
3:45 PM
1:00 PM
Cy-JA-12-19-1 8/19/2009 11:00 AM
Cy-JA-12-20-1 8/19/2009
Cy-JA-12-21-1 8/19/2009
1:30 PM
5:30 PM
Cy-JA-12-22-1 8/20/2009 11:00 AM
Cy-JA-12-23-1 8/20/2009
Cy-JA-12-24-1 8/20/2009
Cy-JA-12-25-1 8/21/2009
2:00 PM
9:30 PM
1:15 PM
Cy-JA-12-26-1 8/22/2009 11:00 AM
Cy-JA-12-27-1 8/24/2009 2:00 PM
Cy-JA-12-28-1 8/25/2009 1:30 PM
Cy-JA-12-30-1 8/28/2009 2:00 PM
334.08
337.83
362.3
386.1
407.6
432.1
436.1
453.6
456.6
464.1
479.8
501.6
0.0
28.3
50.3
72.6
143.8
171.1
194.3
218.3
238.3
241.8
263.6
289.3
312.3
528.6
624.6
0.0
624.6
Cultivation
Time (hr)
Culture pH
120
120
120
120
120
120
60
120
120
120
120
120
Sample volume, includes 20 mL to clear lines (mL)
320
60
320
60
320
60
320
180
180
180
180
180
60
120
120
320
120
9.38
8.79
9.27
9.76
9.38
9.05
9.05
9.55
9.69
9.45
9.28
9.03
9.82
9.42
9.78
9.61
9.6
9.36
8.29
8.61
9.05
9.45
9.68
9.75
9.74
9.56
9.13
8.29
9.13
Medium volume
(mL)
2027
2488
2568
2765
3181
3755
2395
2275
2155
2035
1915
1921
3235
3055
2875
2695
2515
2455
4500
375
4555
4495
4175
4115
3795
3735
3415
4528
4288
4555
4288
Table B 20. Cy-JA-12, Cell density
Cy-JA-12-1-1
Cy-JA-12-1-2
Cy-JA-12-2-1
Cy-JA-12-2-2
Cy-JA-12-3-1
Cy-JA-12-3-2
Cy-JA-12-4-1
Cy-JA-12-4-2
Cy-JA-12-5-1
Cy-JA-12-5-2
Cy-JA-12-6-1
Cy-JA-12-6-2
Cy-JA-12-7-1
Cy-JA-12-7-2
Cy-JA-12-8-1
Cy-JA-12-8-2
Sample# Trial #1
(cells/ mL)
217352
211696
304010
281184
477932
461974
744572
720130
Trial #2
(cells/ mL)
223816
195334
308858
299162
495304
490456
724372
732856
Trial #3
(cells/ mL)
211898
Trial average
(cells/mL)
2.177E+05
Trial +/- 1
S.D.
(cells/mL)
5.966E+03
173316 1.934E+05 1.926E+04
Sample average
(cells/mL)
2.06E+05
Sample +/-
1 S.D.
(cells/mL)
1.84E+04
Total No. cells in reactor
1.00E+09
Total No. cells in reactor +/-
1 S.D.
8.97E+07
310070 3.076E+05 3.207E+03 2.99E+05 1.14E+04 1.46E+09 5.54E+07
289870 2.901E+05 8.991E+03
476316 4.832E+05 1.053E+04 4.84E+05 1.51E+04 2.36E+09 7.37E+07
503788
731038
748612
4.854E+05
7.333E+05
7.339E+05
2.136E+04
1.029E+04
1.427E+04
7.34E+05 1.11E+04 3.58E+09 5.43E+07
1.05E+06 1.09E+06 1.01E+06 1.050E+06 3.751E+04 1.01E+06 4.70E+04 4.94E+09 2.29E+08
9.86E+05 9.64E+05 9.82E+05 9.772E+05 1.184E+04
1.07E+06 1.07E+06 1.05E+06 1.066E+06 1.058E+04 1.09E+06 5.69E+04 5.31E+09 2.77E+08
1.13E+06 1.18E+06 1.02E+06 1.111E+06 8.045E+04
1.13E+06 1.13E+06 1.13E+06 1.129E+06 3.606E+03 1.12E+06 4.09E+04 5.48E+09 1.99E+08
1.19E+06 1.11E+06 1.06E+06 1.118E+06 6.391E+04
1.19E+06 1.11E+06 1.06E+06 1.118E+06 6.391E+04 1.15E+06 5.12E+04 5.59E+09 2.50E+08
1.17E+06 1.17E+06 1.18E+06 1.176E+06 3.786E+03
Cy-JA-12-9-1
Cy-JA-12-9-2
1.16E+06 1.11E+06 1.15E+06 1.140E+06 2.325E+04 1.17E+06 3.61E+04 5.70E+09 1.76E+08
1.19E+06 1.20E+06 1.21E+06 1.199E+06 1.002E+04
Cy-JA-12-10-1 1.21E+06 1.26E+06 1.22E+06 1.230E+06 2.427E+04 1.24E+06 3.00E+04 6.07E+09 1.46E+08
Cy-JA-12-10-2 1.27E+06 1.29E+06 1.23E+06 1.260E+06 3.164E+04
Cy-JA-12-11-1 1.26E+06 1.26E+06 1.27E+06 1.265E+06 7.638E+03 1.27E+06 1.76E+04 6.21E+09 8.58E+07
Cy-JA-12-11-2 1.31E+06 1.27E+06 1.28E+06 1.284E+06 2.065E+04
Cy-JA-12-12-1 1.27E+06 1.28E+06 1.24E+06 1.265E+06 1.950E+04 1.36E+06 1.06E+05 6.63E+09 5.17E+08
Table B 20. Continued
Sample#
Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Sample average
(cells/mL)
Sample +/-
1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
Cy-JA-12-12-2 1.48E+06 1.41E+06 1.46E+06 1.453E+06 3.568E+04
Cy-JA-12-13-1 1.41E+06 1.44E+06 1.43E+06 1.427E+06 1.501E+04 1.46E+06 4.03E+04 7.13E+09 1.97E+08
Cy-JA-12-13-2 1.49E+06 1.50E+06 1.50E+06 1.497E+06 9.866E+03
Cy-JA-12-14-1 1.47E+06 1.41E+06 1.42E+06 1.436E+06 3.223E+04 1.47E+06 6.72E+04 7.19E+09 3.28E+08
Cy-JA-12-14-2 1.60E+06 1.46E+06 1.48E+06 1.513E+06 7.601E+04
Cy-JA-12-15-1 1.49E+06 1.35E+06 1.49E+06 1.443E+06 8.346E+04 1.52E+06 1.84E+05 7.40E+09 8.96E+08
Cy-JA-12-15-2 1.72E+06 1.75E+06 1.31E+06 1.592E+06 2.466E+05
Cy-JA-12-16-1 1.62E+06 1.55E+06 1.52E+06 1.562E+06 5.281E+04 1.61E+06 5.89E+04 7.83E+09 2.87E+08
Cy-JA-12-16-2 1.66E+06 1.65E+06 1.64E+06 1.649E+06 1.401E+04
Cy-JA-12-17-1 1.65E+06 1.69E+06 1.61E+06 1.648E+06 3.907E+04 1.65E+06 2.67E+04 8.06E+09 1.30E+08
Cy-JA-12-17-2 1.66E+06 1.67E+06 1.65E+06 1.658E+06 1.375E+04
Cy-JA-12-18-1 1.64E+06 1.64E+06 1.63E+06 1.636E+06 6.429E+03 1.66E+06 2.79E+04 8.08E+09 1.36E+08
Cy-JA-12-18-2 1.71E+06 1.67E+06 1.66E+06 1.679E+06 2.369E+04
Cy-JA-12-19-1 1.63E+06 1.60E+06 1.62E+06 1.617E+06 1.473E+04 1.61E+06 1.49E+04 7.85E+09 7.27E+07
Cy-JA-12-19-2 1.60E+06 1.62E+06 1.59E+06 1.604E+06 1.436E+04
Cy-JA-12-20-1 1.60E+06 1.49E+06 1.52E+06 1.536E+06 5.353E+04 1.59E+06 7.36E+04 7.85E+09 3.59E+08
Cy-JA-12-20-2 1.69E+06 1.64E+06 1.60E+06 1.641E+06 4.869E+04
Cy-JA-12-21-1 1.65E+06 1.62E+06 1.61E+06 1.629E+06 2.122E+04 1.65E+06 2.92E+04
Cy-JA-12-21-2 1.70E+06 1.65E+06 1.65E+06 1.665E+06 2.730E+04
Cy-JA-12-22-1 1.40E+06 1.41E+06 1.43E+06 1.411E+06 1.769E+04 1.40E+06 1.72E+04
Cy-JA-12-22-2 1.39E+06 1.39E+06 1.39E+06 1.387E+06 1.528E+03
Cy-JA-12-23-1 1.34E+06 1.36E+06 1.41E+06 1.369E+06 3.270E+04 1.38E+06 2.56E+04
Cy-JA-12-23-2 1.39E+06 1.39E+06 1.40E+06 1.395E+06 6.658E+03
8.31E+09 1.42E+08
7.71E+09 8.36E+07
7.72E+09 1.25E+08
119
Table B 20. Continued
Sample#
Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Sample average
(cells/mL)
Cy-JA-12-24-1 1.31E+06 1.34E+06 1.32E+06 1.322E+06 1.664E+04 1.30E+06 2.63E+04
Cy-JA-12-24-2 1.31E+06 1.29E+06 1.26E+06 1.286E+06 2.159E+04
Cy-JA-12-25-1 1.34E+06 1.35E+06 1.35E+06 1.348E+06 6.000E+03 1.34E+06 1.73E+04
Cy-JA-12-25-2 1.31E+06 1.35E+06 1.35E+06 1.333E+06 2.339E+04
Cy-JA-12-26-1 1.25E+06 1.23E+06 1.24E+06 1.240E+06 7.937E+03 1.28E+06 4.57E+04
Cy-JA-12-26-2 1.34E+06 1.31E+06 1.31E+06 1.321E+06 1.562E+04
Cy-JA-12-27-1 1.45E+06 1.42E+06 1.44E+06 1.438E+06 1.453E+04 1.40E+06 4.29E+04
Cy-JA-12-27-2 1.35E+06 1.37E+06 1.36E+06 1.362E+06 8.000E+03
Cy-JA-12-28-1 1.45E+06 1.43E+06 1.46E+06 1.447E+06 1.997E+04 1.51E+06 7.22E+04
Cy-JA-12-28-2 1.61E+06 1.57E+06 1.53E+06 1.569E+06 3.851E+04
Cy-JA-12-30-1 1.97E+06 1.95E+06 2.03E+06 1.982E+06 3.995E+04 2.05E+06 7.69E+04
Cy-JA-12-30-2 2.14E+06 2.10E+06 2.10E+06 2.111E+06 2.685E+04
Sample +/-
1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
7.54E+09 1.28E+08
8.31E+09 8.45E+07
8.68E+09 2.23E+08
9.49E+09 2.09E+08
1.02E+10 3.52E+08
1.39E+10 3.75E+08
Cy-JA-12-1-1
Cy-JA-12-1-2
Cy-JA-12-2-1
Cy-JA-12-2-2
Cy-JA-12-3-1
Cy-JA-12-3-2
Cy-JA-12-4-1
Cy-JA-12-4-2
Cy-JA-12-5-1
Cy-JA-12-5-2
Cy-JA-12-6-1
Cy-JA-12-6-2
Cy-JA-12-7-1
Cy-JA-12-7-2
Cy-JA-12-8-1
Cy-JA-12-8-2
Cy-JA-12-9-1
Cy-JA-12-9-2
Cy-JA-12-10-1
Cy-JA-12-10-2
Cy-JA-12-11-1
Cy-JA-12-11-2
Cy-JA-12-12-1
Cy-JA-12-12-2
Cy-JA-12-13-1
Cy-JA-12-13-2
Cy-JA-12-14-1
Cy-JA-12-14-2
Cy-JA-12-15-1
Cy-JA-12-15-2
Cy-JA-12-16-1
Cy-JA-12-16-2
Cy-JA-12-17-1
Cy-JA-12-17-2
Cy-JA-12-18-1
Cy-JA-12-18-2
Cy-JA-12-19-1
Cy-JA-12-19-2
Cy-JA-12-20-1
A at
360 nm
(mAU)
0.101
0.092
0.074
0.136
0.140
0.083
0.184
0.146
0.085
0.090
0.118
0.088
0.084
0.089
0.094
0.116
0.083
0.071
0.099
0.103
0.132
0.109
0.464
0.499
0.464
0.499
0.269
0.264
0.131
0.124
0.079
0.146
0.063
0.043
0.111
0.045
0.084
Table B 21. Cy-JA-12, Si concentration by spectrophotometer
Sample# Dilution
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
Csi (mmolSi/L)
0.07
0.12
0.12
0.08
0.09
0.08
0.09
0.09
0.07
0.10
0.13
0.11
0.10
0.05
0.07
0.10
Average
Csi
(mmolSi/
L)
0.42
0.42
0.23
0.09
0.08
0.07
0.12
0.12
0.07
0.16
0.13
0.07
0.08
0.10
0.08
0.07
0.08
0.08
0.10
0.07
0.06
0.09
0.09
0.12
0.10
0.12
0.11
0.07
0.13
0.06
0.04
0.10
0.04
0.07
0.41
0.44
0.41
0.44
0.24
0.23
Csi +/-
1S.D.
(mmolSi/L)
0.01
0.00
0.06
0.00
0.02
0.00
0.00
0.02
0.02
0.02
0.04
0.00
0.04
0.01
0.04
0.04
0.02
0.02
0.00
Table B 21. Continued
Sample#
Cy-JA-12-20-2
Cy-JA-12-21-1
Cy-JA-12-21-2
Cy-JA-12-22-1
Cy-JA-12-22-2
Cy-JA-12-23-1
Cy-JA-12-23-2
Cy-JA-12-24-1
Cy-JA-12-24-2
Cy-JA-12-25-1
Cy-JA-12-25-2
Cy-JA-12-26-1
Cy-JA-12-26-2
Cy-JA-12-27-1
Cy-JA-12-27-2
Cy-JA-12-28-1
Cy-JA-12-30-1
Cy-JA-12-30-2
A at
360 nm
(mAU)
0.177
0.159
0.160
0.115
0.149
0.173
0.163
0.134
0.123
0.198
0.156
0.192
0.154
0.181
0.181
0.195
0.201
0.185
Dilution
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
0.14
0.12
0.11
0.17
0.14
0.17
0.14
0.16
0.16
0.17
0.18
0.16
0.16
0.14
0.14
0.10
0.13
0.15
Csi (mmolSi/L)
0.11
0.16
0.15
0.16
0.17
0.17
Average
Csi
(mmolSi/
L)
0.14
0.12
0.15
Csi +/-
1S.D.
(mmolSi/L)
0.01
0.03
0.02
0.00
0.01
0.01
0.00
0.02
0.01
122
Table B 22. Cy-JA-12, Si concentration by ICP
Sample#
Cy-JA-12-1-1
Cy-JA-12-1-2
Cy-JA-12-4-1
Cy-JA-12-4-2
Cy-JA-12-8-1
Cy-JA-12-8-2
Cy-JA-12-13-1
Cy-JA-12-13-2
Cy-JA-12-18-1
Cy-JA-12-18-2
Cy-JA-12-19-1
Cy-JA-12-19-2
Cy-JA-12-20-1
Cy-JA-12-20-2
Cy-JA-12-21-1
Cy-JA-12-21-2
Cy-JA-12-23-1
Cy-JA-12-23-2
Cy-JA-12-26-1
Cy-JA-12-26-2
Cy-JA-12-30-1
Cy-JA-12-30-2
Emission at
251.611 nm
Trial #1
Emission at
251.611 nm
Trial #2
512
725
698
621
661
692
780
839
809
800
831
2770
2474
560
561
569
492
580
644
629
521
657
502
706
706
614
690
713
758
860
806
815
836
2776
2440
533
564
581
507
565
636
627
518
652
Emission at
251.611 nm
Trial #3
Trial
Avg.
515
720
696
635
656
695
769
872
825
811
831
2769
2483
541
575
563
491
606
637
629
515
672
Assay
Conc.
(ppm)
510
717
700
623
669
700
769
857
813
809
833
2772 1.386
2466 1.233
545
567
571
0.272
0.283
0.286
497
584
639
628
518
660
0.248
0.292
0.320
0.314
0.259
0.330
0.255
0.359
0.350
0.312
0.335
0.350
0.385
0.429
0.407
0.404
0.416 dilution
Sample conc.
(ppm)
Si conc.
(mmol
Si/L)
Average
Si conc
(mmol
Si/L)
0.464
Average Si conc +/- 1
S.D.
(mmol
Si/L)
0.039
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
2.548
3.585
3.500
3.117
3.345
3.500
3.845
4.285
4.067
4.043
4.163
13.858
12.328
2.723
2.833
2.855
2.483
2.918
3.195
3.142
2.590
3.302
0.089
0.126
0.123
0.109
0.117
0.123
0.135
0.151
0.143
0.142
0.146
0.491
0.437
0.095
0.099
0.100
0.086
0.102
0.112
0.110
0.090
0.116
0.097
0.093
0.107
0.100
0.102
0.124
0.113
0.129
0.147
0.144
0.003
0.009
0.007
0.014
0.019
0.002
0.006
0.009
0.005
0.003
Table B 23. Cy-JA-12, Lipid data
Trial 1
Dry cell mass (mg)
Total vol lipid extract
(mL)
Extract assay vol
(mL)
Dilution
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L)
Trial 2 wt% lipid
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at
350 nm
Trial 3 calc. palmitic acid in cuvette
(mg/L) wt% lipid average wt% st dev adjusted wt% adj stdev
8.7
18.1
19.2
17.5
19
13
18.8
5
6
5.1
16.5
15.8
17
16.6
17.4
17.1
13.9
17.1
6.5
Cy-JA-12-1-1
Cy-JA-12-1-2
Cy-JA-12-5-1
Cy-JA-12-5-2
Cy-JA-12-7-1
Cy-JA-12-7-2
Cy-JA-12-11-1
Cy-JA-12-11-2
Cy-JA-12-15-1
Cy-JA-12-15-2
Cy-JA-12-19-1
Cy-JA-12-19-2
Cy-JA-12-21-1
Cy-JA-12-21-2
Cy-JA-12-22-1
Cy-JA-12-22-2
Cy-JA-12-24-1
Cy-JA-12-24-2
Cy-JA-12-26-1
13
12.5
12.5
13
13
13
13
12.5
13
13.5
12.75
12.75
11.75
11.75
11.75
11.75
12
13
8
0.05 1.502
0.05 1.483
0.05 1.186
0.05 1.213
0.05 0.992
0.05 1.072
0.05 0.859
0.05 0.896
0.05 0.782
0.05 0.689
0.05 0.78
0.05
0.05 0.491
0.05
0.05
0.50
0.59
0.05
0.05
0.57
0.05
0.05
0.59
1.30
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.98
1.17
1.84
1.69
2.90
2.46
3.63
3.43
4.05
4.57
4.05
5.65
5.59
5.09
5.24
5.13
1.24
0.21 1.494
30.97 1.476
14.18 1.228
13.61 1.244
20.05 1.002
17.42 1.083
24.53 0.822
23.56 0.883
35.01 0.827
34.71 0.71
49.91 0.827
39.05
36.42
37.80
35.86
35.45
30.91
0.485
0.444
0.581
0.584
0.604
1.261
0.14
0.24
1.60
1.52
2.85
2.40
3.84
3.50
3.81
4.45
3.81
5.69
5.91
5.16
5.14
5.03
1.42
0.03 1.452
6.40 1.5
12.40 1.209
12.24 1.209
19.67 0.992
17.00 1.095
25.90 0.791
24.05 0.882
32.87 0.815
33.83 0.697
46.86 0.815
39.27
38.49
38.33
35.19
34.80
35.58
0.496
0.474
0.539
0.569
0.570
1.261
0.37
1.71
1.71
2.90
2.34
4.01
3.51
3.87
4.52
3.87
5.63
5.75
5.39
5.23
5.22
1.42
0.08
38.86
37.42
40.04
35.75
36.09
7.54
13.20 13.24
13.79
20.05 18.45
16.53
27.05 24.86
24.09
33.44 34.04
34.38
47.68 48.15
39.06
37.16
35.45
35.58 40.13
13.38
20.92
0.78
1.64
1.34
0.81
1.58
0.21
1.88
0.65
6.95
7.54
13.54
18.84
24.42
34.09
46.50
39.06
36.93
35.45
40.59
0.30
1.50
0.89
0.56
1.43
0.21
1.32
0.65
5.79
Table B 23. Continued
Trial 1
Cy-JA-12-26-2
Dry cell mass (mg)
Total vol lipid extract
(mL)
Extract assay vol
(mL)
Dilution
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L)
5 12.5 0.2 0.05 1.19 1.82
Trial 2 Trial 3 wt% lipid
Abs at
350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at
350 nm
45.60 1.174 1.90 47.53 1.188 calc. palmitic acid in cuvette
(mg/L)
1.82 wt% lipid
45.60 average wt% st dev adjusted wt% adj stdev
49.91 50.06 1.89 49.32 0.61 Cy-JA-12-27-1
Cy-JA-12-27-2
7.8
7.8
13
13
0.2
0.2
0.05 0.991
0.05 0.98
2.91
2.95
48.44 0.975
49.18 0.933
2.99
3.23
49.91 0.975
53.75 0.983
2.99
2.95 49.18
Cy-JA-12-28-1
Cy-JA-12-28-2
Cy-JA-12-30-1
Cy-JA-12-30-2
4.5
4.5
8
6.5
13
13
12
12
0.2
0.2
0.2
0.2
0.05
0.05
1.25
1.20
0.05
0.05
0.92
1.03
1.51
1.75
3.30
2.70
43.49 1.216
50.63 1.201
49.45 0.928
49.91 1.038
1.67
1.75
3.25
2.65
48.25 1.216
50.63 1.216
48.79 0.943
48.89 1.068
1.67
1.67
3.17
2.48
48.25 48.25
48.25
2.61 48.25
47.55 48.41 1.48 48.41
45.85
2.61
1.48
Table B 24. Cy-JA-13, Cultivation information
Sample# Date
Sample time medium
Inoculum
Cy-JA-13-01-1 1/5/2010
Cy-JA-13-02-1 1/7/2010
Cy-JA-13-03-1 1/9/2010
Cy-JA-13-04-1 1/12/2010
Cy-JA-13-05-1 1/14/2010
Cy-JA-13-06-1 1/15/2010
Cy-JA-13-07-1 1/17/2010
2:00 PM
3:40 PM
3:30 PM
1:00 PM
1:40 PM
4:50 PM
2:30 PM
Cy-JA-13-08-1 1/20/2010 1:15 PM
Cy-JA-13-09-1 1/23/2010 12:40 PM
Cy-JA-13-10-1 1/25/2010 1:00 PM
Cy-JA-13-11-1 1/27/2010 4:15 PM
Cy-JA-13-12-1 1/28/2010 10:00 AM
Cy-JA-13-13-1 1/29/2010 9:45 AM
Cy-JA-13-14-1 1/29/2010 5:15 PM
Cy-JA-13-15-1 1/30/2010 10:15 AM
Cy-JA-13-16-1 1/30/2010 4:15 PM
Cy-JA-13-17-1 1/31/2010 10:30 AM
Cy-JA-13-18-1 1/31/2010 2:15 PM
Cy-JA-13-19-1 2/1/2010 3:00 PM
Cy-JA-13-20-1 2/3/2010
Cy-JA-13-21-1 2/5/2010
Cy-JA-13-22-1 2/6/2010
Cy-JA-13-23-1 2/8/2010
Cy-JA-13-24-1 2/10/2010
4:30 PM
4:45 PM
2:45 PM
4:00 PM
3:00 PM
392.00
409.00
415.00
431.25
435.00
459.75
509.25
557.50
579.50
628.75
675.75
0.0
20.0
45.5
71.8
91.8
118.0
137.8
164.3
235.7
284.0
335.3
356.3
384.5
Cultivation
Time (hr)
Culture pH
140
100
100
100
100
140
140
140
140
140
140
Sample volume, includes 20 mL to clear lines (mL)
320
320
320
140
140
60
140
140
140
140
140
140
140
9.37
9.09
9.01
9.49
8.82
9.49
8.89
9.53
9.24
9.35
9.01
9.05
9.7
8.82
8.57
8.37
9.15
9.64
9.38
9.35
9.80
9.48
Medium volume
(mL)
2732
2946
2931
3129
3068
2928
2788
2688
2588
2488
2388
3245
3105
2965
1969
2267
2715
4500
375
4505
4185
3865
3725
3585
3525
3385
Table B 25. Cy-JA-13, Cell density
Sample# Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Cy-JA-13-01-1
Cy-JA-13-01-2
239168
319564
237754
341968
238280
304414
2.38E+05 7.15E+02
3.22E+05 1.89E+04
Cy-JA-13-02-1
Cy-JA-13-02-2
641956
649632
646198
632260
654884
631856
6.48E+05
6.38E+05
6.59E+03
1.01E+04
Cy-JA-13-03-1 1.238E+06 1.235E+06 1.215E+06 1.23E+06 1.25E+04
Cy-JA-13-03-2 1.202E+06 1.239E+06 1.207E+06 1.22E+06 2.01E+04
Cy-JA-13-04-1 1.686E+06 1.673E+06 1.720E+06 1.69E+06 2.43E+04
Cy-JA-13-04-2 1.584E+06 1.623E+06 1.552E+06 1.59E+06 3.56E+04
Cy-JA-13-05-1 2.002E+06 1.973E+06 2.005E+06 1.99E+06 1.77E+04
Cy-JA-13-05-2 1.937E+06 1.951E+06 1.950E+06 1.95E+06 7.81E+03
Cy-JA-13-06-1 1.979E+06 1.961E+06 1.958E+06 1.97E+06 1.14E+04
Cy-JA-13-06-2 2.060E+06 2.053E+06 2.014E+06 2.04E+06 2.48E+04
Cy-JA-13-07-1 1.997E+06 1.980E+06 1.992E+06 1.99E+06 8.74E+03
Cy-JA-13-07-2 2.212E+06 2.239E+06 2.203E+06 2.22E+06 1.87E+04
Cy-JA-13-08-1 2.585E+06 2.561E+06 2.552E+06 2.57E+06 1.71E+04
Cy-JA-13-08-2 2.482E+06 2.501E+06 2.483E+06 2.49E+06 1.07E+04
Cy-JA-13-09-1 2.655E+06 2.658E+06 2.621E+06 2.64E+06 2.06E+04
Cy-JA-13-09-2 2.742E+06 2.663E+06 2.681E+06 2.70E+06 4.14E+04
Cy-JA-13-10-1 2.732E+06 2.705E+06 2.751E+06 2.73E+06 2.31E+04
Cy-JA-13-10-2 2.842E+06 2.874E+06 2.773E+06 2.83E+06 5.16E+04
Cy-JA-13-11-1 2.732E+06 2.706E+06 2.752E+06 2.73E+06 2.31E+04
Cy-JA-13-11-2 2.793E+06 2.782E+06 2.822E+06 2.80E+06 2.07E+04
Cy-JA-13-12-1 2.268E+06 2.249E+06 2.221E+06 2.25E+06 2.36E+04
Sample average
(cells/mL)
Sample +/-
1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
2.80E+05 4.73E+04 1.352E+09 2.283E+08
6.43E+05 9.34E+03 3.101E+09 4.505E+07
1.22E+06 1.66E+04 5.899E+09 8.031E+07
1.64E+06 6.45E+04 7.911E+09 3.110E+08
1.97E+06 2.87E+04 9.504E+09 1.383E+08
2.00E+06 4.52E+04 9.670E+09 2.182E+08
2.10E+06 1.26E+05 1.015E+10 6.067E+08
2.53E+06 4.42E+04 1.219E+10 2.134E+08
2.67E+06 4.03E+04 1.288E+10 1.945E+08
2.78E+06 6.56E+04 1.341E+10 3.164E+08
2.76E+06 4.26E+04 1.334E+10 2.054E+08
2.25E+06 1.75E+04 1.182E+10 8.438E+07
Table B 25. Continued
Sample#
Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Cy-JA-13-12-2 2.248E+06 2.229E+06 2.257E+06 2.24E+06
Cy-JA-13-13-1 2.0446e 2.055E+06 2.049E+06 2.05E+06
Cy-JA-13-13-2 2.070E+06 2.029E+06 2.021E+06 2.04E+06
Cy-JA-13-14-1 2.054E+06 2.087E+06 2.034E+06 2.06E+06
Cy-JA-13-14-2 2.077E+06 2.063E+06 2.027E+06 2.06E+06
Cy-JA-13-15-1 2.196E+06 2.140E+06 2.202E+06 2.18E+06
Cy-JA-13-15-2 2.073E+06 2.104E+06 2.077E+06 2.08E+06
Cy-JA-13-16-1 1.912E+06 2.037E+06 2.204E+06 2.05E+06
Cy-JA-13-16-2 2.118E+06 2.299E+06 2.099E+06 2.17E+06
Cy-JA-13-17-1 2.094E+06 2.088E+06 2.155E+06 2.11E+06
Cy-JA-13-17-2 2.168E+06 2.078E+06 2.161E+06 2.14E+06
Cy-JA-13-18-1 2.044E+06 2.039E+06 1.993E+06 2.03E+06
Cy-JA-13-18-2 2.271E+06 2.181E+06 2.155E+06 2.20E+06
Cy-JA-13-19-1 2.154E+06 2.181E+06 2.127E+06 2.15E+06
Cy-JA-13-19-2 2.330E+06 2.299E+06 2.241E+06 2.29E+06
Cy-JA-13-20-1 2.158E+06 2.142E+06 2.122E+06 2.14E+06
Cy-JA-13-20-2 2.181E+06 2.131E+06 2.115E+06 2.14E+06
Cy-JA-13-21-1 2.066E+06 2.183E+06 2.266E+06 2.17E+06
Cy-JA-13-21-2 2.281E+06 2.305E+06 2.292E+06 2.29E+06
Cy-JA-13-22-1 2.066E+06 2.183E+06 2.266E+06 2.17E+06
Cy-JA-13-22-2 2.281E+06 2.305E+06 2.292E+06 2.29E+06
Cy-JA-13-24-1 2.030E+06 2.026E+06 1.968E+06 2.01E+06
Cy-JA-13-24-2 2.400E+06 2.202E+06 2.304E+06 2.30E+06
Trial +/- 1
S.D.
(cells/mL)
1.43E+04
4.24E+03
2.63E+04
2.68E+04
2.58E+04
3.42E+04
1.69E+04
1.47E+05
1.10E+05
3.71E+04
5.01E+04
2.81E+04
6.09E+04
2.70E+04
4.52E+04
1.80E+04
3.44E+04
1.00E+05
1.20E+04
1.00E+05
1.20E+04
3.47E+04
9.90E+04
Sample average
(cells/mL)
Sample +/-
1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
2.04E+06 1.98E+04 1.196E+10 9.568E+07
2.06E+06 2.36E+04 1.236E+10 1.136E+08
2.13E+06 5.72E+04 1.356E+10 2.759E+08
2.11E+06 1.34E+05 1.370E+10 6.447E+08
2.12E+06 4.14E+04 1.450E+10 1.999E+08
2.11E+06 1.06E+05 1.459E+10 5.106E+08
2.22E+06 8.16E+04 1.494E+10 3.937E+08
2.14E+06 2.46E+04 1.440E+10 1.187E+08
2.23E+06 9.21E+04 1.501E+10 4.445E+08
2.23E+06 9.21E+04 1.501E+10 4.445E+08
2.16E+06 1.74E+05 1.449E+10 8.404E+08
128
Cy-JA-13-01-1
Cy-JA-13-01-2
Cy-JA-13-02-1
Cy-JA-13-02-2
Cy-JA-13-03-1
Cy-JA-13-03-2
Cy-JA-13-04-1
Cy-JA-13-04-2
Cy-JA-13-05-1
Cy-JA-13-05-2
Cy-JA-13-06-1
Cy-JA-13-06-2
Cy-JA-13-07-1
Cy-JA-13-07-2
Cy-JA-13-08-1
Cy-JA-13-08-2
Cy-JA-13-09-1
Cy-JA-13-09-2
Cy-JA-13-12-1
Cy-JA-13-12-2
Cy-JA-13-14-1
Cy-JA-13-14-2
Cy-JA-13-16-1
Cy-JA-13-16-2
Cy-JA-13-18-1
Cy-JA-13-18-2
A at
360 nm
(mAU)
0.094
0.113
0.108
0.14
0.14
0.16
0.15
0.2
0.19
0.249
0.237
0.433
0.504
0.351
0.301
0.108
0.094
0.103
0.108
0.100
0.098
0.102
0.101
0.099
0.093
0.112
Table B 26. Cy-JA-13, Si concentration by spectrophotometer
Sample# Dilution
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
Csi (mmolSi/L)
0.10
0.13
0.15
0.19
0.23
0.10
0.09
0.10
0.09
0.10
Average
Csi
(mmolSi/
L)
0.45
0.31
0.10
0.09
0.11
0.10
0.13
0.13
0.15
0.14
0.19
0.18
0.24
0.23
0.10
0.10
0.10
0.09
0.10
0.10
0.09
0.09
0.11
0.41
0.48
0.33
0.29
0.10
0.09
Csi +/-
1S.D.
(mmolSi/L)
0.00
0.00
0.01
0.01
0.01
0.00
0.00
0.00
0.00
0.01
0.05
0.03
0.01
Table B 27. Cy-JA-13, Si concentration by ICP
Sample#
Cy-JA-13-01-1
Cy-JA-13-01-2
Cy-JA-13-03-1
Cy-JA-13-03-2
Cy-JA-13-07-1
Cy-JA-13-07-2
Cy-JA-13-09-1
Cy-JA-13-09-2
Cy-JA-13-11-1
Cy-JA-13-11-2
Cy-JA-13-13-1
Cy-JA-13-13-2
Cy-JA-13-15-1
Cy-JA-13-15-2
Cy-JA-13-17-1
Cy-JA-13-17-2
Cy-JA-13-18-1
Cy-JA-13-18-2
Cy-JA-13-20-1
Cy-JA-13-20-2
Cy-JA-13-22-1
Cy-JA-13-22-2
Emission at
251.611 nm
Trial #1
Emission at
251.611 nm
Trial #2
510
748
753
1041
1039
1259
1232
1248
1253
1597
1605
2023
1988
634
622
574
608
567
617
546
505
549
503
734
758
1019
1045
1284
1234
1279
1257
1598
1597
2043
1986
639
628
582
612
567
625
542
510
544
Emission at
251.611 nm
Trial #3
Trial
Avg.
504
757
749
1023
1056
1244
1250
1250
1275
1589
1612
2043
1971
650
618
588
611
570
618
536
505
528
Assay
Conc.
(ppm)
2036 1.018
1982 0.991
641
623
581
0.321
0.311
0.291
610
568
620
541
507
540
0.305
0.284
0.310
0.271
0.253
0.270
506
746
753
0.253
0.373
0.377
1028 0.514
1047 0.523
1262 0.631
1239 0.619
1259 0.630
1262 0.631
1595 0.797
1605 0.802 dilution
Sample conc.
(ppm)
Si conc.
(mmol
Si/L)
Average
Si conc
(mmol
Si/L)
0.45
Average Si conc +/- 1
S.D.
(mmol
Si/L)
0.007
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
2.528
3.732
3.767
5.138
5.233
6.312
6.193
6.295
6.308
7.973
8.023
10.182
9.908
3.205
3.113
2.907
3.052
2.840
3.100
2.707
2.533
2.702
0.088
0.131
0.132
0.181
0.184
0.223
0.218
0.222
0.223
0.282
0.284
0.360
0.351
0.112
0.109
0.101
0.107
0.099
0.108
0.094
0.088
0.094
0.11
0.10
0.10
0.09
0.09
0.13
0.18
0.22
0.22
0.28
0.002
0.004
0.007
0.004
0.004
0.001
0.002
0.003
0.000
0.001
Table B 27. Continued
Sample#
Emission at
251.611 nm
Trial #1
Emission at
251.611 nm
Trial #2
593
563
584
565
Emission at
251.611 nm
Trial #3
Trial
Avg.
583 587
590 573
Assay
Conc.
(ppm)
0.293
0.286 dilution
Sample conc.
(ppm)
10
10
2.933
2.863
Si conc.
(mmol
Si/L)
0.102
0.100
Average
Si conc
(mmol
Si/L)
0.10
Average Si conc +/- 1
S.D.
(mmol
Si/L)
0.002 Cy-JA-13-24-1
Cy-JA-13-24-2
132
Table B 28. Cy-JA-13, Lipid data
Cy-JA-13-01-1
Cy-JA-13-01-2
Cy-JA-13-03-1
Cy-JA-13-03-2
Cy-JA-13-07-1
Cy-JA-13-07-2
Cy-JA-13-09-1
Cy-JA-13-09-2
Cy-JA-13-11-1
Cy-JA-13-11-2
Cy-JA-13-13-1
Cy-JA-13-13-2
Cy-JA-13-15-1
Cy-JA-13-15-2
Cy-JA-13-17-1
Cy-JA-13-17-2
Cy-JA-13-18-1
Cy-JA-13-18-2
Cy-JA-13-20-1
Cy-JA-13-20-2
Trial 1 Trial 2 Trial 3
Dry cell
Total vol Extract lipid assay mass extract vol
(mg)
(mL) (mL)
Dilution Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid average wt% st dev adjusted wt% adj stdev
23.7
24.6
25.9
20.8
22.4
22.2
21.5
23.2
26.9
11.1
13.9
20.8
21
18.8
18.7
13.8
13
21.8
21.3
23.8
12.5
15
15
15
15
15
15
12.5
12.5
12
12
13
12.5
12
13
13
13
16
16
12.5
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.285
0.591
0.619
0.739
0.622
0.619
0.639
0.723
0.63
1.201
1.075
0.879
0.936
0.328
0.389
0.588
0.696
0.433
0.355
0.289
2.20 17.03
2.87 19.45
3.93 20.66
3.62 17.86
6.89 40.01
6.56 41.30
5.49 45.88
4.91 42.90
6.32 41.86
6.74 45.99
7.10 34.02
7.12 34.28
5.47 29.60
5.32 27.24
4.68 29.27
5.31 31.39
5.32 31.78
5.22 32.07
8.65 43.27
8.65 37.32
0.278
0.549
0.558
0.795
0.66
0.586
0.691
0.723
0.642
1.221
1.065
0.907
0.944
0.382
0.41
0.595
0.700
0.447
0.357
0.283
7.16
5.70
5.65
4.38
5.10
5.50
4.94
8.65
8.65
2.09
2.93
3.78
3.58
6.60
6.45
5.45
4.89
6.25
6.73
7.13
34.48
30.98
29.14
27.10
30.02
32.98
30.12
43.27
37.32
15.87
19.91
19.72
17.60
38.16
40.51
45.53
42.68
41.31
45.91
34.19
1.075
0.866
0.952
0.306
0.387
0.616
0.671
0.466
0.438
0.34
0.275
0.584
0.558
0.8
0.653
0.619
0.668
0.732
0.654
2.87
4.00
3.53
7.01
6.57
5.34
5.04
6.15
6.30
6.82
7.17
5.51
5.65
4.35
5.14
5.32
5.06
4.72
20.98
18.34
19.45
21.10 19.05 1.66
17.34
40.76 40.35 1.19
41.37
44.46 44.28 1.31
44.24
40.56 43.05 2.35
42.64
32.58 34.02 0.73
34.56
29.83 29.32 1.22
29.14
26.90 29.16 1.81
30.27
31.78 31.62 0.98
23.7
24.6
25.9
20.8
22.4
22.2
12.5
15
30.98 21.5 15
22.07 34.04 10.06 23.2 12.5
11.1
13.9
20.8
21
18.8
18.7
13.8
13
21.8
21.3
13
13
16
16
23.8 12.5
12
12
13
12.5
12
13
15
15
15
15
133
Table B 28. Continued
Cy-JA-13-22-1
Cy-JA-13-22-2
Cy-JA-13-24-1
Cy-JA-13-24-2
Cy-JA-12-26-2
Cy-JA-12-27-1
Cy-JA-12-27-2
Cy-JA-12-28-1
Cy-JA-12-28-2
Cy-JA-12-30-1
Cy-JA-12-30-2
Trial 1 Trial 2 Trial 3
Dry cell mass
(mg)
Total vol Extract lipid assay extract
(mL) vol
(mL)
Dilution Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid average wt% st dev adjusted wt% adj stdev
7.8
7.8
4.5
4.5
8
6.5
18.2
17.2
24.8
23
5
13
13
13
13
12
12
12.5
12.5
12.5
12.5
12.5
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.991
0.98
1.25
1.20
0.92
1.03
0.83
0.843
0.66
0.748
1.19
8.65 55.16
8.65 58.37
8.65 40.48
8.65 43.65
1.82 45.60
2.91 48.44
2.95 49.18
1.51 43.49
1.75 50.63
3.30 49.45
2.70 49.91
0.975
0.933
1.216
1.201
0.928
1.038
0.801
0.819
0.673
0.682
1.174
2.99
3.23
1.67
1.75
3.25
2.65
8.65
8.65
8.65
8.65
1.90
49.91
53.75
48.25
50.63
48.79
48.89
55.16
58.37
40.48
43.65
47.53
0.975
0.983
1.216
1.216
0.943
1.068
0.796
0.897
0.732
0.733
1.188
4.37
3.83
4.72
4.71
1.82
2.99
2.95
1.67
1.67
3.17
2.48
25.77 46.03 16.72 18.2 12.5
23.32
20.65 35.19 10.76 24.8 12.5
22.23
45.60
49.91 50.06 1.89 49.32 0.61
49.18
48.25 48.25 2.61 48.25 2.61
48.25
47.55 48.41 1.48 48.41 1.48
45.85
17.2
23
12.5
12.5
Table B 29. Cy-JA-14, Cultivation information
Sample# Date
Sample time medium
Inoculum
Cy-JA-14-01-1 1/5/2010
Cy-JA-14-02-1 1/7/2010
Cy-JA-14-03-1 1/9/2010
Cy-JA-14-04-1 1/12/2010
Cy-JA-14-05-1 1/14/2010
Cy-JA-14-06-1 1/15/2010
Cy-JA-14-07-1 1/17/2010
2:00 PM
3:40 PM
3:30 PM
1:00 PM
1:40 PM
4:50 PM
2:30 PM
Cy-JA-14-08-1 1/20/2010 1:15 PM
Cy-JA-14-09-1 1/23/2010 12:40 PM
Cy-JA-14-10-1 1/25/2010 1:00 PM
Cy-JA-14-11-1 1/27/2010 4:15 PM
Cy-JA-14-12-1 1/28/2010 10:00 AM
Cy-JA-14-13-1 1/29/2010 9:45 AM
Cy-JA-14-14-1 1/29/2010 5:15 PM
Cy-JA-14-15-1 1/30/2010 10:15 AM
Cy-JA-14-16-1 1/30/2010 4:15 PM
Cy-JA-14-17-1 1/31/2010 10:30 AM
Cy-JA-14-18-1 1/31/2010 2:15 PM
Cy-JA-14-19-1 2/1/2010 3:00 PM
Cy-JA-14-20-1 2/3/2010
Cy-JA-14-21-1 2/5/2010
Cy-JA-14-22-1 2/6/2010
Cy-JA-14-23-1 2/8/2010
Cy-JA-14-24-1 2/10/2010
4:30 PM
4:45 PM
2:45 PM
4:00 PM
3:00 PM
392.00
409.00
415.00
431.25
435.00
459.75
509.25
557.50
579.50
628.75
675.75
0.0
20.0
45.5
71.8
91.8
118.0
137.8
164.3
235.7
284.0
335.3
356.3
384.5
Cultivation
Time (hr)
Culture pH
140
100
100
100
100
140
140
140
140
140
140
Sample volume, includes 20 mL to clear lines (mL)
320
320
320
140
140
60
140
140
140
140
140
140
140
9.37
9.09
9.01
9.3
9.28
9.49
8.82
9.49
8.89
9.53
9.24
9.35
9.01
9.05
9.7
8.82
8.57
8.37
9.15
9.64
9.38
9.35
9.80
9.48
Medium volume
(mL)
2405
2265
2125
3885
3745
3605
3465
3365
3265
3165
3065
3245
3105
2965
2825
2685
2545
4500
375
4505
4185
3865
3725
3585
3525
3385
Table B 30. Cy-JA-14, Cell density
Sample# Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Cy-JA-14-01-1
Cy-JA-14-01-2
298152
265226
271892
245228
281790
285128
2.84E+05 1.33E+04
2.65E+05 2.00E+04
Cy-JA-14-02-1
Cy-JA-14-02-2
595496
577518
600546
570044
605596
560550
6.01E+05
5.69E+05
5.05E+03
8.50E+03
Cy-JA-14-03-1 1.079E+06 1.017E+06 1.029E+06 1.04E+06 3.29E+04
Cy-JA-14-03-2 1.098E+06 1.109E+06 1.105E+06 1.10E+06 5.57E+03
Cy-JA-14-04-1 1.268E+06 1.252E+06 1.224E+06 1.25E+06 2.23E+04
Cy-JA-14-04-2 1.332E+06 1.375E+06 1.351E+06 1.35E+06 2.15E+04
Cy-JA-14-05-1 1.518E+06 1.545E+06 1.557E+06 1.54E+06 2.00E+04
Cy-JA-14-05-2 1.517E+06 1.510E+06 1.498E+06 1.51E+06 9.61E+03
Cy-JA-14-06-1 1.627E+06 1.623E+06 1.592E+06 1.61E+06 1.92E+04
Cy-JA-14-06-2 1.789E+06 1.735E+06 1.713E+06 1.75E+06 3.91E+04
Cy-JA-14-07-1 1.721E+06 1.728E+06 1.721E+06 1.72E+06 4.04E+03
Cy-JA-14-07-2 1.855E+06 1.830E+06 1.825E+06 1.84E+06 1.61E+04
Cy-JA-14-08-1 2.085E+06 2.085E+06 2.073E+06 2.08E+06 6.93E+03
Cy-JA-14-08-2 2.047E+06 2.112E+06 2.138E+06 2.10E+06 4.69E+04
Cy-JA-14-09-1 2.367E+06 2.346E+06 2.362E+06 2.36E+06 1.10E+04
Cy-JA-14-09-2 2.429E+06 2.382E+06 2.417E+06 2.41E+06 2.44E+04
Cy-JA-14-10-1 2.345E+06 2.393E+06 2.333E+06 2.36E+06 3.17E+04
Cy-JA-14-10-2 2.467E+06 2.491E+06 2.477E+06 2.48E+06 1.21E+04
Cy-JA-14-11-1 2.450E+06 2.398E+06 2.426E+06 2.42E+06 2.60E+04
Cy-JA-14-11-2 2.483E+06 2.453E+06 2.444E+06 2.46E+06 2.04E+04
Cy-JA-14-12-1 2.472E+06 2.387E+06 2.371E+06 2.41E+06 5.43E+04
Sample average
(cells/mL)
Sample +/-
1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
2.75E+05 1.83E+04 1.325E+09 8.832E+07
5.85E+05 1.82E+04 2.822E+09 8.774E+07
1.07E+06 4.01E+04 5.176E+09 1.936E+08
1.30E+06 6.06E+04 6.274E+09 2.923E+08
1.52E+06 2.23E+04 7.354E+09 1.076E+08
1.68E+06 7.72E+04 8.105E+09 3.725E+08
1.78E+06 6.30E+04 8.589E+09 3.038E+08
2.09E+06 3.15E+04 1.008E+10 1.522E+08
2.38E+06 3.27E+04 1.150E+10 1.576E+08
2.42E+06 6.98E+04 1.167E+10 3.370E+08
2.44E+06 2.85E+04 1.178E+10 1.375E+08
2.46E+06 7.83E+04 1.296E+10 3.780E+08
Table B 31. Continued
Sample# Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Cy-JA-14-12-2 2.587E+06 2.494E+06 2.469E+06 2.52E+06 6.22E+04
Cy-JA-14-13-1 2.210E+06 2.237E+06 2.241E+06 2.23E+06 1.69E+04
Cy-JA-14-13-2 2.255E+06 2.235E+06 2.217E+06 2.24E+06 1.90E+04
Cy-JA-14-14-1 2.223E+06 2.294E+06 2.218E+06 2.25E+06 4.25E+04
Cy-JA-14-14-2 2.202E+06 2.226E+06 2.202E+06 2.21E+06 1.39E+04
Cy-JA-14-15-1 2.181E+06 2.213E+06 2.191E+06 2.20E+06 1.64E+04
Cy-JA-14-15-2 2.155E+06 2.171E+06 2.205E+06 2.18E+06 2.55E+04
Cy-JA-14-16-1 2.264E+06 2.264E+06 2.253E+06 2.26E+06 6.35E+03
Cy-JA-14-16-2 2.480E+06 2.424E+06 2.437E+06 2.45E+06 2.93E+04
Cy-JA-14-17-1 2.214E+06 2.172E+06 2.157E+06 2.18E+06 2.95E+04
Cy-JA-14-17-2 2.477E+06 2.432E+06 2.448E+06 2.45E+06 2.28E+04
Cy-JA-14-18-1 2.577E+06 2.567E+06 2.544E+06 2.56E+06 1.69E+04
Cy-JA-14-18-2 2.455E+06 2.428E+06 2.436E+06 2.44E+06 1.39E+04
Cy-JA-14-19-1 2.688E+06 2.631E+06 2.588E+06 2.64E+06 5.02E+04
Cy-JA-14-19-2 2.486E+06 2.529E+06 2.485E+06 2.50E+06 2.51E+04
Cy-JA-14-20-1 2.980E+06 2.994E+06 2.991E+06 2.99E+06 7.37E+03
Cy-JA-14-20-2 3.015E+06 2.998E+06 2.928E+06 2.98E+06 4.61E+04
Cy-JA-14-21-1 3.205E+06 3.202E+06 3.119E+06 3.18E+06 4.88E+04
Cy-JA-14-21-2 3.490E+06 3.527E+06 3.447E+06 3.49E+06 4.00E+04
Cy-JA-14-22-1 3.211E+06 3.135E+06 3.173E+06 3.17E+06 3.80E+04
Cy-JA-14-22-2 3.421E+06 3.438E+06 3.529E+06 3.46E+06 5.81E+04
Cy-JA-14-23-1 3.174E+06 3.138E+06 3.111E+06 3.14E+06 3.16E+04
Cy-JA-14-23-2 3.365E+06 3.331E+06 3.334E+06 3.34E+06 1.88E+04
Sample average
(cells/mL)
Sample +/-
1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
2.23E+06 1.64E+04 1.306E+10 7.933E+07
2.23E+06 3.42E+04 1.338E+10 1.648E+08
2.19E+06 2.16E+04 1.391E+10 1.041E+08
2.35E+06 1.04E+05 1.527E+10 5.017E+08
2.32E+06 1.50E+05 1.581E+10 7.261E+08
2.50E+06 6.88E+04 1.727E+10 3.318E+08
2.57E+06 8.23E+04 1.727E+10 3.973E+08
2.98E+06 2.99E+04 2.007E+10 1.441E+08
3.33E+06 1.76E+05 2.241E+10 8.485E+08
3.32E+06 1.65E+05 2.231E+10 7.943E+08
3.24E+06 1.13E+05 2.180E+10 5.464E+08
136
Table B 32. Continued
Sample# Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Cy-JA-14-24-1 3.144E+06 3.153E+06 3.112E+06 3.14E+06 2.15E+04
Cy-JA-14-24-1 3.363E+06 3.423E+06 3.319E+06 3.37E+06 5.22E+04
Sample average
(cells/mL)
Sample +/-
1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
3.25E+06 1.32E+05 2.187E+10 6.369E+08
137
A at
360 nm
(mAU)
0.094
0.113
0.108
0.14
0.14
0.16
0.15
0.2
0.19
0.249
0.237
0.433
0.504
0.351
0.301
0.108
0.094
0.103
0.108
0.100
0.098
0.102
0.101
0.099
0.093
0.112
Table B 33. Cy-JA-14, Si concentration by spectrophotometer
Sample#
Cy-JA-14-01-1
Cy-JA-14-01-2
Cy-JA-14-02-1
Cy-JA-14-02-2
Cy-JA-14-03-1
Cy-JA-14-03-2
Cy-JA-14-04-1
Cy-JA-14-04-2
Cy-JA-14-05-1
Cy-JA-14-05-2
Cy-JA-14-06-1
Cy-JA-14-06-2
Cy-JA-14-07-1
Cy-JA-14-07-2
Cy-JA-14-08-1
Cy-JA-14-08-2
Cy-JA-14-09-1
Cy-JA-14-09-2
Cy-JA-14-12-1
Cy-JA-14-12-2
Cy-JA-14-14-1
Cy-JA-14-14-2
Cy-JA-14-16-1
Cy-JA-14-16-2
Cy-JA-14-18-1
Cy-JA-14-18-2
Dilution
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
5
Csi (mmolSi/L)
0.10
0.13
0.15
0.19
0.23
0.10
0.09
0.10
0.09
0.10
Average
Csi
(mmolSi/
L)
0.45
0.31
0.10
0.09
0.11
0.10
0.13
0.13
0.15
0.14
0.19
0.18
0.24
0.23
0.10
0.10
0.10
0.09
0.10
0.10
0.09
0.09
0.11
0.41
0.48
0.33
0.29
0.10
0.09
Csi +/-
1S.D.
(mmolSi/L)
0.00
0.00
0.01
0.01
0.01
0.00
0.00
0.00
0.00
0.01
0.05
0.03
0.01
Cy-JA-14-01-1
Cy-JA-14-01-2
Cy-JA-14-03-1
Cy-JA-14-03-2
Cy-JA-14-05-1
Cy-JA-14-05-2
Cy-JA-14-07-1
Cy-JA-14-07-2
Cy-JA-14-09-1
Cy-JA-14-09-2
Cy-JA-14-10-1
Cy-JA-14-10-2
Cy-JA-14-12-1
Cy-JA-14-12-2
Cy-JA-14-13-1
Cy-JA-14-13-2
Cy-JA-14-14-1
Cy-JA-14-14-2
Cy-JA-14-15-1
Cy-JA-14-15-2
Cy-JA-14-16-1
Cy-JA-14-16-2
Table B 34. Cy-JA-14, Si concentration by ICP
Sample#
Emission at
251.611 nm
Trial #1
Emission at
251.611 nm
Trial #2
655
868
862
989
1010
1399
1383
1727
1682
1433
1518
1941
2014
632
590
694
730
769
796
776
762
660
681
898
874
1002
1023
1411
1376
1720
1654
1452
1529
1953
1995
633
613
682
743
778
810
775
704
674
Emission at
251.611 nm
Trial #3
Trial
Avg.
1943 1946
1943 1984
642
624
636
609
671 682
722 732
757 768
801 802
771 774
713 726
672 669
672 669
860 875
860 865
978 990
1030 1021
1407 1406
1396 1385
1697 1715
1673 1670
1461 1449
1526 1524
Assay
Conc.
(ppm)
0.335
0.438
0.433
0.495
0.511
0.703
0.693
0.857
0.835
0.724
0.762
0.973
0.992
0.318
0.305
0.341
0.366
0.384
0.401
0.387
0.363
0.334
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10
10 dilution
Sample conc.
(ppm)
3.35
4.38
4.33
4.95
5.11
7.03
6.93
8.57
8.35
7.24
7.62
9.73
9.92
3.18
3.05
3.41
3.66
3.84
4.01
3.87
3.63
3.34
Si conc.
(mmol
Si/L)
0.12
0.15
0.15
0.17
0.18
0.25
0.25
0.30
0.30
0.26
0.27
0.35
0.35
0.11
0.11
0.12
0.13
0.14
0.14
0.14
0.13
0.12
Average
Si conc
(mmol
Si/L)
0.45
Average Si conc +/- 1
S.D.
(mmol
Si/L)
0.005
0.11
0.12
0.14
0.13
0.12
0.15
0.18
0.25
0.30
0.26
0.003
0.006
0.004
0.006
0.000
0.001
0.004
0.003
0.006
0.010
Cy-JA-14-17-1
Cy-JA-14-17-2
Cy-JA-14-18-1
Cy-JA-14-18-2
Cy-JA-14-20-1
Cy-JA-14-20-2
Cy-JA-14-22-1
Cy-JA-14-22-2
Cy-JA-14-24-1
Cy-JA-14-24-1
Table B 35. Continued
Sample#
Emission at
251.611 nm
Trial #1
Emission at
251.611 nm
Trial #2
1931
1839
1585
1864
1497
1488
1229
1251
1031
1041
1948
1836
1583
1893
1532
1509
1264
1251
1028
1042
Emission at
251.611 nm
Trial #3
Trial
Avg.
1946 1942
1856 1844
1582 1583
1864 1874
1526 1518
1540 1512
1237 1243
1256 1253
1027 1029
1048 1044
Assay
Conc.
(ppm)
0.971
0.922
0.792
0.937
0.759
0.756
0.622
0.626
0.514
0.522
10
10
10
10
10
10
10
10
10
10 dilution
Sample conc.
(ppm)
9.71
9.22
7.92
9.37
7.59
7.56
6.22
6.26
5.14
5.22
Si conc.
(mmol
Si/L)
0.34
0.33
0.28
0.33
0.27
0.27
0.22
0.22
0.18
0.18
Average
Si conc
(mmol
Si/L)
0.34
Average Si conc +/- 1
S.D.
(mmol
Si/L)
0.012
0.31
0.27
0.22
0.18
0.037
0.001
0.001
0.002
140
Cy-JA-14-01-1
Cy-JA-14-01-2
Cy-JA-14-03-1
Cy-JA-14-03-2
Cy-JA-14-07-1
Cy-JA-14-07-2
Cy-JA-14-09-1
Cy-JA-14-09-2
Cy-JA-14-11-1
Cy-JA-14-11-2
Cy-JA-14-13-1
Cy-JA-14-13-2
Cy-JA-14-15-1
Cy-JA-14-15-2
Cy-JA-14-17-1
Cy-JA-14-17-2
Cy-JA-14-20-1
Cy-JA-14-20-2
Cy-JA-14-22-1
Cy-JA-14-22-2
Table B 36. Cy-JA-14, Lipid data
Trial 1 Trial 2 Trial 3
Dry cell mass
(mg)
Total vol Extract lipid extract
(mL) assay vol
(mL)
Dilution Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid average wt% st dev
26.3
11.6
10.8
21.5
22
24.6
22.9
17
20.9
11.6
11.7
21.3
21.9
13.7
13.6
20
17
20.3
20.6
26.7
12
13
13
12
12
13
13
13
12.5
12.5
12.5
12.5
13
13
13
12.5
12.5
13
12.5
12
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
1.085
0.794
0.859
0.757
0.794
0.469
0.628
0.687
0.759
0.455
0.479
1.118
1.13
0.858
0.821
1.028
1.056
1.08
0.966
2.77 23.43
4.33 22.05
3.98 20.23
4.53
4.33
6.08
5.23
4.91
4.52
6.16
37.54
35.92
34.40
34.19
27.75
23.94
25.08
6.03 24.87
2.59 22.59
2.53 23.48
3.99 19.05
4.19 19.70
3.08 13.21
2.92 13.33
2.80 16.97
3.41 16.94
1.093
0.789
0.859
0.829
0.783
0.513
0.585
0.696
0.769
0.471
0.428
1.075
1.118
0.889
0.827
1.028
1.058
1.039
1
2.73
4.36
3.98
4.15
4.39
5.84
5.46
4.86
4.47
6.07
6.30
2.82
2.59
3.82
4.16
3.08
2.91
3.02
3.23
22.97
22.21
20.23
33.87
36.48
32.93
35.89
27.44
23.62
24.69
26.12
25.18
24.26
18.12
19.53
13.21
13.27
18.66
15.85
1.096
0.752
0.817
0.753
0.823
0.519
0.552
0.71
0.69
0.504
0.46
1.107
1.130
0.858
0.845
1.038
1.048
1.07
0.966
2.71
4.56
4.21
4.55
4.18
5.81
5.63
4.78
4.89
5.89
6.13
2.65
2.53
3.99
4.06
3.02
2.97
2.85
3.41
23.06
22.80
23.38 21.61 1.22
21.57
37.75 36.00 1.59
34.43
32.72 34.55 1.73
37.19
26.95 25.98 1.79
26.19
23.89 25.00 0.74
25.34
23.25 23.71 0.90
23.48
19.05 19.08 0.55
19.00
12.92 13.25 0.21
13.58
17.39 17.13 0.91
16.94
141
Table B 36. Continued
Cy-JA-14-24-1
Cy-JA-14-24-1
Trial 1 Trial 2 Trial 3
Dry cell
Total vol lipid mass extract
(mg)
(mL)
Extract assay vol
(mL)
Dilution
Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid average st dev wt%
19.7
20.1
12
12.5
0.2
0.2
0.05
0.05
1.005
1.006
3.20 15.98
3.19 16.28
1.005
1.015
3.20 15.98
3.15 15.98 0.987 3.30 16.91
16.23 0.41
142
Table B 37.Cy-JA-15, Cultivation information
Sample# medium
Inoculum
Date
Sample time
Cy-JA-15-01-1 5/14/2010 11:30 AM
Cy-JA-15-02-1 5/15/2010 2:00 PM
Cy-JA-15-03-1 5/16/2010 3:15 PM
Cy-JA-15-04-1 5/17/2010 1:30 PM
Cy-JA-15-05-1 5/18/2010 2:30 PM
Cy-JA-15-06-1 5/19/2010 3:30 PM
Cy-JA-15-07-1 5/20/2010 2:00 PM
Cy-JA-15-08-1 5/21/2010 11:30 AM
Cy-JA-15-09-1 5/22/2010 2:00 PM
Cy-JA-15-10-1 5/23/2010 12:30 PM
Cy-JA-15-11-1 5/24/2010 4:00 PM
Cy-JA-15-12-1
Cy-JA-15-13-1
5/25/2010
5/25/2010
1:30 PM
4:00 PM
Cy-JA-15-14-1 5/26/2010 4:00 PM
Cy-JA-15-15-1 5/27/2010 4:00 PM
0.0
26.5
51.8
74.0
99.0
124.0
146.5
168.0
194.5
217.0
244.5
266.0
292.0
316.50
340.50
Cultivation
Time (hr)
8.61
9.02
9.12
9.15
9.20
9.23
9.01
9.37
9.45
9.63
9.56
9.26
9.2
9.18
Culture pH
Sample volume, includes 20 mL to clear lines (mL)
320
320
60
40
40
40
40
220
60
320
60
100
100
100
100
Medium volume
(mL)
4500
375
2670
2350
2290
2250
2210
2170
2130
1910
1850
1530
1470
1392
1908
1808
1708
Table B 38. Cy-JA-15, Cell density
Sample#
Cy-JA-15-01-1
Cy-JA-15-01-2
Cy-JA-15-02-1
Cy-JA-15-02-2
Cy-JA-15-03-1
Cy-JA-15-03-2
Cy-JA-15-04-1
Cy-JA-15-04-2
Cy-JA-15-05-1
Cy-JA-15-05-2
Cy-JA-15-06-1
Cy-JA-15-06-2
Cy-JA-15-07-1
Cy-JA-15-07-2
Cy-JA-15-08-1
Cy-JA-15-08-2
Trial #1
(cells/ mL)
298960
300172
518918
536704
752854
768408
771842
766590
7.87E+05
7.37E+05
9.12E+05
9.48E+05
1.05E+06
1.12E+06
1.20E+06
1.20E+06
Trial #2
(cells/ mL)
295324
291486
593870
555086
768610
750228
747400
781336
7.86E+05
7.24E+05
9.05E+05
9.36E+05
1.00E+06
1.12E+06
1.20E+06
1.20E+06
Trial #3
(cells/ mL)
305222
284214
502758
565792
743966
781336
762146
739320
7.98E+05
7.28E+05
9.08E+05
9.34E+05
1.05E+06
1.13E+06
1.01E+06
1.17E+06
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
3.00E+05 5.01E+03
2.92E+05 7.99E+03
5.39E+05 4.86E+04
5.53E+05 1.47E+04
7.55E+05 1.25E+04
7.67E+05 1.56E+04
7.60E+05 1.23E+04
7.62E+05 2.13E+04
7.90E+05
7.30E+05
9.08E+05
9.40E+05
1.03E+06
1.12E+06
1.14E+06
1.19E+06
6.61E+03
6.28E+03
3.74E+03
7.56E+03
2.54E+04
5.13E+03
1.09E+05
1.64E+04
Sample average
(cells/mL)
2.96E+05
Sample +/-
1 S.D.
(cells/mL)
7.36E+03
Total No. cells in reactor
7.90E+08
Total No. cells in reactor +/-
1 S.D.
1.97E+07
5.46E+05 3.30E+04 1.28E+09 7.76E+07
7.61E+05 1.41E+04 1.74E+09 3.24E+07
7.61E+05 1.56E+04 1.71E+09 3.51E+07
7.60E+05 3.35E+04 1.68E+09 7.41E+07
9.24E+05 1.79E+04 2.00E+09 3.87E+07
1.08E+06 5.04E+04 2.29E+09 1.07E+08
1.16E+06 7.62E+04 2.22E+09 1.45E+08
Cy-JA-15-09-1 1.15E+06 1.10E+06 1.11E+06 1.12E+06 2.82E+04
Cy-JA-15-09-2 1.10E+06 1.12E+06 1.08E+06 1.10E+06 1.70E+04
Cy-JA-15-10-1 1.12E+06 1.09E+06 1.12E+06 1.11E+06 1.61E+04
Cy-JA-15-10-2 1.20E+06 1.20E+06 1.17E+06 1.19E+06 1.64E+04
1.11E+06
1.15E+06
2.42E+04
4.70E+04
2.05E+09
2.07E+09
4.47E+07
7.20E+07
Cy-JA-15-11-1 1.66E+06 1.69E+06 1.64E+06 1.66E+06 2.51E+04
Cy-JA-15-11-2 1.52E+06 1.51E+06 1.48E+06 1.50E+06 1.82E+04
1.58E+06 8.85E+04 2.32E+09 1.30E+08
Cy-JA-15-12-1 1.89E+06 1.86E+06 1.87E+06 1.87E+06 1.59E+04 1.83E+06 4.55E+04 2.55E+09 6.33E+07
Table B 39. Continued
Sample# Trial #1
(cells/ mL)
Trial #2
(cells/ mL)
Trial #3
(cells/ mL)
Trial average
(cells/mL)
Trial +/- 1
S.D.
(cells/mL)
Sample average
(cells/mL)
Sample +/-
1 S.D.
(cells/mL)
Total No. cells in reactor
Total No. cells in reactor +/-
1 S.D.
Cy-JA-15-12-2 1.79E+06 1.82E+06 1.78E+06 1.80E+06 2.57E+04
Cy-JA-15-13-1 2.10E+06 2.03E+06 2.11E+06 2.08E+06 4.36E+04 2.06E+06 5.15E+04 3.93E+09 9.82E+07
Cy-JA-15-13-2 2.01E+06 2.11E+06 2.00E+06 2.04E+06 5.99E+04
Cy-JA-15-14-1 2.21E+06 2.20E+06 2.11E+06 2.17E+06 5.51E+04 2.21E+06 5.15E+04 4.00E+09 9.31E+07
Cy-JA-15-14-2 2.20E+06 2.25E+06 2.31E+06 2.25E+06 5.51E+04
Cy-JA-15-15-1 2.20E+06 2.25E+06 2.31E+06 2.25E+06 5.51E+04
Cy-JA-15-12-2 2.21E+06 2.20E+06 2.11E+06 2.17E+06 5.51E+04
2.21E+06 5.00E+04 4.00E+09 8.54E+07
145
Table B 40. Cy-JA-15, Si concentration by spectrophotometer
Sample#
A at
360 nm
(mAU)
Cy-JA-15-01-1 0.413
Cy-JA-15-01-2 0.424
Cy-JA-15-02-1 0.116
Cy-JA-15-02-2 0.115
Cy-JA-15-03-1 0.097
Cy-JA-15-03-2 0.087
Cy-JA-15-07-1 0.155
Cy-JA-15-07-2 0.096
Cy-JA-15-08-1 0.129
Cy-JA-15-08-2 0.120
Cy-JA-15-10-1 0.142
Cy-JA-15-10-2 0.124
Cy-JA-15-11-1 0.13
Cy-JA-15-11-2
Cy-JA-15-12-1
0.13
0.14
Cy-JA-15-12-2
Cy-JA-15-13-1
Cy-JA-15-13-2
Cy-JA-15-14-1
Cy-JA-15-14-2
Cy-JA-15-15-1
Cy-JA-15-15-2
0.12
0.11
0.10
0.12
0.11
0.1
0.9
Dilution
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
5.0
0.13
0.08
0.11
0.10
0.12
0.11
0.11
0.11
0.11
0.35
0.36
0.10
0.10
0.08
0.07
0.10
0.09
0.08
0.10
0.09
0.09
0.77
Csi (mmolSi/L)
Average
Csi
(mmolSi/
L)
0.38
0.10
0.08
0.11
0.11
0.11
0.11
0.11
0.09
0.10
0.10
Csi +/-
1S.D.
(mmolSi/L)
0.04
0.01
0.01
0.01
0.01
0.00
0.01
0.00
0.01
0.01
0.00
Table B 41.Cy-JA-15, Lipid data
Cy-JA-15-02-1
Cy-JA-15-02-2
Cy-JA-15-08-1
Cy-JA-15-08-2
Cy-JA-15-10-1
Cy-JA-15-10-2
Cy-JA-15-12-1
Cy-JA-15-12-2
Cy-JA-15-15-1
Cy-JA-15-15-2
Trial 1 Trial 2 Trial 3
Dry cell mass
(mg)
Total vol Extract lipid extract
(mL) assay vol
(mL)
Dilution Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid
Abs at 350 nm calc. palmitic acid in cuvette
(mg/L) wt% lipid average wt% st dev
20.4
20.1
13.7
23
16.5
17.1
21
20
22.3
21.8
8.5
8.5
10.8
10.5
11.8
11.8
11
11
13
13
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.2
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
0.05
1.316
1.024
1.06
1.125
1.001
0.989
0.899
0.923
0.889
0.87
3.005 12.521
5.925 25.056
5.565 43.870
4.915 22.438
6.155 44.018
6.275 43.301
7.175 37.583
6.935 38.143
7.275 42.4103
7.515 44.8142
1.264
1.41
1.095
1.1
1.021
0.993
0.902
1.035
0.932
0.972
3.525 14.688
2.065 8.733
5.215 41.111
5.165 23.579
5.955 42.587
6.235 43.025
7.145 37.426
5.815 31.983
6.845 39.904
6.445 38.433
1.226
1.213
1.12
1.031
1.026
1.031
0.912
0.953
0.912
0.874
3.905 16.271 15.72 5.47
4.035 17.063
4.965 39.140 32.81 9.60
5.855 26.729
5.905 42.230 42.59 1.24
5.855 40.403
7.045 36.902 36.42 2.25
6.635 36.493
7.045 41.070 41.818 2.4923
7.425 44.278
Appendix C – Calibration Curves
UV Spectrophotometric Calibration: Silicon
Figure C 1 . Calibration data for Silicon dioxide detection at 360 nm on 11/15/2008
Solution
Concentration
(mg/L)
0.000
0.000
0.025
0.025
0.025
0.050
0.050
0.050
0.100
0.100
0.100
0.200
0.200
0.200
0.300
0.300
0.300
Absorbance
0.0227
0.0175
0.1833
0.2151
0.2250
0.3291
0.3566
0.3641
0.6527
0.6478
0.6487
1.2459
1.2300
1.2333
1.7667
1.6982
1.7167
0.350
0.300
0.250
0.200
0.150
0.100
0.050
0.000
0.000
0.500
1.000
1.500
Absorbance at 360 nm
2.000
Figure C 2. Calibration curve for silicon detection at 360 nm.
149
Figure C 3.Statistical analysis for 11/18/2008 calibration curve
SUMMARY OUTPUT
Regression Statistics
Multiple R 0.998
0.997 R Square
Adjusted R
Square
Standard
Error
0.934
0.009
Observations 17
ANOVA
Regression
Residual
Total
Intercept
X
Variable
1 df
1
16
17
Coefficients
0 #N/A #N/A #N/A #N/A
0.167
SS MS
0.428 0.428
8.64E
0.001 -05
0.429
F
4956.
448
Significance
F
2.54E-20
Standard
Error
0.002 t Stat
70.40
2
Pvalue
0.000
Lower
95%
0.162
Upper
95%
#N/A
0.172
Lower
95.0%
#N/A
0.162
Upper
95.0%
#N/A
0.172
Figure C 4 . Calibration data for Silicon detection at 360 nm on 6/11/2009
Solution
Concentration
(mg/L)
0
0
0.0625
0.0625
0.0625
0.125
0.125
0.125
0.25
0.25
0.25
0.3
Absorbance
0.007
0.021
0.343
0.318
0.332
0.667
0.657
0.679
1.267
1.251
1.251
0.25
0.2
0.15
0.1
0.05
0
0 0.5
1 1.5
Absorbance
Figure C 5. Calibration curve for silicon detection at 360 nm.
150
151
SUMMARY OUTPUT
Regression Statistics
Multiple R
R Square
Adjusted R
Square
Standard
Error
0.998
0.997
0.934
0.009
Observations 17
ANOVA
Regression
Residual
Total
Intercept
X Variable 1 df
1
16
17
Coefficients
SS MS
0.428 0.428
8.64E
0.001 -05
0.429
F
4956.
448
Significance
F
2.54E-20
Standard
Error t
Stat
Pvalue
0 #N/A #N/A #N/A
70.40
0.167 0.002 2 0.000
Lower
95%
#N/A
0.162
Upper
95%
Lower
95.0%
Upper
95.0%
#N/A #N/A #N/A
0.172 0.162 0.172
Figure C 6. Statistical analysis for 6/11/2009 calibration curve
Solution
Concentration
(mg/L)
0.1
0.05
0.05
0
0
0.3
0.3
0.25
0.25
0.15
0.15
0.1
Absorbance
1.347
1.347
1.192
1.185
0.73
0.73
0.447
0.461
0.195
0.205
0.015
0.001
Figure C 7. Calibration data for Silicon detection at 360 nm on 12/15/2009
0.35
0.3
0.25
0.2
0.15
0.1
0.05
0
0 0.5
Absorbance at 360 nm
1 1.5
Figure C 8. Calibration curve for silicon detection at 360 nm.
152
153
SUMMARY OUTPUT
Regression Statistics
Multiple R 0.998
0.996 R Square
Adjusted R
Square
Standard
0.996
Error 0.007
Observations 12.000
ANOVA df
Regression
Residual
Total
Intercept
X Variable 1
1.000
10.000
11.000
Coefficie nts
0.000
0.216
SS
0.134
0.000
0.134
MS
0.13
4
0.00
0
Standard
Error
0.003
0.004
F
Significance
F
2731.
237 0.000 t
Stat
0.12
Pvalue
Lower
95%
Upper
95%
Lower
95.0%
1 0.906 -0.007 0.008 -0.007
52.2
61 0.000 0.207 0.225 0.207
Upper
95.0%
0.008
0.225
Figure C 9. Statistical analysis for 12/15/2009 calibration curve
Solution
Concentration
(mg/L)
0.15
0.15
0.10
0.10
0.10
0.05
0.05
0.05
0.00
0.00
0.30
0.30
0.30
0.20
0.20
0.20
0.15
Absorbance
1.6740
1.6914
1.7090
1.1857
1.2020
1.1512
0.9002
0.9074
0.8644
0.5680
0.6078
0.6078
0.2764
0.2744
0.3336
-0.0030
0.0030
Figure C 10. Calibration data for Silicon detection at 360 nm on 7/13/2009
0.35
0.30
0.25
0.20
0.15
0.10
0.05
0.00
0.0
0.5
1.0
Absorbance at 360 nm
1.5
2.0
Figure C 11. Calibration curve for silicon detection at 360 nm
154
155
SUMMARY
OUTPUT
Regression Statistics
Multiple R 0.999
0.997 R Square
Adjusted R
Square
Standard
Error
0.997
0.005
Observations 17
ANOVA df SS
Regression
Residual
Total
1
15
16
MS
Significance
F F
0.156 0.156 5624 9.857E-21
0.000 2.77E-05
0.156
Intercept
X Variable
1
Coeffici ents
Standar d Error t Stat P-value
Lower
95%
Upper
95%
Lower
95.0%
Upper
95.0%
-0.004 0.002 -1.518 0.150 -0.008 0.001 -0.008 0.001
9.857E-
0.176 0.002 74.994 21 0.171 0.181 0.171 0.181
Figure C 12. Statistical analysis for 7/13/2009 calibration curve
UV spectrophotometer Calibration: Lipid Analysis
Palmitic Acid
Concentration (mg/L) Absorbance at 350 nm
0
0
0
0.7815
0.7815
0.7815
1.5625
1.5625
1.5625
3.125
3.125
3.125
6.25
6.25
6.25
12.5
12.5
12.5
1.452
1.457
1.285
1.287
1.265
0.974
1.011
0.945
0.372
1.594
1.639
1.598
1.548
1.53
1.53
1.457
0.415
0.376
Figure C 13. Calibration lipid detection at 350 nm on 2/19/2009
1.8
1.6
1.4
1.2
1
0.8
0.6
0.4
0.2
0
0 5
Palmitic Acid (mg/L)
10 15
Figure C 14. Calibration curve for lipids at 350 nm
156
157
SUMMARY
OUTPUT
Regression Statistics
Multiple R
R Square
Adjusted R
Square
Standard Error
Observations
0.999
0.998
0.998
0.022
18.000
ANOVA
Regression
Residual df
1.000
SS
3.184
16.000 0.007
Total 17.000 3.191
Intercept
X Variable 1
MS
3.184
0.000
F
6873
Significance
F
0.000
Coefficient s
1.603
-0.098
Standard
Error
0.007 t
Stat
230.
Pvalue
160 0.000
Lower
95%
Upper
95%
Lower
95.0%
Upper
95.0%
1.589 1.618 1.589 1.618
0.001 -82.1 0.000 -0.101 -0.096 -0.101 -0.096
Figure C 15. Statistical analysis for 2/19/2009 calibration curve
Palmitic Acid
Concentration
(mg/L)
Absorbance at 350 nm
5
7.5
7.5
12.5
12.5
0
0
0
2.5
2.5
5
1.614
1.622
1.57
1.416
1.385
1.155
1.131
0.804
0.785
0.349
0.399
Figure C 16. Calibration data for lipids at 350 nm on 6/17/2009
1.8
1.6
1.4
1.2
1
0.8
0.6
0.4
0.2
0
0 2 4 6 8
Palmitic Acid (mg/L)
10
Figure C 17. Calibration curve for lipid analysis at 350 nm.
12 14
158
159
SUMMARY
OUTPUT
Regression Statistics
Multiple R 0.996
R Square
Adjusted R
Square
Standard
Error
0.992
0.991
0.045
Observations 11.000
ANOVA
Regression
Residual
Total
Intercept
X Variable 1 df
1.000
9.000
10.000 2.183
Coeffici ents
1.617
-0.101
SS
2.165
0.018
MS F
2.165 1072 0.000
0.002
Significa nce F
Standar d Error t Stat
Pvalue
Lower
95%
Upper
95%
0.021 78.790 0.000 1.570 1.663
Lower
95.0%
1.570
Upper
95.0%
1.663
0.003 -32.753 0.000 -0.108 -0.094 -0.108 -0.094
Figure C 18. Statistical analysis for lipid calibration data on 6/17/2009
Palmitic Acid
Concentration
(mg/L)
Absorbance at 350 nm
0
0
0
2.5
2.5
5
1.614
1.622
1.57
1.416
1.385
1.155
5
7.5
1.131
0.804
7.5 0.785
12.5
12.5
0.349
0.399
Figure C 19. Calibration data for lipid detection at 350 nm on 8/15/2009
1.8
1.6
1.4
1.2
1
0.8
0.6
0.4
0.2
0
0 2 4
Palmitic Acid (mg/L)
6 8
Figure C 20. Calibration curve for lipids on 8/15/2009
160
161
SUMMARY
OUTPUT
Regression Statistics
Multiple R 0.991
R Square
Adjusted R
Square
0.982
0.980
Standard Error 0.074
11.00
Observations 0
ANOVA on
Regressi
Residual
Total df
1.000
9.000
10.000
SS MS F
2.711 2.711 497.274
0.049 0.005
2.760
Significance
F
0.000
Coefficients
Standard
Error t Stat
Pvalue
Lower
95%
Upper
95%
Lower
95.0%
Upper
95.0%
Intercept 1.520 0.033 46.345 0.000 1.446 1.594 1.446 1.594
X Variable 1 -0.182 0.008 -22.300 0.000 -0.200 -0.163 -0.200 -0.163
Figure C 21. Statistical analysis of lipid calibration data from 8/15/2009
UV spectrophotometer Calibration: Nitrogen Analysis
Solution Concentration Absorbance at 530 nm
0 µmol/L
0 µmol/L
0 µmol/L
100 µmol/L
100 µmol/L
100 µmol/L
150 µmol/L
150 µmol/L
150 µmol/L
200 µmol/L
200 µmol/L
0.004
0.002
0.001
0.212
0.188
0.183
0.289
0.262
0.258
0.323
0.326
Figure C 22. Calibration data for nitrogen detection at 530 nm
350
300
250
200
150
100
50
0
0 0.1
0.2
0.3
0.4
Absorbance at 530 nm
0.5
Figure C 23. Calibration curve for nitrogen analysis at 530 nm.
0.6
162
163
SUMMARY OUTPUT
Regression Statistics
Multiple R
R Square
Adjusted R
Square
Standard
Error
Observatio ns
0.987
0.974
0.972
17.241
15
ANOVA
Regression
Residual
Total df
1
13
14
SS
146136
3864
MS
14613
F
Significan ce F
6 492 1.03E-11
297
150000
Intercept
X Variable
1
Coefficien ts
-13.82
662.15
Standar d Error t Stat
Pvalue
8.63 -1.60 0.13
29.86 22.17
1.03E
-11
Lower
95%
Uppe r 95%
-32.45 4.82
597.63
726.6
6
Lower
95.0%
-
32.45
597.6
3
Upper
95.0%
4.82
726.66
Figure C 24. Statistical analysis of nitrogen calibration data
ICP Calibration: Silicon Dioxide Analysis
Figure C 25. Calibration data for ICP analysis of Si
Solution Concentration
(mg/L)
0
1
2
4
5
8
Absorbance at 530 nm
0.004
0.002
0.001
0.212
0.188
0.183
7
6
5
4
3
9
8
2
1
0
0 5000 10000
Intensity counts
15000
Figure C 26 .
Calibration curve for silicon dioxide analysis on ICP.
20000
164
GC-FID Calibration: C16:0 and C19:0 calibration
Conc.
(mg/mL)
0.25
0.25
0.25
0.125
0.125
0.125
0
0
0
0.5
0.5
0.5
0.0625
0.0625
C16:0
Peak
Area
0
0
0
333903
373494
338049
141589
156992
163904
75560
80569
83216
30895
39292
C19:0 Peak
Area
0
0
0
1082405
1113198
984730
419084
477755
495704
235578
246659
251366
111982
117224
Figure C 26. Calibration data for C16:0 and C19:0
1200000
1000000
800000
600000
C16:0
C19:0
400000
200000
0
0 0.2
Conc. (mg/mL)
0.4
Figure C 27. Calibration curve for C16:0 and C19:0
0.6
165
166
SUMMARY
OUTPUT
Regression Statistics
Multiple R
R Square
Adjusted R
Square
Standard
Error
0.997
0.995
0.895
15586
Observations 11
ANOVA
Total
Regression
Residual df SS MS
1
10 2429091407 242909141
F
Significa nce F
4.56E+11 4.56E+11 1877 9.28E-12
11 4.58E+11
Coeffici ents
Standard
Error t Stat P-value
Lower
95%
Upper
95%
Lower
95.0%
Upper
95.0%
Intercept 0 #N/A #N/A #N/A
X Variable 1 677946 15647 43 1.03E-12
#N/A #N/A #N/A
643082 712809 643082
#N/A
712809
Figure C 28. Statistical analysis for C16:0
167
SUMMARY
OUTPUT
Regression Statistics
Multiple R
0.99
7
0.99
R Square
Adjusted R
Square
Standard Error
4
0.89
4
487
10
Observations 11
ANOVA
Regression
Residual
Total df SS MS F
Signific ance F
1 4.21E+12 4.21E+12 1775 0.00
10 2.E+10 2.E+09
11 4.24E+12
Intercept
Coefficie nts
Standar d Error t Stat P-value
0 #N/A #N/A #N/A
Lower
95%
#N/A
Upper
95%
#N/A
Lower
95.0%
#N/A
Upper
95.0%
#N/A
X Variable 1 2060425 48901 42 1.36E-12 1951466 2169384 1951466 2169384
Figure C 30. Statistical analysis of C19:0 calibration data
168
Appendix D – Lipid Composition Data
Table D 1. 72 h perfusion GC data
Sample No. C
IS
(mg/mL) GC RT (min) RRT Compound Peak Area
C i
(mg/mL)
% of total
Conc.
Cy-JA-11-11-01 #1 0.319 11.999
13.921
0.460
0.534 14:0
7898
48852
0.01
0.07
0.89
5.53
15.211
16.433
16.682
16.784
0.583
0.630
0.640
0.644
16:3
16:1
9144
66161
413523
9890
0.01
0.10
0.61
0.01
1.04
7.49
46.84
1.12
17.016
17.458
18.962
21.245
21.655
22.36
26.073
0.653
0.670
0.727
0.815
0.831
0.858
1.000
16:0
18:1
18:0
19:0
219215
8954
5164
6495
6703
7973
656566
0.32 24.83
0.01 1.01
0.01
0.01
0.01
0.01
0.58
0.74
0.76
0.90
Cy-JA-11-11-01 #2 0.353
27.501
11.972
13.885
15.164
16.379
16.678
1.055
0.459
0.534
0.584
0.630
0.642
20:5
14:0
16:3
16:1
72822
9063
55840
9412
84360
461575
0.11
0.01
0.08
0.01
8.25
0.92
5.66
0.95
0.12 8.56
0.68 46.81
16.959
17.394
18.209
21.15
21.573
22.279
25.075
25.978
27.395
0.653
0.670
0.701
0.814
0.830
0.858
0.965
1.000
1.055
16:0
18:1
18:0
19:0
20:5
235889
7617
6209
8717
7864
10811
5643
726412
83004
0.35 23.92
0.01 0.77
0.01
0.01
0.63
0.88
0.01
0.02
0.01
0.80
1.10
0.57
0.12 8.42
169
Table D 1. Continued
Sample No. C
IS
(mg/mL) GC RT (min) RRT Compound Peak Area C i
(mg/mL)
% of total
Conc.
Cy-JA-11-18-01 #1 0.167
Cy-JA-11-18-01 #2 0.171
11.966
13.873
15.154
16.704
16.359
16.594
16.921
17.933
21.128
22.262
25.824
27.314
11.962
13.866
15.145
16.323
16.585
16.696
16.912
22.249
25.816
0.632
0.642
0.647
0.655
0.862
1.000
0.813
0.857
0.994
1.051
0.463
0.537
0.463
0.534
0.583
0.643
0.630
0.639
0.651
0.690
14:0
18:1
18:0
19:0
20:5
16:3
16:1
16:0
14:0
16:3
16:1
16:0
18:0
19:0
20:5
13816
32481
6660
6266
50361
325870
149812
5078
5193
10072
343232
51864
14519
32287
6683
53911
332027
6817
152779
10478
352266
0.08
0.02
0.05
0.01
0.08
0.49
0.01
0.23
0.02
0.07
0.48
0.22
0.01
0.02
0.05
0.01
0.01
0.01
0.01
2.10
4.94
1.01
0.95
7.66
49.56
22.79
0.77
0.79
1.53
Cy-JA-11-28-01 #1 0.377
27.303
11.957
13.858
16.301
16.517
16.866
22.238
25.933
1.058
0.461
0.534
0.629
0.637
0.650
0.858
1.000
14:0
16:3
16:1
16:0
18:0
19:0
53141
10565
9666
17292
72859
49363
8684
777492
0.08
0.02
0.01
0.03
0.11
0.07
0.01
7.89
2.19
4.87
1.01
8.14
50.11
1.03
23.06
1.58
8.02
5.80
5.31
9.49
39.99
27.10
4.77
27.258 1.051 20:5 13752 0.02 7.55
170
Table D 1. Continued
Sample No. C
IS
(mg/mL) GC RT (min) RRT Compound Peak Area C i
(mg/mL)
% of total
Conc.
Cy-JA-11-28-01 #2 0.360
Cy-JA-11-28-01 #2 0.360
11.957
13.856
16.313
16.512
16.862
22.233
25.925
27.25
11.957
13.856
16.313
0.461
0.534
0.629
0.637
0.650
0.858
1.000
1.051
0.461
0.534
0.629
14:0
16:3
16:1
16:0
18:0
19:0
20:5
14:0
16:3
11042
9545
17157
71203
49088
8321
741770
12653
11042
9545
17157
0.02
0.02
0.01
0.03
0.02
0.01
0.03
0.11
0.07
0.01
6.17
5.33
9.58
39.78
27.42
4.65
7.07
6.17
5.33
9.58
16.512 0.637 16:1 71203 0.11 39.78
Cy-JA-09-08-02 #2
Table D 2. 48 h perfusion GC data
Sample No.
Cy-JA-09-08-02 #1
C
IS
(mg/mL)
0.256
22.266
25.885
16.707
16.932
17.373
17.922
19.561
20.169
20.416
21.116
22.246
23.923
25.745
27.36
27.4
11.964
13.868
15.144
15.753
16.08
16.337
16.589
GC RT
(min)
11.972
13.874
15.15
15.764
16.068
16.345
16.602
16.718
16.944
17.379
17.939
18.173
19.533
20.188
20.433
21.136
0.118
0.860
1.000
0.649
0.658
0.675
0.696
0.760
0.783
0.793
0.820
0.864
0.929
1.000
1.063
1.059
0.462
0.539
0.588
0.612
0.625
0.635
0.644
RRT
0.463
0.536
0.585
0.609
0.621
0.631
0.641
0.646
0.655
0.671
0.693
0.702
0.755
0.780
0.789
0.817
171
Cy-JA-09-14-02 #1
Cy-JA-09-14-02 #2
Cy-JA-09-22-02 #1
Table D 2. Continued
Sample No.
0.170
C
IS
(mg/mL) GC RT (min) RRT
0.163 8.411
11.961
0.327
0.464
13.859
15.136
0.538
0.587
16.311
16.536
0.633
0.641
16.882
17.89
0.655
0.694
21.11
22.238
0.819
0.863
23.914
25.78
0.928
1.000
27.285 1.058
0.174 8.414
11.972
0.326
0.464
13.87
15.15
0.537
0.587
16.327
16.555
0.633
0.642
16.901
17.798
21.131
21.473
22.254
25.805
27.316
11.963
1.059
0.464
0.655
0.690
0.819
0.832
0.862
1.000
13.862
16.343
16.565
16.882
17.913
21.109
21.535
22.243
25.788
27.288
0.538
0.634
0.642
0.655
0.695
0.819
0.835
0.863
1.000
1.058
16:0
18:1
18:0
19:0
14:0
16:3
16:1
20:5
Compound
16:0
18:1
18:0
14:0
16:3
16:1
19:0
20:5
18:1
18:0
14:0
16:3
16:1
16:0
19:0
20:5
22035
52148
147076
101347
10257
6751
8607
8444
349775
63538
5401
21811
30884
5689
41626
176058
126177
6504
12228
5140
21061
358130
67442
7385
Peak Area C i
(mg/mL) % of total
5167
13395
0.008
0.020
1.066
2.765
26054
5274
38689
168937
0.038
0.008
0.057
0.249
5.377
1.089
7.985
34.867
121980
8457
10362
12491
6729
336430
66978
0.180
0.012
0.015
0.018
0.010
0.099
25.176
1.745
2.139
2.578
1.389
13.824
0.033
0.077
0.217
0.149
0.015
0.010
0.013
0.012
0.094
0.186
0.010
0.018
0.008
0.031
0.008
0.032
0.046
0.008
0.061
0.260
0.099
0.011
5.153
12.196
34.397
23.702
2.399
1.579
2.013
1.975
14.860
24.264
1.251
2.351
0.988
4.050
1.039
4.194
5.939
1.094
8.005
33.856
12.969
1.727
172
Table D 2. Continued
Sample No.
Cy-JA-09-22-02 #2
C
IS
(mg/mL) GC RT (min) RRT
0.173 11.958
13.856
16.335
16.527
16.874
17.778
21.097
21.523
22.234
25.775
27.27
30
0.464
0.538
0.634
0.641
0.655
0.690
0.819
0.835
0.863
1.000
1.058
1.16
Compound Peak Area C i
(mg/mL) % of total
16:0
18:1
14:0
16:3
16:1
18:0
19:0
20:5
7977
21885
52714
147643
102956
10485
7159
8485
8871
356083
62444.00
7279.00
0.012
0.032
0.078
0.218
0.152
0.015
0.011
0.013
0.013
0.092
0.011
1.822
4.998
12.038
33.716
23.511
2.394
1.635
1.938
2.026
14.260
1.662
173
Figure D 1. GC-MS scans
Figure D 1. Continued
Figure D 1. Continued
Figure D 1. Continued
177
Appendix E. – Light Attenuation
Light attenuation constant k c measurements
Materials
Glass Petri dish
Equipment
Li-cor quantam sensor
Fluorescent light
Procedure
1.
Add 180 mL ASM to 96 mm glass culture dish with opaque sides. By calculation the culture depth ( z ) in culture dish should be 2.5 cm.
2.
Place a fluorescent lamp directly above the culture dish , and the Li-Cor SA 190
PAR quantum sensor underneath the dish facing the lamp.
3.
Measure the incident irradiance to the liquid suspension surface ( I o
) from 15 to 152
-2 s
-1
by moving the light source closer to the dish. Record the distance required for each incident irradiance.
4.
At each incident light intensity, measure the average light intensity exiting the wellmixed culture suspension ( I z
). Swirl the medium between measurements to keep the culture suspended. The measured I z values will fluctuate a lot during measurements, so it is desirable to record at least twenty I z values in the notebook and report the average value for a given I o
.
5.
Add known volume of Cyclotella sp. to culture dish to achieve desired cell density.
6.
Place a fluorescent lamp directly above the culture dish , and the Li-Cor SA 190
PAR quantum sensor underneath the dish facing the lamp.
7.
Measure the incident irradiance to the liquid suspension surface ( I o
) from 15 to 152
-2 s
-1
by moving the light source closer to the dish. Record the distance required for each incident irradiance.
179
8.
At each incident light intensity, measure the average light intensity exiting the wellmixed culture suspension ( I z
). Swirl the medium between measurements to keep the culture suspended. The measured I z values will fluctuate a lot during measurements, so it is desirable to record at least twenty I z values in the notebook and report the average value for a given I o
.
9.
Repeat steps 5-8, increasing the cell density over a given range to obtain the dependence of k’ on the cell density.
10.
At each new cell density, the light attenuation constant k’
was estimated from the least squares slope of I z vs. I o data with the intercept forced to zero.
I z
I o e k ' z
11. Plot k’
vs. X (cell density), the k c
will be the slope of the plot. The apparent light attenuation constant ( k’
) is a linear function of cell density X k ' k o k c
X where k c
is the specific light attenuation constant of the biomass, and k o
is the light attenuation constant of the cell-free medium.
k c
180
Table E 2. Light attenuation data
I o
(Light through blank culture)
Cell denisty
(cells/mL) z (cm)
0.28
0.4
0.6
I z
98
96.5
94
1.06E+06
102
-ln I z
/I
0.040
0.055
0.082 o
I z
94
92
88
1.62E+06
-ln I z
/I
0.082
0.103
0.148 o
I z
93
89
84
2.28E+06
-ln I z
/I
0.092
0.136
0.194 o
I z
91
85
74
3.56E+06
-ln I z
/I
0.114
0.182
0.321 o
0.7
0.8
92.75
91.5
0.095
0.109
86
84
0.171
0.194
81
78
0.231
0.268
70
66
0.376
0.435
I z
89
81
68
4.76E+06
-ln I z
/I
0.136
0.231
0.405 o
63
58
0.482
0.565 k' 0.1364 cm
-1
0.2473 cm
-1
0.3314 cm
-1
0.526 cm
-1
0.6726 cm
-1
1.44E-07 mL/cells-cm
181
0.6
0.5
0.4
0.3
1.06E+06
1.62E+06
2.28E+06
3.56E+06
4.76E+06
0.2
0.1
0.0
0 0.2
0.4
0.6
Z, culture depth (cm)
Figure E 1. k’, apparent light attenuation constant
0.8
y = 0.6726x
R² = 0.9601
y = 0.526x
R² = 0.9695
y = 0.3314x
R² = 0.9976
y = 0.2473x
R² = 0.9778
y = 0.1364x
R² = 0.9986
1
0.8
0.7
0.6
0.5
0.4
0.3
y = 1.435E-07x + 1.955E-03
R² = 9.956E-01
0.2
0.1
0
0.E+00 1.E+06 2.E+06 3.E+06 4.E+06 5.E+06
Cell Density (cells/mL)
Figure E 2. k c
, specific light attenuation constant (mL/cells-cm)
Appendix F – Correlation of cell number to dry cell mass
Table F 2. Tabulated Data
Cell density no.
(cells/mL)
Sample vol (mL)
Number of cells
Dry cell mass (mg)
1.54E+06
1.59E+05
5.45E+05
1.21E+06
1.32E+06
1.18E+06
1.46E+06
1.57E+05
3.01E+05
3.66E+05
5.72E+05
8.93E+05
1.05E+06
1.20E+06
1.25E+06
1.51E+06
1.27E+06
200 3.13E+07 25.1
200 6.02E+07 28.2
200 7.32E+07 35
200 1.14E+08 30.3
200 1.79E+08 37.5
200 2.10E+08 54.2
200 2.39E+08 61.2
100 1.25E+08 14.2
100 1.51E+08 17.6
160 2.03E+08 21.2
180 2.76E+08 57.1
200 3.18E+07 34.6
200 1.09E+08 40.7
100 1.21E+08 15
100 1.32E+08 20
160 1.89E+08 22.3
180 2.63E+08 56.2
182
3.00E+08
2.50E+08
2.00E+08
1.50E+08
1.00E+08
5.00E+07
0.00E+00
0 y = 4.03E+06x
10 20 30 40
DCW (mg)
Figure F 1. Correlated data
50 60 70
183