Quantitative soform Profiling & Isoform Convergence
advertisement

Quantitative soform Profiling & Isoform Convergence
by
Chris Varma
B.S. Computer Science, Dept. of Computer Science,
M.S. Computational Biology, Dept. of Computer Science,
M.S. Management, Dept. of Management,
Stanford University, 2001
Submitted to the Harvard-MIT Division of Health Sciences and Technology
In Partial Fulfillment of the Requirements for the Degree of
Doctor of Philosophy in Medical Engineering
at
Harvard Medical School
&
MASSATS
r
iNSTFU'E
OF TEOHNO)cQY
Massachusetts Institute of Technology
May 2005
JUN 3
2005
LIBRAR
IES
© 2005 Chris Varma. All rights reserved.
The author hereby grants to Harvard Medical School and to MIT permission to
reproduce and to distribute publicly paper and electronic copies of this-thesis document
in whole o/in part.
Signature of Author:
Harvard-MIT Division of Health Sciences & Technology
May 13, 2005
Ci by
Certified by
· z7 ~ c2~
PeterSzolovits,Ph.D.
Professor of Computer Science & Engineering / Health Sciences & Technology, MIT
Thesis Chairman
Certified by
George M. Church, Ph.D.
Professor of Genetics, Harvard Medical School
Thesis Supervisor
Accepted by
I
WV
Martha L. Gray
Edward Hood Chaplin Professor of Medic al and Electrical Engineering
-
Co-director, Harvard-M.I.T. Division of Health Sciences and Technology
ARCHIVES
Quantitative Isoform Profiling & Isoform Convergence
by
Chris Varma
Submitted to the Harvard-MIT Division of Health Sciences & Technology on May
13, 2005 in Partial Fulfillment of the Requirements for the Degree of Doctor of
Philosophy in Medical Engineering
Abstract
Alternative pre-messenger RNA splicing is a crucial step in eukaryotic gene
expression, and therefore it is subject to tight regulation. Given its importance in
conferring protein diversity, alternative splicing is sensitive to changes in cellular
states including malignancy. We present a new paradigm by which to
quantitatively study the alternative splicing of any molecule through the
presented methods of quantitative exon profiling and quantitative isoform profiling
which take advantage of a single-molecule based technology [Mit99].
Furthermore, we extend this paradigm to include a novel unified platform-called
Isoform Convergence-to qualify particular isoforms as candidate diagnostic
markers, potential therapeutic targets, and perhaps even as precursor
therapeutics themselves. We apply this paradigm to quantitatively investigate
the alternative
splicing of CD44 in two leukemias.
CD44 is an alternatively
spliced cell surface receptor, which is generally implicated in cancer though the
specifics are mired in controversy. In this work, we suggest several corrections
to previously made claims about the presence of specific CD44 exons and of
specific
CD44
isoforms
in leukemia
as
well
as
in non-diseased
cells.
Furthermore, we provide not only the first comprehensive characterization of
CD44's (or any molecule's) alternative exon splicing in human cells, but also its
resulting quantities of exons and isoforms to an average resolution on the order
of 1.E+06 molecules. Finally, we identify specific isoforms in each leukemia that
may serve as candidate markers or possibly as therapeutic targets.
Thesis Supervisor: George M. Church, Professor of Genetics, Harvard Medical
School
Varma, Chris
2 of 65
Ph.D. Thesis
Table of Contents
OVERVIEW .................................................................................................................................................. 6
CONTRIBUTIONSOF THIS THESIS ...............................................................................................................
6
CD44 CONTROVERSY............................ ......................................................................................................7
BACKGROUND
...........................................................................................................................................
0.1. PRE-MRNA AL TERNA TIVE SPLICING...........................................................................
8
8
Overview& M ajor Players....................................................................................................................
8
AlternativeSplicing,Disease, & TherapeuticIntervention.................................................................
9
0.2. HCD44 ....................................................................
10
11...............................
I
Structure& Function ....................................................................
CD44 Alternative Splicing ................................
.................................................................................
11
CD44 & Implications in Disease ....................................................................
13
CD44 in B-cell & T-cell Development ....................................................................
14
0.3. SPLIC
IN-INSIGHTS
D
RIVEN THERAPY ..............................................................................................
14
0.4. LEUKEMI
A .......................................................................................................................................... 16
A L L ..................................... ................................................................................................................. 16
AML........................................
17
Diagnosis of Leukemia .......................................................................................................................
17
Treatment of Leukemia ......................................................................................................................
18
CHAPTER 1: PROFILING OF ALTERNATIVE EXON SPLICING ..................................................19
1.1. INTRODUCTION....................................................................
1.2. POLONY TECHNOLOGYFOR EXONPROFILING ....................................................................
1.3. OVERVIEW OFEXPERIMENTAL METHODS .................................
....................................
1.4. PRIMER DESIGN....................................................................
1.5. POLONY SLIDE CREATION& SUBSEQUENTPREPARATION................................................................
1.6. SINGLE BASEEXTENSIONS & SCANNING....................................................................
1. 7. REAL-TIE QPCR .....................................................................
1.8. CONCLU .. N........................
............................................................................................................
19
19
20
21
22
22
23
23
CHAPTER 2: CONSTRUCTION OF EXPRESSION PROFILES ...................................................... 24
2.1. INTRODCTION ....................................................................................................................
2.2. OVERVIEW OF SOFTWARE ........................................................
2.3. IMAGE PROCESSING
.......................................................
24
24
25
2.4. THRESHOLDINC
....................................................................................................................
25
2.5. POLONY IDENTIFICATION .....................................................................................................
26
2.6. ISOFORMCONSTRUCTION
.......................................................
27
2. 7. SPECIAL CASES & OTHER SOFTWARE .......................................................
27
2.8. PERFORMANCE OF SOFTWARE.......................................................
28
2.9. CONCLUSION...................................................................................................................................... 28
CHAPTER 3: CHARACTERIZATION OF SAMPLES ......................................................
28
3.1. INTRODUCTION.....................................................................................................................
3.2. OBTAINING SAMPLES.......................................................
3.3. SEMI-QUINTITA TIVEANALYSIS OF CD44 .......................................................
3.4. QUANTIFYING VARIANTEXONS ......................................................
28
29
29
31
3.5. CONCLUSION
........................................................
33
CHAPTER 4: EXON EXPRESSION PROFILES .......................................................
34
4.1. INTRODUCTION.......................................................
34
4.2. QUANTITTIVE COMPARISON
OFAML TONORMAL......................................................
34
35
4.4. QUANTITATIVE COMPARISONOFAML TO ALL ...............................................................
Varma, Chris
3 of 65
Ph.D. Thesis
35
4.5. QUANTITATIVE COMPARISONOFALL TO NORMAL ...........................................................
4.6. QUANTITA TIVE COMPARISONOF CANCER TO NORMAL ............................................................
36
4.7. CONCLUSION
........................... .................................
37
CHAPTER 5: ISOFORM EXPRESSION PROFILES ..............................................................
5.1. INTROD CTIO N ..................................................................................................................................
5.2. COMMON ISOFORMS ........................................................................................................
5.3. QUANTITATIVE COMPARISONOFAML TO NORMAL ...........................................................
5.4. QUANTIrA
TIVE COMPARISON
OFAML TOALL
.................................................................................
5.5. QUANTITATIVE COMPARISONOF ALL TONORMAL ............................................................
5.6. QUANTI:A TIVE COMPARISONOF CANCER TO NORMAL ............................................................
5.7. CONCLUSION.......................... ..................................
38
38
38
40
42
43
44
46
CHAPTER 6: IN PURSUIT OF EXCLUSIVE CONVERGING-ISOFORMS .....................................47
6.1. INTROD CTI ON ................................................................................................................
6.2. INTRODUCINGCISOFORM
CONVERGENCE........................................
6.3. FINDING CONVERGENCE.................................................................................................
47
4...................................4
49
6.4. CONVERGI
NG-ISOFORMS
.................................................................................................
50
6.5. EXCLUSIVE CONVERGING-ISOFORMS................................................................................................
6.6. IDENTIFICATION OFPOSSIBLE CANDIDATE TARGETS
.........................................
6.7. CONCLU.ION........................... .................................
51
52
52
CONCLUSION
...............................................................
54
SUPPLEMENTARY RESULTS .............................................................
55
S.. TM-EXONSKIPPI
NG ........................................................................................................
Initial Observation,Identification & Background......................................... ....................
Isoform ExpressionProfiles................................
.............................
UniversalExpression.............................................................
55
55
57
59
S.2. E ONEXPRESSIONPRFILE OFCELLLINES ..................................................................................59
Obtaining, Samples .............................................................
59
QuantitativeComparisons.............................................................
Varma, Chris
4 of 65
60
Ph.D. Thesis
Acknowledgments
I am indebted to George Church, Peter Szolovits, and Srini Devadas for their
invaluable help, advice, support, and mentorship in all aspects of my thesis and
graduate career. I would like to thank Jun Zhu for his significant guidance and
support. In addition, I would like to acknowledge the contribution of members of
the Church lab - particularly Jay Shendure and Kun Zhang - for providing useful
feedback.
Varma, Chris
5 of 65
Ph.D. Thesis
Overview
Contributions of This Thesis
The end goal of our research is to enable meaningful quantitation in biology that
leads to the generation of new therapeutic targets and new markers for the
diagnosis and prognosis of malignant disease. Since alternative splicing and
general splicing are fundamental processes, it is conceivable that some diseases
or abnormalities may be associated with changes or defects in the cellular
splicing machinery [Kra97], resulting in inappropriately spliced transcripts given
the cell type, physiologic conditions, environment, etc. It is also conceivable that
consistent abnormalities in alternative splicing reflect general dysfunction of
cellular machinery due to a cell's compromised condition or cancerous state. In
fact, 1) alternative splicing is a crucial step in gene expression, 2) thus alternative
splicing is subjected to tight regulation, 3) given its importance in conferring
diversity, alternative splicing is very sensitive to the changes in cellular states
including malignant or abnormal states. Therefore, alternative splicing provides a
unique angle to study disease, particularly malignant disease.
CD44 defects are strongly associated with malignant disease.
CD44 is a cell
surface molecule that is involved in numerous cell processes including cellular
communication and cell-matrix interaction, which is important for tumor
progression. CD44 is alternatively spliced via 10 variant exons in most species
(though only 9 in humans-not
including the tail region), enabling 1024
theoretical isoforms, and specific isoforms are correlated with particular
malignant states including leukemic states. In fact, peripheral blood lymphocytes
(PBLs) of patients with AML, CML, and ALL have significantly elevated CD44
alternatively-spliced isoforms, which are undetected in normal patients [Kha96].
However, even semi-quantitative evaluation of these values has not been
successfully obtained which has lead to significant controversy in the current
CD44 literature. Therefore, CD44 serves a model molecule on which to begin to
study the alternative splicing in malignant disease.
In this work, we present a new paradigm by which to quantitatively study the
alternative splicing of any molecule in any cell type through a single-molecule
based technology [Mit99] and our methods of quantitative exon profiling and
quantitative isoform profiling. Furthermore, we extend this paradigm to include a
novel unified platform-called Isoform Convergence-to qualify particular
isoforms as candidate diagnostic markers, potential therapeutic targets, and
perhaps even as precursor therapeutics themselves.
Thus our purpose and goals are the following:
Varma, Chris
6 of 65
Ph.D. Thesis
1. To evaluate our proposed methods of quantitative exon profiling,
quantitative isoform profiling, and Isoform Convergence for their ability to
generate new and biologically meaningful discoveries
2. To provide unprecedented quantitative insight into the splicing of CD44 in
two human leukemias while resolving controversy in the relevant
literature-through novel methods of quantitative exon profiling and
quantitative isoform profiling
3. To investigate if alternative spicing of CD44 confers a unique and
consistent signature in two human leukemias-through
a novel method
called Isoform Convergence
CD44 Controversy
According to [HerOO], "The type of action of CD44 on a given cell will depend on
the isoform pattern of CD44 expressed...despite a flood of more than 2,000
papers on CD44, the correlation of its detection with disease progression has
remained controversial, and fundamental questions on the function of the CD44
proteins have not been answered." Several studies on the correlation of CD44
isoforms with particular diseases have inconsistent results and conclusions. For
example, [Hei93] and [Mu194]demonstrated that CD44v6 (exon-v6-containing
CD44) correlated with colorectal carcinoma, whereas [Fin95] showed that
CD44v6 does not correlate with colorectal carcinoma. As another example,
[Kau95] reported that expression of CD44v3, v5, and especially v6 epitopes were
found in most primary breast cancer tumor samples but were not present in
normal mammary ductal epithelial cells, and the expression of variant isoforms
especially CD44v6 was correlated with shorter length of survival. In direct
contrast, [Foe99] reported that the expression of CD44v6 may be a marker to
identify patients with a relatively favorable prognosis. We speculate that the
controversy in the literature is mainly due to the semi-quantitative nature of
assays used and the inability to distinguish individual splicing isoforms. We
believe that a single-molecule-based technique (i.e. Polony Technology) along
with our methods of quantitative exon profiling, quantitative isoform profiling, and
Isoform Convergence can help to resolve such controversy by enabling direct
quantification of splicing isoforms. Furthermore, by being able to consider the
complete isoform expression of CD44 at the pre-mRNA level we can elucidate
statistical profiles of alternatively spliced transcripts in a quantitative manner that
together implicate disease. Therefore, this study is not only important as a basic
biological study of alternative splicing regulation but also as a possible
development program for the identification of potential diagnostic markers and
therapeutic targets.
Varma, Chris
7 of 65
Ph.D. Thesis
Background
o. 1. Pre-mRNA
Alternative Splicing
Pre-messenger RNA (pre-mRNA) splicing is a crucial step for mammalian gene
expression. The removal of introns has to be highly precise in order to produce
the appropriate message (i.e. mRNA) for protein production. Splicing can also
be alternatively regulated which is one of the major mechanisms to give rise to
proteome diversity (see Figure 1). Given the importance of pre-mRNA splicing in
gene expression, it is conceivable that splicing is under tight spatial and/or
temporal control [Has01]. Alternative splicing refers to splicing of variable exons
at the pre-mRNA step which generate different messages through exon inclusion
or exon skipping (see Figure 1) resulting in functionally diverse protein isoforms
in a spatially and temporally regulated manner [Has01].
~.
~.
s v
C.' C.2 C.3C.4C=
••
...u
secreled II mRNA
M Exons
••
membrane II mRNA
Figure 1. Alternative splicing of the mouse immunoglobulin \J heavy chain results in two distinctive
mRNAs, one that is secreted as an antibody and another that is membrane bound (Note: introns
are represented by the straight line in the pre-mRNA step. The mRNAs contain no introns.
Figure from htto:/Iwww.blc.arizona.edu/martv/411/Modules/altsolice.html.
)
Overview & Major Players
Splicing occurs in the nucleus of cells and is executed by the spliceosome, which
consists of five small nuclear ribonucleoprotein (snRNP) complexes U1, U2, U4,
U5, and U6 and many non-snRNP proteins [MarOO]. The spliceosome precisely
excises introns and joins exons in a sequential order (see Figure 2) through
numerous RNA-RNA, RNA-protein and protein-protein interactions [Has01].
Varma, Chris
8 of 65
Ph.D. Thesis
Figure 2. Cartoon depiction of the spliceosome as it splices out introns and splices in/out variable
exons. This process is not well understood and this depiction may not be accurate. Figure from
htto:/Iwww.blc.arizona.edu/martv/411/Modules/soliceo.html.
There are five major modes of alternative splicing, namely alternative 5' or 3'
splice-site choice, exon skipping, intron retention, and mutually exclusive exons.
It has been estimated that at least 59% of human genes are alternatively spliced,
and approximately 80% of them result in changes in the encoded protein
[Wau03, Has01]. Alternative splicing has a plethora of functional effects on
mammalian gene expression. Generating several mRNA variants from a single
gene allows functionally diverse protein isoforms to be expressed according to
different regulatory programs. The regulatory programs are cell specific and the
splicing pathways are modulated according to the type of cell, development
stage, gender of organism, external stimuli, etc [Wau03]. Alternative splicing
results in structural variation by the insertion or removal of amino acids, shifting
of the reading frame, and introduction of termination codons. This enables gene
expression variability by the removal or insertion of regulatory elements that
control translation, mRNA stability, or localization [Wau03].
Alternative Splicing, Disease, & Therapeutic Intervention
Although alternative splicing accounts for much genetic variability by generating
multiple protein isoforms, aberrant splicing-- which is alternative splicing that has
lost regulation due to a particular defect--can result in disease [BleOO]. For
example, Thalassemia is an inherited condition, which results in impaired
production of either the alpha or beta hemoglobin chain resulting in severe
anemia and possible organ and bone defects. Patients with Thalassemia carry
mutations in the HBB (beta-globin) gene, which activates cryptic splice sites in
beta-globin pre-mRNA, resulting in a deficiency of adult hemoglobin A [Has01].
There are several other cases of defects in splicing leading to disease (see Table
1) [ArsOO], [Liu01].
Varma, Chris
9 of 65
Ph.D. Thesis
Disorder
intcrmriitent
Aclite
porphyna
Breastandovariancancer
Carbohydratedeficient
glycoprotein syndi'ome
Cne
Translationallysilent
R28R(C -i,G3)
Nonsense
Missense
Pmphobilinogen
deaminase
BRCAI
PMM2
Refs
[69]
1701
1711
E139K(G-E 18)
E139K(G-A. 5)
typela
Sterol 27 hydroxylase
Cysticfibrosis
CFTR
E60X (G-T.: 3): R75X
Ehlers-Danlos synd:ome
type V'I
Fanconianenfia
Fronlatoemporal den:ntia
(IFTDP-17)
HemophiliaA
Lysyl hydroxylase
1 ): W1228X (G -A. 20)
Y511X (C-A 14)
IlPRT deficiency
Hypoxanthine phosphon- G40V (G-T. 2): R4811
(G-A. 3): A161E (C--A.
bosyltransferase
6): P1841. (C T. 8):
D194Y (G-T. 8): E197K
xanthntlatosis
FANCG
Tau
1721
G112G(G-T 2)
Cerbrotendinous
[651
(C-T 3): R553X(C-:
Q356X (C -T. 8)
S305N(G -A. 10): N297K
(T--G. 10)
E1987X (G-I:
R2116X (C -T, 22)
Facta ViL
[731
[74]
1.2841.(T C. 10): S305S [75.76]
(T--C. 10)
1651
19):
FI99F(C-T. 8)
[65]
G1850 (A -G. 6)
[77]
51)
178]
[79]
(G-A. 8): Ei197V(A-T.
enleigh's
cephalomyelopathy
Marfan syndrome
Mctachroraltic leukodystrophy(juvenileform)
Neurofibromatosis
type I
8)
Pyruvate dehydhogenase
ElIx
Fibrillin
T4)91 (C -T. 8)
ArylsulfataseA
121181(C-.
180I
R304X (C-T. 7): Q756X
14): Y2264X
(C -'.
NIF
(C-A. 37):
Ornithine carbanloyltrans
ferase
Uporphyrogen
Porphyriacutanea tardi
decarboxylase
P404L (C-T: 1])
Hexosaninidase
Sandhoffdisease
R142Q(G A. 5)
Severe cbined
inmlun- Adenosinedeanunasc
odeficiency
SMN1
Spinalmuscle atrohpy
SMN2
Spinal muscleatrophy
Fumaryl acetoacetate hy- Q279R(A-G. 8)
Tyrosinemnia
type
drolase
OCT deficiency
[81
T.9)
L304F (G--
[82]
E314E (G-A, 9)
1831
[84]
R142X(C -T. 5)
185]
[86]
[87.88]
WI02X (G-A. 3)
F280F (C -T. 7)
N232N (C-T. 8)
Table 1. The effects of defective alternative splicing leads to various disease states. Table from:
Caceres JF and Kornblihtt AR, Trends Genet 18: 186-93 (2002).
In the case of Thalassemia, scientists reported successful treatment of these
patients' erythroid progenitor cells using antisense oligonucleotides targeted to
the cryptic splice sites, which restored appropriate splicing of the HBB gene and
increased hemoglobin production to near normal levels [LacOO]. There are
numerous other recent examples of such therapeutic interventions of aberrant
splicing induced disease states with similar beneficial effects [CarO3],[SkoO3].
0.2. hCD44
Human CD44 (hCD44) is a cell surface glycoprotein receptor for the glycosamino
glycan hyaluronan (HA), which is a major component of extracellular spaces.
CD44 is expressed on many types of cells including most hematopoietic cells
(e.g. B-cells, T-cells, and myloid cells), keratinocytes, chondrocytes, many
epithelial cell types, and some endothelial and neural cells [Bio98-2]. The
functional role of CD44 (in different cell types or developmental stages) is
regulated by alternative splicing and by post-translational modification such as
glycosulation [AbbOO].
Varma, Chris
10 of 65
Ph.D. Thesis
Structure & Function
Human CD44 is an 80-250 kDa transmembrane glycoprotein [Tan94]. Its
primary ligands are osteopontin, fibronectin, collagen (I, IV23), and hyaluronan
(HA)-HA has been CD44's most important ligand in terms of disease implication
[Uku01]. However, more recently osteopontin has been implicated in disease
progression. For example, the work of [Kha02] has demonstrated that binding of
osteopontin to variant isoforms of CD44 has anti-apoptotic effects, which may
provide immunity to cancerous cells.
CD44 is a multifunctional receptor involved in cell-cell and cell-ECM (extracellular matrix) interactions, cell trafficking, T-cell and B-cell adhesion, lymph
node homing, presentation of chemokines and growth factors to traveling cells,
transmission of growth signals, uptake and intracellular degradation of HA, and
transmission of signals mediating hematopoiesis and apoptosis. The structure of
CD44's HA-binding domain is shown in Figure 3a and 3b. (Crystal structures of
complete CD44 are not available-see the Protein Data Bank). The cytoplasmic
domain of CD44 (approximately 70 amino acids long) is highly conserved (>
900/0)in most of the CD44 isoforms [Bi098-2] .
..
CD44 Alternative Splicing
Human CD44 encodes 20 total exons (see Figure 4). (There are two different
nomenclatures in the hCD44 literature for designation of exons; here we use the
original designation.) Exons 1-5, 15 & 16 are constant exons and they code for
the extracellular domain. Exons 6a,b-14 are variably spliced, which are also
designated as V1 to V10. (Note: Exon 6a / V1 is thought not to be expressed in
humans.) Exon 17 codes for the transmembrane segment (21 amino acids
long)-thus this exon enables CD44 to be expressed on the cell surface. Exons
18 and 19 code for the cytoplasmic tail (72 amino acids long) and are mutually
exclusive.
Varma, Chris
11 of 65
Ph.D. Thesis
In tra-
Extracellular
CcUuJar
Domain
Domalru;
IIMI
I
1
2
J
2
J
4
I
5
6a 6b 7 8 9 10111213
14 15 16
I
17 18 19
Elton
Numbering
~1
4
5 6 7 8 91011 12131415 16
17 18
1t 20
Figure 4. Exons of hCD44 in the 5' to 3' direction. TM denotes transmembrane.
Exon 6a
includes a stop codon and is thought not to be expressed in humans. Note: hCD44 has two sets
of nomenclatures-the original nomenclature is on top and the new nomenclature is below.
The common or standard form of Human CD44 is the hematopoietic form
designated as CD44H or as CD44s ('s' for standard). CD44H does not encode
any variant exons (see Figure 5), includes exon 19 as the tail segment, and is
approximately 270 amino acids long. Besides the common form, at least 45
alternatively spliced variants have been found to exist [Van93]. In the murine
Eph4 cell line, 69 distinct CD44 isoforms have been reported [Zhu03]. Exons are
alternatively spliced on the extracellular proximal domain of CD44 (see Figure 5)
which is involved in ligand binding.
Varma, Chris
12 of 65
Ph.D. Thesis
Varlanl non aJlnn.lh~
Spl,",
SII ..
--.
Trnn!lmembnme
Domain
}
}
C,1oPlasmic
Domain
Figure 5. Cartoon representation of CD44 including location of variant exons.
The results of [Sle97] indicate that alternative splicing regulates the ligand
binding specificity of CD44 and suggests that structural changes in the CD44
protein have a profound effect on the range of Iigands to which CD44 can bind
with potentially wide-ranging functional consequences. Specifically, they report
that isoforms containing exons v6 and v7 enable direct binding to chondroitin
sulfate, heparin, and heparin sulfate in addition to HA. It has also been
established that splicing isoforms of CD44 affect the affinity of binding of
particular growth factors and growth-promoting proteins because the larger
isoforms better stabilize clusters of CD44 molecules [HerOO]. In addition, there is
evidence that CD44's function is enhanced or manipulated by clustering of CD44
molecules [Yu99] and one way found to enhance CD44 clustering is by inclusion
of alternative-splicing isoforms [Sle96]. Interestingly, the effect of including
alternative isoforms on overall CD44 function is analogous to HA's binding to and
stimulation of CD44 [Sle96].
CD44 & Implications in Disease
Both the standard form of CD44 as well as CD44 variants have been implicated
in disease in a plethora of human cancers. The work of [Bou97] indicates that
CD44s and p185-HER2 are physically linked to each other via interchain disulfide
bonds on the surface of ovarian carcinoma cells. Further, HA stimulates CD44sassociated p185-HER2 tyrosine kinase activity which leads to an increase in the
growth of the ovarian carcinoma [Bou97].
In [Shu01] the authors show that CD44v4-v10 confer enhanced in vitro rolling,
enhanced in vivo local tumor growth, and lymph node invasion by lymphoma
cells. Further, a site-directed point mutation at the HA-binding site of the variants
Varma, Chris
13 of 65
Ph.D. Thesis
resulted in loss of these enhanced functions. In another study [Kat99], the
authors demonstrate that several different variants bind osteopontin (OPN), but
CD44s does not. Moreover, the expression of OPN and CD44 variants has been
correlated with tumorigenesis and metastasis. Further investigation has shown
that OPN binding by CD44 variants promotes cell spreading, motility, and
chemotactic behavior.
The work of [Kha96] elucidates the expression of CD44 variants in lymphomas
and leukemia.
In the study, peripheral blood lymphocytes
(PBLs) of 30 normal
patients and PBLs of 183 patients with hematologic disorders revealed that only
in patients with malignant disorders did a measurable proportion of PBLs express
CD44 variant isoforms, mostly exons v5, v6, v7 and less frequently v10.
Elevated levels of these variant isoforms were present in the following
percentage of patients for each hematologic disorder: acute myeloid leukemia
(AML) 16%, chronic myeloid leukemia (CML) 25%, acute lymphoid leukemia
(ALL) 23%, Hodgkin's disease 17%, non-Hodgkin's disease 54%, and multiple
myeloma 22%. In addition, expression of CD44v in PBLs was not linked to the
histological grading or clinical staging of disease.
CD44 in B-cell & T-cell Development
Antibodies to CD44 (isoform non-specific) block development of B-cell precursors
in the marrow-both myeloid and lymphoid cells-in culture, but it is not known how
CD44 and hyaluronan (HA) function in the bone marrow [Jan96]. Therefore,
CD44 is present throughout the development of B-cells.
During T-cell development, progenitor T-cells migrate from the marrow to the
thymus-thus they are now called thymocytes. Initially thymocytes express CD44
for a short time, but as they mature they lose expression of CD44. Since this is a
negative selection process, it is highly unlikely that these cells could become
neoplastic [Jan96]. After development, both effector T-cells and memory T-cells
significantly express CD44 [AbbOO]. The property of CD44 binding to HA is
responsible for the retention of T-cells in extravascular tissues at sites of infection
and for the binding of effector and memory T-cells to endothelium at sites of
inflammation and in mucosal tissues [AbbOO].
0.3. Splicing-Insights Driven Therapy
Although a particular CD44 antibody that identifies an epitope that is aberrantly
expressed (presumably through alternative exon splicing) and that is significantly
upregulated on acute lymphoblastic leukemia (ALL) cells has been found
[BenO3], a rational method of target identification via insights gained from
alternative splicing has not yet been developed. We theorize this is due to the
semiquantitative nature of RT-PCR and antibody-based assays as well as to the
inability to study exon combinatorics until recently.
Varma, Chris
14 of 65
Ph.D. Thesis
However, there are several cases where insights gained from study of the
aberrant alternative-exon splicing of a particular gene in a disease state resulted
in re-establishing correct splicing which subsequently re-enabled normal function.
By using antisense oligonucleotides to correct splicing, normal gene expression
has been established in cellular models of -Thalassemia [Suw02], cystic fibrosis
[Fri99], and Duchenne muscular dystrophy [Wil99].
Antisense oligonucleotides
can interact with mRNA or its precursors in a
sequence specific fashion thereby affecting the expression of the transcript. As
we have seen (see Table 1) it is frequently the case that mutations causing
disease act by affecting pre-mRNA splicing, often resulting in unique transcripts.
Antisense oligonucleotides can be targeted against these unique transcripts to
restore correct splicing [SieOO, Sie99].
carry mutations in the HBB (beta-
For example, patients with l-Thalassemia
globin) gene, which activates cryptic splice sites in beta-globin pre-mRNA,
resulting in a deficiency of adult hemoglobin A [HasOl]. In [Suw02] correct
human beta-globin mRNA was restored in erythroid cells from transgenic mice
carrying the human gene by correcting the splicing of thalassemic human B-
. .F.
globin pre-mRNA via an oligonucleotide targeted to the aberrant 5' splice site.
Aberrantly (ab) and correctly (c) spliced mRNAs are shown in Figure 6.
IL
-
human 13-globinIVS2-654 pre-mRNA
aigonucleotide
b
a
I
ss
III
b
a
b
0
4
]
ab mRNA
|------I-
ab IVS2-654 (367)
Z
c
I
ss
I
I
c mRNA
I
I
C
I
c IVS2-654 (294)
c IVS2-654 (231)
ab IVS2-654 (304)
Figure 6. Correction of splicing of thalassemic human B-globin pre-mRNA by oligonucleotide
targeted to the aberrant 5' splice site [Suw02]. Aberrantly (ab) and correctly (c) spliced mRNAs
are shown in Figure 6. Forward primers a and b were used for patient and murine RNA,
respectively.
Varma, Chris
15 of 65
Ph.D. Thesis
This was accomplished in a dose- and time-dependent manner by free uptake of
morpholino oligonucleotide antisense to the aberrant splice site at position 652 of
intron 2 in beta-globin pre-mRNA. Under optimal conditions of oligonucleotide
uptake, the maximal levels of correct human beta-globin mRNA and hemoglobin
A in patients' erythroid cells were 77 and 54%, respectively. These levels of
correction were equal to, if not higher than, those obtained by syringe loading of
the oligonucleotide into the cells. The effectiveness of the free antisense
morpholino oligonucleotide in restoration of correct splicing suggests the
applicability of this or similar compounds in vivo experiments and possibly in
treatment of thalassemia.
0.4. Leukemia
Leukemia is a disease of the reticuloendothelial system which involves
uncontrolled proliferation of leukocytes (i.e. white blood cells). Leukemia is
generally thought to be an acquired (i.e. non-inherited) cancer, though genetic
abnormalities (e.g. The Philadelphia Chromosome) may also play a role in the
development of this condition [HeaO2]. Leukemia originates in an early cell in the
blood-forming marrow or in the portion of the lymphoid system in the marrow.
The major forms of leukemia are divided into four categories. Myelogenous (i.e.
myeloid) and lymphocytic (i.e. lymphoid) refer to the progenitor cell type involved.
Myeloid cells differentiate into erythrocytes, granulocytes, macrophages,
monocytes, and platelets [Jan96]. Lymphoid cells differentiate into two types of
lymphocytes: B-cells and T-cells. Acute and chronic refer approximately to the
rate of progression of the disease. Thus, the four major types of leukemia are
acute myelogenous leukemia (AML), chronic myelogenous leukemia (CML),
acute lymphocytic leukemia (ALL), and chronic lymphocytic leukemia (CLL). (In
our work, we will examine only AML and ALL.) The term acute lymphocytic
leukemia is synonymous with acute lymphoblastic leukemia. The latter term is
more frequently used to denote cases in children [HeaO4].
Acute leukemia is a rapidly progressing disease that affects mostly cells that are
not yet fully developed or differentiated. These immature cells cannot carry out
their normal functions. In addition, cells of acute leukemia strongly impede
production of normal blood cells because of their uncontrolled multiplication,
which crowds out normal cells. In contrast, chronic leukemia progresses slowly
and does not significantly impede the production and development of normal
blood cells, at least initially. In addition, these more mature cells can carry out
some of their normal functions [LeuO4].
ALL
Most lymphoid neoplasms (i.e. 80 to 85%) including ALLs are precursor B-cell in
origin, whereas the remaining are primarily T-cell in origin [Cot99]. The majority
of ALLs (85%) manifest as childhood leukemia with extensive bone marrow
involvement [Cot99]. Clinically and morphologically, pre-B and pre-T
lymphoblastic: malignancies are indistinguishable, therefore differentiation
requires immunophenotyping [Cot99]. To further complicate the clinical picture,
Varma, Chris
16 of 65
Ph.D. Thesis
not only do pre-B and pre-T lymphoblastic malignancies present similarly, but
they present with similar clinical features to AML--even though ALL and AML are
immunophenotypically and genotpyically distinct diseases. This has been
attributed to the fact that in both cases there is an accumulation of neoplastic
blast cells in the bone marrow suppressing normal hematopoiesis by physical
crowding and other mechanisms [Cot99]. This manifests the states of anemia,
thrombocytopenia (i.e. low platelet count), and neutropenia (i.e. low neutophil
count) which are shared by both ALL and AML.
AML
Acute myelogenous leukemia is characterized by the rapid proliferation of
precursor myeloid cells (i.e. blood forming cells) resulting in the clinical picture
shared with ALL which includes deficiency of red cells, decreased numbers of
platelets, and reduced count of normal white cells (especially neutrophils) in the
blood [LeuO4]. Also, the rapid proliferation of these precursor myloid cells, along
with a reduction in their ability to undergo programmed cell death (apoptosis),
results in their accumulation in various organs, most commonly the spleen and
liver [EmeO4].
Diagnosis of Leukemia
Diagnosis of a particular leukemia usually involves the following steps:
· Medical history and physical examination
·
Complete blood counts
·
·
Bone marrow examination for presence of leukemic blast cells
Cytogenetics: Patient tissue is used in the process of analyzing the
number and shape of the chromosomes of cells
Immunophenotyping: A method that uses the reaction of antibodies with
cell antigens to determine a specific type of cell in a sample of blood cells,
marrow cells, or lymph node cells
·
To diagnose leukemia including the particular type of leukemia, the blood and
marrow cells must be examined. In addition to low red blood cell and platelet
counts, examination of the stained (dyed) blood cells with a light microscope will
usually show the presence of leukemic blast cells. This is confirmed by
examination of the marrow which usually show leukemia cells. The blood and/or
marrow cells are also used for studies of the number and shape of chromosomes
(cytogenetic examination), immunophenotyping, and other special studies, if
required. Blood and bone marrow samples are used to diagnose and classify the
disease. The following tests are used in the further classification of the disease.
Examination of leukemic cells by cytogenetic techniques permits identification of
chromosomes or gene abnormalities in the cells. The immunophenotype and
chromosome abnormalities in the leukemic cells are very important guides in
determining the approach to treatment and the intensity of the drug combinations
to be used [LeuO4].
Varma, Chris
17 of 65
Ph.D. Thesis
CML (and less frequently, ALL) is a particularly unique case in that a specific
molecular test exists for its diagnosis. The Philadelphia Chromosome is a
translocation of chromosomes 9 and 22 [t(9:22)(q34;q 1)], which is diagnostic for
95% of chronic myelogenous leukemia (CML) and a subset of acute
leukemia(-l-13% of ALL). Molecular detection of the BCR-ABL translocation is
performed using Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
analysis. This is currently performed at hospital labs such as Barnes-Jewish
Hospital in St Louis, Missouri which is associated with Washington University in
St Louis [Bar97]. In general however, molecular techniques are just beginning to
enter the clinical setting for diagnosing leukemia.
Treatment of Leukemia
Leukemia is usually treated by chemotherapy and radiation therapy which aim to
reduce the growth of the leukemic cells in the bone marrow and by
transplantation of the bone marrow [MedO2]. For CML in particular, there is a
drug called imatineb (Gleevec) which slows the proliferation of precursor myeloid
cells by targeting the BCR-ABL proto-oncogene. However, resistance to
Gleevec has become a common problem due to approximately 20 point
mutations arising in BCR-ABL. There are currently several next generation
imatineb-like drugs in late-stage development for the treatment of CML.
Varma, Chris
18 of 65
Ph.D. Thesis
Chapter 1: Profiling of Alternative Exon Splicing
Note: a subset of the CD44 SBE and polony amplification primers as shown in
Table 1 were originally designed and ordered by Jun Zhu, formerly a post-doc in
the laboratory of George Church at Harvard Medical School.
1.1. Introduction
In this chapter, we discuss the experimental methods developed and used for the
profiling of alternative exon splicing: quantitative exon profiling and quantitative
isoform profiling. However, this is just the first part of creating such profilesChapter 2 will introduce the necessary computational algorithms and
computational methods to provide quantification.
To profile exons, a method for in situ polymerase chain reaction in acrylamide
gels followed by subsequent querying by single base extensions (specific for
each exon of interest) is presented. The enabling wetlab technology for this
method was previously developed by [Mit99] and improved by [Zhu03]. Our
method-taken as the sum of both the wetlab and computational components (as
presented in Chapter 2)-significantly improves on previous work as well as
enables a higher level of robust quantitation.
1.2. Polony Technologyfor Exon Profiling
Polymerase colony (polony) technology enables parallel amplification of millions
of DNA or RNA molecules via performing polymerase chain reaction (PCR)
within an acrylamide gel on a thin glass slide. This process of solid-phase PCR
results in each template giving rise to a unique and distinct colony of amplified
products known as polonies-and thus, each polony is monoclonal [Mit99, MitO3].
During PCR amplification, when two polonies come into contact they tend to form
a distinct border excluding each another rather than overlapping or invading each
other-thus they are spatially distinct [AacO4]. Therefore, each polony is
effectively an independent PCR reaction on the order of nanoliters to femtoliters
in size. Furthermore, because an acrydite modification is included at the end of
one of the amplification primers, a strand of each amplicon is covalently linked to
the acrylamide matrix of the gel and serves as a template for single base
extensions (SBEs) and for probe hybridizations (see Figure 1). For more
information on Polony Technology, please visit the polony website at
http://arep.med.harvard.edu/Polonator/
Varma, Chris
19 of 65
Ph.D. Thesis
- "_/=-8--
Figure 1-1. The core of Polony Techology.
/ I -/
~a~~
It has been shown that combinatorial patterns
I pm- I
oeq.-ced
I
I
pour acrylamide gel
of exon inclusion or exclusion can be
with DNA and PCR
reagents
determined across multiple polonies in parallel
through an exon profiling method [Zhu03].
be!onl
single
DNA
__
Since each polony arises from a single
molecule
molecule and because of the digital nature of
I in-gel PeR
..
amplification
polony technology, interrogation of polony
slides enables sensitive and accurate
poIony=
-'••
quantification of individual mRNA isoforms. In
PeA colony:
•••
fact, in cases of complex alternative splicing
after
image 01
(Le. many alternative exons), polony
poIaniea
technology is the only practical method to
quantify specific isoforms. Furthermore, it is theorized that polony technology
has even less bias than traditional PCR because 1) it is solid-phase, thus it is
analogous to thousands or millions of separate femtoliter reactions and 2) in
traditional PCR amplicons of different sizes compete for primers-and larger
transcripts compete better (thus creating a bias).
A
DNA '0 be
prvne< B
l
EJ---
!
••
ampkflC:Blion
•••
ampl#lc:alion
1.3. Overview ~f Experimental Methods
Assume following pre-mRNA:
Figure 1-2. Simple conceptual example of creating
a polony slide and querying it by single base
extensions (SBEs).
A polony slide is created by first polymerizing
acrylamide in a solution containing standard
PCR reagents and a substantially low
concentration of cDNA template - this
composes the gel [Mit99]. Before the gel is
polymerized, it is added to a slide which is
bind saline treated so that the gel can attach
to the glass of the slide. Additionally, the
reverse primer is Acrydite (Ac) modified on
one end so that it can be covalently attached
to the gel matrix. Thus, after the slide is
thermal cycled, a single cDNA template has
given rise to a PCR colony or 'polony'
because each particular cDNA template was
immobilized by virtue of being bound to the
Acrydite modified reverse primer and products
Varma, Chris
20 of 65
S'
3'
Patient cDNA
~
1. Pour gel wI PCR reagents & DNA
j
Single Molecule
__
/"..
At
2. In gel PCR amplification
3. Query Exon C2 by SSE
!
4. Query V1& V2 by SSE
S. Aggregate
[. .0
°0
Profile
Ph.D. Thesis
of the reaction remain localized near their respective templates. Now, each
polony can be queried for its signature by single base extensions in parallel (see
Figure 2). This involves designing primers that uniquely bind to each exon of
interest. The base subsequent to the last base of the primer is where
fluorescent-labeled
deoxynucleotides
(dNTP) are attached and thus can be
uniquely queried via an integrated array scanner.
1.4. Primer Design
All primers were designed using the Primer3 software package [RozOO]. Primers
along with name and brief purpose are listed in Table 1. Primers to quantify
amounts of CD44s and CD44v, L_5,17 and L_5,14 respectively, were designed
such that approximately 1/4 of the bases flank the end of exon 5 and the rest of
the primer flanks the beginning of either exon 17 or exon 14, respectively. This
was done to reduce false positives during realtime QPCR experiments. All
primers were checked for appropriate PCR product.
NAME
SEQUENCE
PURPOSE
L_5,17
L 5,14
R_ 19(s,v)
hCD44ETE 19R2
hCD44ETE4F
TCACTGTTCCTGATTGCTCA
ATCACTGCTGATTCCACCTC
AGCACAAAAGGTGAAGATCG
TTTCCTGAGACTTGCTGGCCTCTCC
ACAGACCTGCCCAATGCCTTTGATG
Quantify CD44s
Quantify CD44v
Quantify CD44s/CD44v
Quantify total CD44
Quantify total CD44
BACT F
BACT R
TCACCCACACTGTGCCCATCTACGA
CAGCGGAACCGCTCATTGCCAATGG
Quantify B-actin
Quantify B-actin
hCD44ETE19Rac2
hCD44ETE18Racl
hCD44ETE4F
hCD44SBE5F
hCD44SBE6F
TTTCCTGAGACTTGCTGGCCTCTCC
ACAGCCCATGTGTCAGTTCTAGCGA
ACAGACCTGCCCAATGCCTTTGATG
ACACCCCATCCCAGACGAAGACAG(tccc)
GAGGCAAGAAACCTGGGATTGGTTT(tca)
Polony amplification
Polony amplification
Polony amplification
Single Base Extension
Single Base Extension
hCD44SBE7AF
hCD44SBE7BF
GTACGTCTTCAAATACCATCTCAGCAGG(ct)
TGGATCAGGCATTGATGATGATGAAG(attt)
Single Base Extension
Single Base Extension
hCD44SBE8F
hCD44SBE9F
CCACGGGCTTTTGACCACACAAA(aca)
GAAGCACACCCTCCCCTCATTCAC(cat)
Single Base Extension
Single Base Extension
hCD44SBE1OF
hCD44SBE1 1F
AGAAGGAACAGTGGTTTGGCAACAGA(tgg)
AGGACAACACCAAGCCCAGAGGAC(agtt)
Single Base Extension
Single Base Extension
hCD44SBE12F
TCCAAACACAGGTTTGGTGGAAGATT(tgg)
Single Base Extension
hCD44SBE13F
TACATCACATGAAGGCTTGGAAGAAGA(taaa) Single Base Extension
hCD44SBE14F
GCAGGACCTTCATCCCAGTGACCT(cag)
Single Base Extension
hCD44SBE15F
GGGGGTCCCATACCACTCATGGA(tct)
Single Base Extension
hCD44SBE17F
CTTGGCCTTGGCTTTGATTCTTGC(agt)
Single Base Extension
Table 1-1. Primers used for quantification of CD44v, CD44s, total CD44, and B-actin via
realtime PCR and primers used for polony amplification
Varma, Chris
21 of 65
and single base extensions.
Ph.D. Thesis
1.5. Polony Slide Creation & Subsequent Preparation
Polony amplification was performed similar to [Mit99, MitOl, Zhu03], however
with significant modifications. 5ul of cDNA sample was added to gel mix (7.5%
acrylamide, 0.35% DATD, 0.035% Bis-acrylamide, 0.71% each of two 100 uM
acrydite modified reverse primers, 3.5% Rhinohide PCR gel strengthener, 0.2%
BSA) along with ammonium persulfate (APS) and TEMED to a final
concentration of 0.087%. 18 ul of this solution was added to a bind saline
(Pfizer) treated, partially Teflon coated (Erie Scientific) glass microscope slide
(Fisher Scientific). The partial coat of Teflon was designed to leave an oval
shaped recessed center where the solution was added. The glass microscope
slide was covered with a glass coverslip (No.2-Fisher Scientific). The gel
polymerized under argon gas for 24 minutes. The slide was washed without its
coverslip for 18 minutes in dH20, then dried under the hood for 16 minutes. 28ul
of polony amplification mix (0.67% BSA, 1.0% 100 uM forward primer, 2.5%
10mM dNTP, 10% 1Ox Jumpstart Taq Buffer, 6.67% Jumpstart Taq) was added
to the slide and it was covered with a coverslip. The slide was covered with 65 ul
frame-seal chamber (MJ Research). The chamber was filled with 550 ul of
mineral oil. 'The slides were cycled as follows: denaturation (94°C for 3 minutes),
59 cycles (94°C for 30s, 56°C for 30s, 72°C for 2 minutes).
Subsequently, the polony slide was denatured in 70% formamide pre-heated at
70°C for 15 minutes (in order to remove the excess template) and washed three
times in wash buffer 1E (10 mM Tris-HCI pH 7.5, 50 mM KCI, 2 mM EDTA,
0.01% Triton X-100).
1.6. Single Base Extensions & Scanning
A frame seal chamber (MJ Research) was attached to each slide, and 100 ul of
annealing mix (0.5% 100 uM SBE primer and 10% 10x Jumpstart Taq Buffer)
was added to the gel. The slide was heated at 94°C for 6 minutes and cooled to
60°C for 15 minutes. The slide was washed three times in wash buffer 1E. The
slide was equilibrated with Klenow extension buffer (50 mM Tris-HCI pH 7.3, 5
mM MgCI 2, 0.01% Triton X-100). Single base extension reactions were
conducted by adding fluorescence-labeled dNTP, Klenow Polymerase (NEB),
Klenow buffer, and single stranded binding protein (SSB). The slide was
incubated at room temperature for 2 minutes. After washing with wash buffer IE,
slide was scanned using a GenePix 4100B Integrated Array Scanner (Axon
Laboratories) at 10 um resolution using 635 nm (Cy5 detection) and 532 nm (Cy3
detection) lasers. Sixteen-bit values per pixel are obtained.
Each slide that was created is queried with single base extensions for each of 14
exons (5 - 14, 17 - 19) followed by scanning. After each cycle of single base
extension, the slide is denatured and scanned again to obtain a valid background
image. This results in 7 cycles of single base extensions,14 scans on two
Varma, Chris
22 of 65
Ph.D. Thesis
channels, and 28 images per slide (including a background image for each exon
queried). (Note: Exon 7a was also queried here - data not reported).
1.7. Real-Time QPCR
Real-Time PCR was done to quantify the amounts of total CD44, amounts of a
specific variant isoform of CD44 - that which only contains variant exon 14, and
amounts of B-Actin in each sample. Experiments were done in duplicate.
An Opticon 2 Real-Time QPCR machine (Bio-Rad Laboratories) and SyBr Green
dye were used to perform real-time PCR experiments. Each sample was first
diluted 10x. Serial dilutions of 1x, 5x, and 25x were performed for each sample
for each primer pair. Each well contained 20 ul of solution (2.5% appropriately
diluted sample, 50% SyBr Green Real-Time PCR Mix, 3% 10 uM reverse primer,
3% 10 uM forward primer). A blank well (i.e. dH20 substituted for sample),
associated with each sample for each primer pair, was used for background
subtraction. The plates were cycled as follows: denaturation (94°C for 6
minutes), 47 cycles (94°C for 30s, 58°C for 30s, Plate Read, 72°C for 1:30
minutes, Plate Read), obtain melting curve (72°C for 8 minutes, melting curve
from 65°C to 90°C, read every 0.2°C, hold
s).
1.8. Conclusion
The experimental methods of polony slide creation and single base extensions
as described here were used to generate images which capture the digital
representation of the exons present on a slide. Chapter 2 discusses how we
obtain data from this large set of images.
Varma, Chris
23 of 65
Ph.D. Thesis
Chapter 2: Construction of Expression Profiles
2. 1. Introduction
Process
Figure 2-1. Summary of raw data obtained.
18
We introduce a software program for the
computational processing and
subsequent construction of quantitative
exon profiles and construction of
quantitative isoform profiles of all sample
slide images acquired in Chapter 1. The
need for computational processing is
significant due to the large amounts of
data needing to be processed for this
work (see Figure 1). Furthermore,
computational processing allows for
standardization of processing which
reduces inconsistencies among images
analyzed.
g
x
1
[= = -
54x
1
756 x
1512 x
•
•
II
.tit
-15 M polonies, >11 GB ot data
2.2. Overview of Software
The goal of such a software program is quite simple (see Figure 2). We are
attempting to determine the number and location of polonies present on each
image. The numbers by themselves serve as early values for exon counts which
when further analyzed are used to create quantitative exon profiles. To obtain
the specific isoforms present, we essentially want to remember the locations of
each polony present on each image of a sample and string these images
together (in the correct order) to construct the isoform signatures.
Varma, Chris
24 of 65
Ph.D. Thesis
Assume following pre-mRNA:
Figure 2-2. Simple conceptual example of
the goal ofthe software -- to determine the
number of exons on each polony image of
each slide and to construct an isoform
profile from this information. This figure is
consistent with Figure 1 of Chapter 1.
These isoform signatures are then
used to create the quantitative isoform
profiles. Of course, the significant
challenges of image-processing wetlab
generated gels, identifying what is truly
the structure of a polony for
recognition, and appropriately
constructing an isoform signature
require sophisticated techniques and
algorithms.
5'
3'
Exons queried by SBEs :
Exon
C1
C2
V1
V2
C3
Count
N/A
5
2
3
N/A
Assume C2 -> C1 -> C3, then isoforms:
V1+V2 ~
Count1
V2
Count 2
V1
Count 1
Zero
Count 1
2.3. Image Processing
Figure 2-3. Original Image - pre processing.
Raw 16-bit images obtained via scanning (see
Figure 3) in Chapter 1 were subtracted from a pre.defined mask that removed the pixels residing on
the non-gel portions of the image. The images
were then filtered (along with their associated
background images) with a 3 X 3 median filter. The
image was then subtracted from its background
image. The border of the image was then cleaned
and smoothed to reduce noise that is often found at
the border of images. These images were then
pre-processed using a specific combination of
image openings and closings including a top-hat
transformation.
2.4. Thresholding
Next, the images were thresholded. Because polonies have florescence on an
image within at least the same order of magnitude which is different than the
Varma, Chris
25 of 65
Ph.D. Thesis
florescence of particle junk (e.g. dust) and remaining background, we are able to
usually apply a fully automated means-based thresholding method (Figure 4).
T
= 0.5*(double(min(f(:)))
=
+ double(max(f(:))));
done false;
while -done
g = f >=T;
Tnext = 0.5*(mean(f(g)) + mean(f(-g)));
done abs(T - Tnext) < 0.5;
T Tnext;
end
=
=
Figure 2-4. Primary threshold method applied.
Figure 2-5. Image after thresholding.
In some cases, this thresholding method did not
yield good results across a sample. For these
cases we either applied the matlab function
'thresh' or we used a semi-automated technique.
The resulting thresholded images (see Figure 5)
were used for subsequent processing.
2.5. Polony Identification
Candidate polonies were derived via matlab functions that yielded the connectedcomponents on the thresholded image. These candidate polonies were
evaluated for true polonies based on their size and shape. Size range was
determined by area (Le. number of pixels). Shape was determined by range of
allowed eccentricity (roughly from a oval to a circle) and by a bounding box
configuration. These minimal yet comprehensive parameters enabled a single
parameter set to be used for the successful processing of all images of all
samples-this was a major goal of our work. True polonies were then saved into
a pre-initialized structure.
Varma, Chris
26 of 65
Ph.D. Thesis
2.6. Isoform Construction
In the previous section, we knew the number of types of exons to expect and
thus memory allocation was simply handled by pre-initialization. In isoform
construction, since the number of isoforms cannot be predicted, more advanced
memory allocation methods were used to enable minimum runtime. The key
portion of the novel algorithm used to construct isoforms is shown in Figure 6.
_
%Determine the isoforms
for i = 1:(NUM_SLIDES - 1) %Comparing the last slide to itself makes no sense, check separately
if (Exons(i).ThereArePolonies == 1) %save computational time by not looping when not required
forj =-1:Exons(i).NumPolonies
if (Exons(i).Polonies(j).AlreadyCounted == 0)
Numlsoforms = Numlsoforms + 1;
Norkinglsoform(i) = 1;
Exons(i).Poloniesoj).AlreadyCounted = 1;
for k = i+1 :NUM_SLIDES %Compare only against slides numbered greater sequentially
if (Exons(k).ThereArePolonies == 1)
for I = :Exons(k).NumPolonies
if (Exons(k).Polonies(l).AlreadyCounted == 0)
NumPixels....
if (comparepolonies(Exons(i).Polonies().Pixels,Exons(i).Polonies(j).
Exons(k). Polonies(l).Pixels, Exons(k). Polonies(l). Num Pixels) == 1)
Workinglsoform(k) = 1;
Exons(k).Polonies(l).AlreadyCounted = 1;
break %break the for loop as only 1 polony can overlap per slide
end
end
end
end
end
[Isoforms, NumUniquelsoforms] = addisoform(lsoforms,NumUniquelsoforms,Working
soform);
Workinglsoform = linspace(O,O,NUM_SLIDES); %reset to all O's
end
end
end
end
Figure 2-6. The main algorithm by which isoforms were constructed.
The function comparepolonies was enabled to allow for a small amount of gel
movement between images because a gel tends to shrink over repeated SBE's
and this may result in slight movement of polonies. The results of the algorithm
were randomly checked by hand.
2.7. Special Cases & Other Software
In order to determine exon 17 skipping, image of exon 17 was subtracted from
image of exon 5, followed by a necessary array of image post-processing
Varma, Chris
27 of 65
Ph.D. Thesis
algorithms which included opening and closing of the images, top-hat
transformations, and guassian filtering.
Software was also written for necessary processing of quantitative exon and
isoform profiles, as well as for the novel process of Isoform Convergence
(introduced in Chapter 5).
2.8. Performance of Software
Software was developed on Matlab (R12.5) with extensive use of the image
processing toolbox functions.
A single sample is represented by 28 images - one for each alternative exon
(including exon 18 and 19), exon 5, exon 17, and one background image for
each. Each 16-bit .tiff image is on average 7.5MB in size - variation in size is
primarily due to number of polonies and gel variation. To completely process
one sample, it takes on average 15 minutes of processing time (assuming cases
where no semi-automated
interference is required) on a Pentium 4 machine with
1GB of RAM. For cases where a semi-automated intervention is required, time
can increase significantly.
2.9. Conclusion
The software described in this Chapter was an integral component of quantitative
exon and isoform profiling.
In fact, it is not likely that these profiles can be
constructed without the algorithms developed due to the large quantity of data,
necessity to maintain rigid consistency of parameters across all images of all
samples, and requirement of fidelity.
Chapter 3: Characterization of Samples
3.1. Introduction
In this chapter, we conduct both semi-quantitative analysis of CD44 as well as
detailed variant exon quantitation. Since much of the previous work on
alternative splicing of CD44 in leukemia has attempted only semi-quantitative
variant exon quantitation, we compare our findings to this previous work. In
several cases, we identify exons that were not thought to be expressed in certain
cell types as well as previously unreported findings.
Varma, Chris
28 of 65
Ph.D. Thesis
3.2. Obtaining Samples
Samples of blasts from human peripheral blood and from human bone marrow
derived from patients with acute myelogenous leukemia (AML) and B-cell acute
lymphocytic leukemia (ALL) are kindly provided by Dr. Linda Bendall, at The
Westmead Institute for Cancer Research, Westmead Millennium Institute,
University of Sydney, Westmead, NSW, Australia. Each sample is obtained from
a different individual and was provided as 20 to 50 ul of cDNA. The percentage of
leukemic cells in all samples is greater than 90%.
Samples of human peripheral blood purified B cells, human cord blood purified B
cells, and human adult whole bone marrow cells derived from normal (i.e. non-
diseased) individuals were purchased from ALLCELLS, LLC, Berkeley, California
USA. Each sample is obtained from a different individual and was provided as
20 ul of cDNA.
A sample of human breast tissue cells derived from a breast tumor was used as
a positive control. This sample was provided by Jun Zhu, Assistant Professor,
Duke University, Durham, NC USA.
The samples that will be analyzed are listed in Table 1.
Sample No.
Designation
Source
318
Normal (NM)
Human cord blood purified B cells
794
984
1072
Normal (NM)
Normal (NM)
Normal (NM)
Human adult bone marrow cells
Human adult bone marrow cells
Human peripheral blood purified B cells
1139
397
Normal (NM)
AML
Human peripheral blood purified B cells
Human peripheral blood
505
656
735
AML
AML
AML
Human adult bone marrow
Human adult bone marrow
Human adult bone marrow
1601
391
572
AML
ALL
ALL
Human peripheral blood
Human peripheral blood
Human adult bone marrow
596
616
0 (originally 'breast tumor')
ALL
ALL
B.Tumor
Human adult bone marrow
Human adult bone marrow
Human solid breast tumor
Table 3-1. Samples analyzed in this work. Sample No. designations
come from the
sources. Normal implies non-diseased.
3.3. Semi-Quantitative Analysis of CD44
All samples were quantified for total CD44, for total B-Actin, and for CD44v10 -an abundant variant isoform of CD44 -- via Real-Time QPCR (as discussed
previously). The quantified values of CD44v10 (not shown) were then used as a
surrogate for overall variant CD44 expression in order to derive the value of total
CD44 present on each polony sample slide. This was necessary because large
Varma, Chris
29 of 65
Ph.D. Thesis
quantities of molecules (> 1.E+04) on a polony slide cannot be accurately
quantified via computational methods due to signal saturation. Additionally,
serial dilutions of starting template for polony amplification were used to verify
derived values of total CD44 (results not shown), and %RNA template ratio
correlates strongly (r=0.99, 95% confidence interval) with %Polony count ratio
[Zhu03]. Total CD44 had the following ranges for each type of sample: AML
4.E+4 to 5.E+5, ALL 5.E+3 to 1.E+5, NM 7.E+3 to 2 E.+5 (see Table 2).
Desig.
Sample No.
#CD44
#CD44s
#CD44v
CD44v/CD44
CD44/Bactin
AML
AML
AML
AML
AML
397
505
656
735
1601
2.E+05
4.E+05
4.E+04
4.E+05
5.E+05
2.E+05
4.E+05
4.E+04
4.E+05
5.E+05
431
977
272
1005
519
2.E-03
2.E-03
8. E-03
2. E-03
9.E-04
3.E-01
3.E-01
2.E-01
1.E-01
3.E-01
3. E-03
4.E-03
2. E-02
3. E-03
9.E-03
2. E-01
2.E-01
3.E-01
1.E-01
2.E-01
3. E-03
3.E-02
6. E-02
4.E-03
2.E-03
2.E-03
2. E-01
4.E-01
4.E-02
4.E-02
4.E-01
6.E-02
2. E-02
2. E-01
ALL
ALL
ALL
ALL
NM
NM
NM
NM
NM
SUM
2.E+06
2.E+06
3204
A VERAGE
391
572
596
616
SUM
AVERAGE
318
794
984
1072
1139
3.E+05
6.E+04
3.E+04
1.E+05
5.E+03
2.E+05
1.E+05
7.E+03
8.E+03
9.E+04
5.E+04
2.E+05
3. E+05
6.E+04
3.E+04
1.E+05
5.E+03
2.E+05
1.E+05
7.E+03
8.E+03
9.E+04
5.E+04
2.E+05
641
290
541
361
49
1241
310
199
520
329
89
340
SUM
3.E+05
3.E+05
1477
AVERAGE
6.E+04
6.E+04
295
Table 3-2. # CD44 represents the total number of CD44 molecules on a slide of each
sample - this value was obtained using the value for CD44v10 (not shown) for each
sample as a surrogate for overall CD44v expression which enabled linking Real-Time PCR
data to Polony Technology data. # CD44s represents the total number of standard (i.e.
without alternative exons) CD44. # CD44v represents the total number of CD44molecules
that include at least one alternative exon - this value was obtained by alternative exon
profiling. Note that CD44v includes isoforms that are exon 5 cryptically-spliced and
includes isoforms that express exon 17-skipping - and these are analyzed separately. The
counts for exon 5 cryptically-spliced isoforms for each of AML, ALL, NM are 362, 440, 337
respectively. The counts for exon 17-skipping isoforms for each of AML, ALL, NM are 300,
151, 203 respectively. CD44v/CD44 represents the ratio of the total number of CD44
molecules that include at least one alternative exon to the total number of molecules of
CD44. CD44/Bactin represents the ratio of number of molecules of CD44 to number of
molecules of the house-keeping gene B-Actin in each sample.
Based on the averaged values of #CD44 (see Table 2) for the various sets of
samples, total CD44 is up-regulated approximately one order of magnitude in
both AML and ALL compared to Normal (NM). However, the ratio of total CD44
to total B-Actin is constant among AML, ALL, and NM. Since B-Actin is a well
known house-keeping gene [Alb94] and may be applied as a surrogate for overall
Varma, Chris
30 of 65
Ph.D. Thesis
gene transcription, it is likely that overall gene transcription (including that of
CD44) is up-regulated by one order of magnitude in the leukemia samples as
compared to Normal.
However, the differences in the averaged ratios of total variant CD44 (obtained
via alternative exon profiling) to total CD44 in the samples suggests that
expression of the variant exons of CD44 is up-regulated by approximately one
order of magnitude in AML and ALL versus normal.
3.4. Quantifying
Variant Exons
Variant exons were queried via alternative exon-profiling (as previously described
in Chapter 1) and appropriate SBE primers. Sample were aggregated by sample
type. In Normal samples, ALL samples, and AML samples (see Figure 1), counts
of exons generally increase in the 5' to 3' direction as observed by [Zhu03].
Exon Counts of Samples Aggregated by Designation
1200
(J)
+-'
1000
C
::J
o
o
c
o
x
800
600
W
400
200
o
V2
V3
V4
V5
V6
V7
V8
V9
V10
Variant Exon
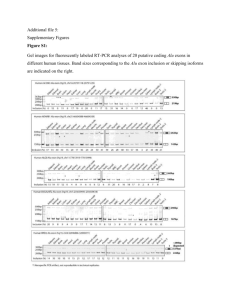
Figure 3-1. Exons are represented by their variant exon designation. Counts of each
variant exon were aggregated within like designations of AML, ALL, and Normal (NM).
In normal samples (see Table 3), we found that all samples expressed variant
exon 7 (V3), all samples expressed variant exon 10 (V6), and all samples
expressed exons 12 (V8) through 14 (V10) as is consistent with previous work
[Aks02, BenOO, Ben04]. We also found that no normal sample expressed
variant exon 11 (V7). Other work has also found lack of CD44v7 mRNA
expression as well via RT-PCR and southern blotting methods [BenOO].
Varma, Chris
31 of 65
Ph.D. Thesis
We also detected a small quantity (0.40/0) of both exon 8 (V4) and exon 9 (V5) in
2 of 5 samples (one of which is human whole bone marrow purified cells and the
other is human peripheral blood purified B-cells). This differs from previous work
in which these mRNA variants were not found and were thought not to be
expressed in normal peripheral blood lymphocytes or normal whole bone marrow
[BenOO]. We also found low expression (0.5%) of exon 6b (V2) in 3 of 5 normal
samples which has not previously been reported to the author's knowledge.
VariantExon
EXON 6B
(V2)
EXON 7
(V3)
EXON 8
(V4)
EXON 9
(VS)
EXON 10
(V6)
EXON 11
(V7 )
EXON 12
(va)
EXON 13
(V9)
EXON 14
(V10)
Totals
AML
%of
#of
Total
Samples
0.7% 20f5
2.9% 40f5
0.4% 30f5
0.4% 50f5
8.1% 50f5
1.1% 30f5
21.6% 40f5
36.2% 50f5
28.7% 50f5
100.0%
ALL
%of
#of
Total
Samples
1.5% 40f4
5.8% 40f4
0.3% 20f4
2.6% 30f4
14.9% 40f4
4.6% 30f4
24.6% 40f4
25.1% 40f4
20.7% 40f4
100.0%
Normal (NM)
%of
#of
Total
Samples
0.5% 30f5
3.3% 50f5
0.4% 20f5
0.4% 20f5
7.2% 50f5
0.0% o of5
25.1% 50f5
33.4% 50f5
29.7% 50f5
100.0%
Table 3-3. % of aggregated samples that express each variant exon and number of
samples expressing each variant exon. % of totalis the percent of polonies identified
from the summation of allsamples of each sample type for each variant exon - thus these
results are pooled from data acquired on independent samples.
In AML samples (see Table 3),we found that a majority of samples expressed
variant exons 7 (V3) and 10 (V6) to 14 (V10) as is consistent with previous work
[Leg98, Aks02, BenOO]. However, we also detected a small quantity (0.40/0) of
both exon 8 (V4) and exon 9 (V5) in 3 of 5 and 5 of 5 samples, respectively. This
differsfrom previous work in which these variants were reported not to be found
[BenOO]. We also found low expression (0.7%) of exon 6b (V2) in 2 of 5 AML
samples which has not previously been reported to the authors knowledge.
In ALL samples (see Table 3), we found that 4 of 4 samples expressed exons 7
(V3), 10 (V6), and 12 (V8) to 14 (V10) which are expressed in higher ratiosthan
previously found [Ben04, Aks02, BenOO]. Further, we found that exon 8 (V4),
exon 9 (V5), and exon 11 (V7) were present in 2 of 4, 3 of 4, and 3 of 4 of the
samples in small to medium amounts: 0.3%, 2.6%, and 4.6%, respectively. This
differsfrom previous work in which these mRNA variants were reported not to be
found [Ben04]. We also found some expression (1.5%) of exon 6b (V2) in all
ALL samples which has not previously been reported to the author's knowledge.
Varma, Chris
32 of 65
Ph.D. Thesis
3.5. Conclusion
Although this is just the beginning of the quantitative profiling ability of our
methods, we have already corrected several incorrect claims in the published
CD44-leukemia literature as well as gained insight previously not reported.
Varma, Chris
33 of 65
Ph.D. Thesis
Chapter 4: Exon Expression Profiles
Note that in order to derive more robust and universally accepted results, we
compare the leukemias against a heterogeneous mix of normal samples
(containing independent samples of human peripheral blood purified B-cells,
human cord blood purified B-cells and, human adult whole bone marrow purified
cells) designated as NM.
4. 1. Introduction
We now apply quantitative methods to obtain insight when looking for significant
differences between exon profiles of the various sample types - we are
specifically interested in statistically significant differences between AML and
normal, ALL and normal, and AML and ALL. Therefore, samples of each type
were aggregated. Here we only consider the variant exons 6 - 14 and the
exclusively splicing exons of the CD44 tail: exon 18 and exon 19. We compare
our results to published work when available and provide new findings. The level
of quantitative analysis provided here significantly exceeds what has previously
been identified in variant exon profiling of CD44 in the leukemias.
4.2. Quantitative Comparison of AML to Normal
Variant exon counts of AML and normal samples were aggregated and
compared (see Table 1). After comparison of the ratios of each variant exon to
the total number of counts for all variant exons of each sample type, we found
that AML is down-regulated with respect to exon 12 (p < .009) and AML is upregulated with respect to exon 13 (p < .05). Since NM does not express exon 11,
AML was up-regulated with respect to this exon (p < .0004). Exon 11 (V7)
enables direct binding to chondroitin sulfate, heparin, and heparin sulfate in
addition to HA [Sle97].
Exon
AML
AML Ratio
NM
NM Ratio
P.value
EXON 6B (V2)
EXON 7
(V3)
EXON B
(V4)
EXON 9
(V5)
EXON 10
(V6)
EXON 11
(V7)
EXON 12
(VB)
EXON 13
(V9)
EXON 14
(VI0)
Varma, Chris
34 of 65
Ph.D. Thesis
Table 4-1. Individual exon counts were based on the aggregation of 5 independent
samples of AML and 5 independent samples of normal. NM = normal samples. X Ratio
(where X = sample type) = ratio between counts of each exon out of total count of exons.
C.f. Ratio = ratio between AML and NM (negative values indicate down-regulation
in AML).
Statistically significant up-regulation of AML is highlighted in light blue and downregulation is highlighted in light green. The p-values were calculated by a standard largesample test procedure for evaluating the difference between proportions obtained in two
different populations [Dev91].
4.4. Quantitative
Comparison
of AML to ALL
Variant exon counts of AML and ALL were aggregated and compared (see Table
2). After comparison of the ratios of each variant exon to the total number of
counts for all variant exons of each sample type, we found that ALL is upregulated vs. AML for a majority of the exons (6 of 9): 6b (p < .02), 7 (p <
.00006), 9 (p « .05), 10 (p « .05), 11 (p« .05), and 12 (p < .04). Exon 10 (V6)
has been reported to be preferentially expressed on ALL cells [Mag01, Ben04].
and our data is consistent with this result. AML was up-regulated as compared
to ALL for only 2 of 9 exons: 12 (p «.05) and 14 (p < .000006). However, AML
expressed almost 10x as much exon 13 than ALL.
Exon
EXON
EXON
EXON
EXON
EXON
EXON
EXON
EXON
EXON
AML
6B (V2)
7 (V3)
a (V4)
9 (V5)
10 (V6)
11 (V7)
12 (va)
13 (V9)
14 (VI0)
20
82
-
12
AML Ratio
ALL
0.0070
0.0288
11
43
0.0042
2
~9
:111'
1:8 ,., «_.~ ::iO.OO3-S
rzam <.;; -,~:; t.;O.OIOB"
5'2
0.0142
614
1029
816
0.2158
0.3617
0.2868
"
,
"
34
183
187
154
ALL
Ratio
0.0148
0.0578
C.f.
Ratio
-2.10
-2.01
2.E-02
6.E-05
0.0027
1.57
3.E-01
0.0255
0.1492
0.0457
0.2460
0.2513
0.2070
-7.27
-1.85
<<5.E-02
<<5.E-02
«5.£:-02
4.E-02
«5.E-02
6.E-06
-4.06
-1.14
1.44
1.39
P-value
Table 4-2. Individual exon counts were based on the aggregation of 5 independent
samples of AML, and 4 independent samples of ALL. X Ratio (where X = sample type) =
ratio between counts of each exon out of total count of exons. C.f. Ratio = ratio between
AML and ALL (negative values indicate down-regulation
in AML or up-regulation
in ALL).
Statistically significant up-regulation of AML is highlighted in light blue and up-regulation
of ALL is highlighted in light green. The p-values were calculated by a standard largesample test procedure for evaluating the difference between proportions obtained in two
different populations [Dev91].
4.5. Quantitative
Comparison
of ALL to Normal
Simiar to section 4.3, variant exon counts of ALL and normal samples were
aggregated and compared (see Table 3). After comparison of the ratios of each
Varma, Chris
35 of 65
Ph.D. Thesis
variant exon to the total number of counts for all variant exons of each sample
type, we found that ALL is up-regulated with respect to exons 6 (p < .02), 7 (p <
.005) and ALL is significantly up-regulated with respect to exons 9 (p < .00002)
and 10 (p «.05).
Exon 6 has been reported to be preferentially expressed on
ALL cells [Mag01, Ben04], and our data is consistent with this result.
Conversely, ALL is significantly down-regulated with respect to exons 13 (p <
.00008) and 14 (p < .000008). Since NM does not express exon 11 and ALL
expresses it in almost 5% of total CD44v (see Table 3, Chapter 2), ALL was
significantly up-regulated with respect to this exon (p «.05).
Exon 11 (V7)
enables direct binding to chondroitin sulfate, heparin, and heparin sulfate in
addition to HA [Sle97].
C.f.
P-value
Ratio
2.69
EXON 6B (V2)
11
0.0148
6
0.0055
2.E-Q2
EXON 7 (V3)
1.75
36
0.0330
5.E-Q3
43
0.0578
EXON B (V4)
4
-1.36
4.E-01
2
0.0037
0.0027
EXON 9 (V5 )
19
4
6.97
2.E-Q5
0.0037
0.0255
EXON 10 (V6)
111
79
0.0724
2.06
«5.E-Q2
0.1492
EXON 11 (V7)
N/A
0
0.0000
«5.E-02
34
0.0457
EXON 12 (VB)
274
-1.02
183
0.2511
4.E-01
0.2460
EXON 13 (V9)
-1.33
187
364
0.3336
8.E-Q5
0.2513
EXON 14 (VI0)
-1.43
154
324
0.2970
0.2070
8.E-Q6
Table 4-3. Individual exon counts were based on the aggregation of 4 independent
samples of ALL and 5 independent samples of normal. NM normal samples. X Ratio
(where X sample type) ratio between counts of each exon out of total count of exons.
C.f. Ratio ratio between ALL and NM (negative values indicate down-regulation in ALL).
Statistically significant up-regulation of Cancer is highlighted in light blue and downregulation is highlighted in light green The p-values were calculated by a standard largesample test procedure for evaluating the difference between proportions obtained in two
different populations [Dev91].
Exon
ALL Ratio
ALL
=
=
4.6. Quantitative
NM
=
=
Comparison
NM Ratio
of Cancer to Normal
Variant exon counts of AML and of ALL were aggregated and designated as
"Cancer." They were then compared to aggregated values of normal samples
(see Table 4). After comparison of the ratios of each variant exon to the total
number of counts for all variant exons of each sample type, we find that no exon
is statistically up-regulated in Cancer vs. normal except for exon 11 (p < .000003)
which was found not to be expressed in normal cells (in Chapter 3). This is
interesting in regard to [Kha96] which reports that the authors found significant
expression of CD44v7 (Le. any isoform containing V7) in 10 healthy volunteers
via staining for anti-CD44v7. However, this is not possible if there is no
expression of CD44v7 mRNA. Other work has also found lack of CD44v7 mRNA
expression via RT-PCR and southern blotting methods [BenOO].
Varma, Chris
36 of 65
Ph.D. Thesis
Exon
Cancer
6B (V2)
7 (V3 )
C. Ratio
NM
31
125
14
29
341
NM Ratio
C.f.
Ratio
1.57
1.06
1.06
2.20
1.31
P-value
0.0086
6
0.0055
2.E-01
0.0348
36
0.0330
4.E-01
a (V4)
0.0039
4
0.0037
5.E-01
9 (VS)
0.0081
4
0.0037
6.E-02
10 (V6)
0.0950
79
0.0724
1.E-02
11 (V7)
66
0.0184
0.0000
N/A
3.E-06
0
12 (va)
0.2221
797
274
0.2511
-1.13
2.E-02
13 (V9)
1216
0.3388
364
0.3336
4.E-01
1.02
14 (VI0)
970
0.2703
324
0.2970
4.E-02
-1.10
Table 4-4. Individual exon counts were based on the aggregation of 5 independent
samples of AML and of 4 independent samples of ALL, versus the aggregation of 5
independent samples of normal. NM
normal samples. X Ratio (where X sample type)
ratio between counts of each exon out of total count of exons. C.f. Ratio ratio between
(AML+ALL) and NM (negative values indicate down-regulation in the leukemias). Upregulation of Cancer is highlighted in light blue. The p-values were calculated by a
standard large-sample test procedure for evaluating the difference between proportions
obtained in two different populations [Dev91].
EXON
EXON
EXON
EXON
EXON
EXON
EXON
EXON
EXON
=
=
=
=
4.7. Conclusion
From this early analysis, we can begin to consider several exons as potentially
important transcriptional elements that may help to distinguish AML and ALL
from normal-based
on their statisticallysignificantup or down regulation. Exon
11 (V7) may perhaps be the most interesting of these since normal cells from
various sources (peripheral blood, bone marrow, and cord) were found not to
express itat an average resolution of on the order of 1.E+06.
However, looking only at the exons present or absent in transcripts does not
provide an accurate or comprehensive view of the actual transcripts that are
present since this data does not account for combinatorial diversity-we explore
that in the next chapter.
Varma. Chris
37 of 65
Ph.D. Thesis
Chapter 5: Isoform Expression Profiles
Note that in order to derive more robust and universally accepted results, we
compare the leukemias against a heterogeneous mix of normal samples
(containing independent samples of human peripheral blood purified B-cells,
human cord blood purified B-cells and, human adult whole bone marrow purified
cells) designated as NM.
5. 1. Introduction
We now apply quantitative methods to obtain insight when looking for significant
differences between isoform profiles of the various sample types - we are
specifically interested in statistically significant differences between AML and
normal, ALL and normal, and AML and ALL. Here we only consider the exons 6
- 14 and the exclusively splicing exons of the CD44 tail: exon 18 and exon 19.
We compare our results to published work when available - although it is quite
sparse in the case of isoform profiles and identification of particular isoforms.
Therefore, the results provided here firstly represent an unprecedented level of
quantitative detail into the exact nature of CD44's (or any protein's) alternative
exon splicing in AML, ALL, and normal cells of the immune system-thus,
many
of the isoforms presented here have previously never been identified.
5.2. Common Isoforms
Quantitative isoform profiling (as described in Chapter 1 and Chapter 2) was
used to determine the common isoforms in aggregated samples of AML, ALL,
and NM. Common isoforms are identified by the following procedure:
1) Aggregate the isoforms (and their associated occurrences) of all samples -including those of different sample types
2) Order the resulting isoforms from greatest occurrence to least
3) Select isoforms such that the number of occurrences (of the aggregated
alternatively spliced isoforms) is greater than 10 and that the last isoform
selected represents at least 1% of the number of alternatively spliced isoforms in
an average sample of the aggregated set.
In our case, we identify 17 common isoforms since the
18 th
candidate common
isoform has only 8 occurrences (data not shown), but the 1 7 th candidate common
isoform has 11 occurrences with an expression of 3.36% in the average sample
of the aggregated alternatively spliced isoforms. This simple process gives us
some degree of qualitative confidence that the isoforms are "real" and are very
Varma, Chris
38 of 65
Ph.D. Thesis
unlikely to be due to errors from polony gel creation, SSE's, or computational
profile construction.
Looking at Table 1 which shows the 17 common isoforms identified, AML clearly
expresses many more total isoforms as well as more total different isoforms (data
not shown) than either ALL or NM. Interestingly, ALL expresses fewer total
isoforms than NM - even when correcting for the difference in number of
samples used for aggregated values.
Desig.
Isoform Signature
AML
ALL
NM
#1
#2
#3
---------------13------19
------------------14---19
------------12---------19
------10---------------19
------------1213------19
------------12---14---19
-7---------------------19
---------------1314---19
-7------------13 ------19
---------11------------19
------------121314---19
6----------------------19
-----9-----------------19
------10------13------19
----------------------18----8-------------------19
---------111213------19
Other
Troe Totals
Theorized Totals
840
701
410
222
110
65
63
27
16
10
0
13
7
1
282
249
185
72
23
35
16
10
16
0
26
6
3
4
1
3
0
7
2542
126
122
137
81
14
13
29
8
9
26
8
11
19
19
11
2
0
15
650
2542
813
#4
#5
#6
#7
#8
#9
#10
#11
#12
#13
#14
#15
#16
#17
Other
9
11
11
26
938
938
Table 5-1. Counts of the 17 most common CD44 alternatively spliced isoforms in
aggregated samples of AML, ALL, and NM were obtained by quantitative isoform profiling
as described in Chapter 1 and Chapter 2. Isoform Signatures describe the known
expressed variant exons where '6' refers to '6b' and we include exons 18 and 19 which are
the mutually exclusive tails. Note that "Other" only contains other isoforms that are NOT
exon 5 cryptically spliced and are NOT exon 17-skipping - these are analyzed separately.
The counts for exon 5 cryptically spliced isoforms for each of AML, ALL, NM are 362, 440,
337 respectively.
The counts for exon 17-skipping isoforms for each of AML, ALL, NM are
300, 151, 203 respectively.
Note that adding these values back into the "True Totals"
results in the values as presented in Table 3-2. Also, notice that whereas AML and NM are
a composite of 5 samples, ALL is only a composite of 4 samples. Thus, we provide a
theorized total for ALL which assumes that a fifth sample would express a number of
isoforms that can be approximated as an average of the reported 4 samples, or 163
isoforms.
There are several orders of magnitude difference in the expression of various
alternatively spliced isoforms of CD44 (see Figure 1). Also, the fall-off between
the most prevalent and less prevalent isoforms is steep. AML encompasses the
Varma, Chris
39 of 65
Ph.D. Thesis
most expansive range between most common alternatively spliced isoform and
most rare alternatively spliced isoform.
Isoform Counts of Samples Aggregated by Designation
900
800
700
.AML
(J)
......
c:
:J
a
600
.ALL
()
ONM
E 500
Sa
(J)
-
400
300
200
100
o
#1
#2
#3
#4
#5
#6
#7
#8
#9
#10 #11 #12 #13 #14 #15 #16 #17 Other
Alternatively Spliced Isoform
Figure 5-1. Counts of the 17 most common alternatively spliced CD44 isoforms in
aggregated samples of AML, ALL, and NM were obtained by quantitative isoform profiling
as described in Chapter 1 and Chapter 2.
5.3. Quantitative Comparison of AML to Normal
Aggregated alternative splicing isoforms of AML samples were compared to
those of NM samples (see Table 2). 4 of the 17 most common isoforms were
found to be up-regulated in AML with statistical significance (from p < .05 to p <
.005). 5 of the 17 isoforms were found to be down-regulated in AML with
statistical significance (from p < .03 to p «.05).
AML was found to express 2
common isoforms that are not present in NM samples: #10 (p < .03) and #17 (p <
.02). Both of these isoforms contain exon 11 (V7) which was found not to be
expressed in NM samples (see Table 3, Chapter 3). Exon 11 (V7) enables direct
binding to chondroitin sulfate, heparin, and heparin sulfate in addition to HA
[Sle97]. Interestingly, NM was found to significantly express one common
Varma, Chris
40 of 65
Ph.D. Thesis
isoform that was not expressed by AML: #11 (p « .05). This isoform contains all
three of the most prevalent exons-12 (VB), 13 (V9), and 14 (V10). However,
upon further investigation of less common alternative splicing isoforms (data not
shown), we identified three isoforms in the aggregated AML samples that
expressed these exons (12,13,14) but always along with exon 11 (V7).
Desig.
Isoform Signature
AML
AML
Ratio
0.3304
0.2758
0.1613
0.0873
0.0433
0.0256
0.0248
0.0106
NM
NM
Ratio
0.3006
0.2655
0.1972
0.0768
0.0245
0.0373
0.0171
0.0107
0.0171
0.ססoo
C.f.
Ratio
1.10
1.04
-1.22
1.14
1.76
-1.46
1.45
-1.00
-2.71
P-value
---------------13------19
5.E-02
840
282
------------------14---19
3.E-01
701
249
------------12---------19
410
6.E-03
185
------1 0---------------19
2.E-01
222
72
------------1213------19
5.E-03
110
23
------------12---14---19
3.E-02
65
35
-7---------------------19
9.E-02
63
16
---------------1314---19
27
5.E-01
10
-7------------13 ------19
#9
0.0063 16
2.E-03
16
---------11------------19
#10
N1A
3.E-02
10
0.0039
0
------------1213 14---19
#11
«5.E-02
0.02n 0.00
0
0.ססoo
26
6----------------------19
0.0051
0.0064
3.E-01
#12
13
6
-1.25
-----9-----------------19
0.0028
#13
7
0.0032
-1.16
4.E-01
3
------10------13 ------19
#14
1
0.0004
4
0.0043
-10.84
4.e-03
----------------------18-9
0.0035
1
0.0011
1.E-01
3.32
#15
---8-------------------19
11
0.0043
#16
3
0.0032
3.E-01
1.35
---------111213------19
#17
11
0.0043
2.e-02
0
0.0000
N/A
Other
26
7
Other
2542
Totals
938
Table 5-2. Quantitation of the 17 most common CD44 alternatively spliced isoforms in
aggregated samples of AML and NM were obtained by quantitative isoform profiling as
described in Chapter 1 and Chapter 2. Isoform Signatures describe the known expressed
variant exons where '6' refers to '6b' and we include exons 18 and 19 which are the
mutually exclusive tails. X Ratio (where X = sample type) = ratio between counts of each
exon out of total count of exons. C.f. Ratio = ratio between AML and NM (negative values
indicate down-regulation
in AML). Statistically significant up-regulation of AML is
highlighted in light blue and down-regulation
is high-lighted
in light green. We report the
following novel isoforms for AML (highlighted in yellow): #1 - #3, #5 - #10, and #12 - #17.
The p-values were calculated by a standard large-sample test procedure for evaluating the
difference between proportions obtained in two different populations [Dev91].
#1
#2
#3
#4
#5
#6
#7
#8
Taking into account the less common alternative splicing isoforms of AML (data
not shown), we can account for almost all of the isoforms found in previous work
[Leg9B, Aks02, BenOO,Kha96]. However, we have also identified previously
unreported and in fact unexpected isoforms. We report the following novel
isoforms for AML: #1 - #3, #5 - #10, and #12 - #17.
Through a comprehensive RT-PCR-based analysis of CD44 transcripts
expressed in 70 AML patient samples, [Leg98] concluded that exon 13 (V9) was
always associated with exon 12 (VB). In contrast, we have found four common
isoforms where this is not the cas~ne
that is quite prevalent (33%): #1, and 3
Varma, Chris
41 of 65
Ph.D. Thesis
that are not: #8, #9, #14. [Leg98] also reported that the following exons tend to
be found in the same isoform together: 12(V8), 13(V9), and 14(V10). However,
we found no instance of this to an average resolution of 1.E.+6 molecules (CD44
mRNA transcripts) - even though isoform #11 is present in NM. In addition,
[Leg98] reported identifying isoforms with the following exons: 10 (V6) and 11
(V7), 10 (V6) and 12 (V8) to 14 (V10), and 10 (V6) to 14 (V10) - however, we
found no instance of these isoforms. We concur with [Leg98] in finding that
isoforms with exon 10 (V6) exist in directly spliced versions - see isoform #4.
There is controversy as to the existence of isoforms containing exons 8(V4) and
exons 9(V5) in AML. [Leg98] reported that exons 8 (V4) and 9 (v5), although
rare, are expressed. In contrast, after performing a RT-PCR-based analysis (and
Southern blotting) of 24 AML patient samples, [BenOO] reported not finding these
variant exons expressed. Here we concur with the results of [Leg98] - see
isoforms #16 and #13.
As another example, [BenOO]found that exon 7 (V3) was not detected in
combination with any other variant exon. However, we found that isoform #9
expresses exon 7 (V3) as well as exon 13 (V9).
5.4. Quantitative
Comparison
of AML to ALL
Aggregated alternative splicing isoforms of AML samples were compared to
those of ALL samples (see Table 3). 4 of the 17 common isoforms were found to
be up-regulated in AML with statistical significance (from p < .05 to P < .005). 10
of the 17 isoforms were found to be down-regulated in AML (or up-regulated in
ALL) with statistical significance (from p < .03 to p « .05).
Desig.
Isoform Signature
#1
---------------13------19
------------------14---19
------------12---------19
------10---------------19
------------1213 ------19
------------12---14---19
-7--------------------19
--------------1314---19
-7------------13------19
---------11----------19
------------121314---19
6----------------------19
-----9-----------------19
------10------13------19
#2
#3
#4
#5
#6
#7
#8
#9
#10
#11
#12
#13
#14
#15
AML
P-value
----------------------18--
Varma, Chris
42 of 65
Ph.D. Thesis
11
0.0043
2
0.0031
1.41
3.E-01
11
0.0043
0.0000
5.E-02
N/A
26
15
Totals
2542
650
Table 5-3. Quantitation of the 17 most common CD44 alternatively spliced isoforms in
aggregated samples of AML and ALL were obtained by quantitative isoform profiling as
described in Chapter 1 and Chapter 2. Isoform Signatures describe the known expressed
variant exons where '6' refers to '6b' and we include exons 18 and 19 which are the
mutually exclusive tails. X Ratio (where X = sample type) = ratio between counts of each
exon out of total count of exons. C.f. Ratio = ratio between AML and ALL (negative values
indicate up-regulation of ALL). Statistically significant up-regulation
of AML is highlighted
in light blue and down-regulation
is highlighted in light green. The p-values were
calculated by a standard large-sample test procedure for evaluating the difference
between proportions obtained in two different populations [Dev91].
#16
#17
Other
---8-------------------19
---------111213 ------19
a
AML was found to express one common isoform that was not present in
(common or less common) NM samples: #17 (p < .05). This isoforms contains
exon 11 (V7) as well as exons 12 (VB) and 13 (V9). ALL was found to express
one common isoform that was not expressed by AML: #11 (p «.05).
This
isoform contains all three of the most prevalent exons-12 (VB), 13 (V9), and 14
(V10). Upon further investigation of less common alternative splicing isoforms
(data not shown), we identified three isoforms in the aggregated AML samples
that expressed these exons (12,13,14) but always along with exon 11 (V7).
5.5. Quantitative
Comparison
of ALL to Normal
Aggregated alternative splicing isoforms of ALL samples were compared to those
of NM samples (see Table 4). 7 of the 17 most common isoforms were found to
be up-regulated in ALL with statistical significance (from p < .02 to p «.05).
4 of
the 17 isoforms were found to be down-regulated in ALL with statistical
significance (from p < .02 to P < .0000009). ALL was found to express one
common isoform that was not present in NM samples: #10 (p« .05). This
isoform contains exon 11 (V7) which was found not to be expressed in NM
samples (see Table 3.3). Exon 11(V7) enables direct binding to chondroitin
sulfate, heparin, and heparin sulfate in addition to HA [Sle97]. NM was not found
to express any common isoform that was not expressed by ALL.
Desig.
Isoform Signature
ALL
ALL
Ratio
#1
#2
#3
#4
#5
#6
#7
#8
#9
---------------13------19
------------------14---19
------------12---------19
------10---------------19
------------1213------19
------------12---14---19
-7---------------------19
---------------13 14---19
-7------------13------19
126 ..
1).'1IA.' 1",28.2
!2i19"
Varma, Chris
NM
12!"/'j 1(:.Gjf$~
137
81
14
13
29
8
9
0.2108
0.1246
0.0215
:::'
9:GIiI)
0.04!46
0.0123
0.0138
43 of 65
185
72
23
3S
1'~..
16
10
16
NM
Ratio
0.3006
0:.2655
0.1972
0.0768
0.0245
0.0313
0.0171
0.0107
0.0171
C.f.
P-value
Ratio
..(;.1.55 ,,,9~E-Dl
-'1.4'1, 2.1£-04
1.07 3.E-01
1.62 7.E-04
-1.14 3.E-01
-1.87 2.E-Q2
2.62 6.!-04
1.15 4.E-01
-1.23 3.E-01
Ph.D. Thesis
#10
#11
#12
#13
#14
#15
#16
#17
Other
---------11------------19
------------121314---19
6----------------------19
-----9-----------------19
------10------13------19
----------------------18----8-------------------19
---------111213------19
26
8
11
19
19
11
2
a
0.0400
0.0123
0.0169
0.0292
0.0292
0.0169
0.0031
0.0000
0.0000
0.0277
0.0064
0.0032
0.0043
0.0011
0.0032
0.0000
0
26
6
3
4
1
3
a
N/A
-2.25
2.65
9.14
6.85
15.87
-1.04
N/A
«5E-02
2.E-02
2.E-02
6.E-06
2.E-05
2.E-04
5.E-01
N/A
15
7
Totals
650
938
Table 5-4. Quantitation of the 17 most common CD44 alternatively spliced isoforms in
aggregated samples of ALL and NM were obtained by quantitative isoform profiling as
described in Chapter 1 and Chapter 2. Isoform Signatures describe the known expressed
variant exons where '6' refers to '6b' and we include exons 18 and 19 which are the
mutually exclusive tails. X Ratio (where X sample type)
ratio between counts of each
exon out of total count of exons. C.f. Ratio = ratio between ALL and NM (negative values
indicate down-regulation
in ALL). Statistically significant up-regulation of ALL is
highlighted in light blue and down-regulation
is high-lighted
in light green. We report the
following novel isoforms for ALL (highlighted in yellow): #1 - #3, #5 - #10, and #12 - #17.
The p-values were calculated by a standard large-sample test procedure for evaluating the
difference between proportions obtained in two different populations [Dev91].
=
=
Taking into account the less common alternative splicing isoforms of ALL (data
not shown), we can account for all of the isoforms found in previous work
[Ben04]. However, we have also identified previous unreported and in fact
unexpected isoforms. For example, as previously discussed in Chapter 3, we
have identified exon 8 (V4), exon 9 (V5), and exon 11 (V7) that were previously
not found in ALL. The exact isoforms that include them are as follows: #16, #13,
and #10 respectively. We report the following novel isoforms for ALL: #1 - #3, #5
- #10, and #12 - #17.
5.6. Quantitative
Comparison
of Cancer to Normal
Alternative splicing isoforms of ALL and of AML samples were aggregated and
designated as 'Cancer.' They were then compared to aggregated isoforms of
NM samples (see Table 5). 15 of the 17 most common isoforms were found to
be up-regulated in Cancer with statistical significance (p «.05).
1 of the 17
isoforms was found to be down-regulated in Cancer (or up-regulated in NM) with
statistical significance (p < .000002). Cancer was found to express 2 common
isoforms that are not present in NM samples: #10 (p « .05) and #17 (p « .05).
Both of these isoforms contain exon 11 (V7) which was found not to be
expressed in NM samples (see Table 3, Chapter 3). NM was not found to
express any common isoform that was not expressed by Cancer.
Varma, Chris
44 of 65
Ph.D. Thesis
Desig.
#1
#2
#3
Isoform
Cancer
NM
NM
Ratio
0.3006
0.2655
0.1972
0.0768
0.0245
0.0373
0.0171
0.0107
0.0171
0.0000
1"'0.0217
0.0064
0.0032
0.0043
0.0011
0.0032
0.0000
C.f.
Ratio
1.01
4.77
4.27
6.07
1.78
3.22
8.30
5.05
2.25
P-value
282
5.E-01
«5E-02
249
185
«5E-02
#4
«5E-02
72
#5
23
«5E-02
#6
«5E-02
35
#7
16
«5E-02
#8
«5E-02
10
#9
16
«5E-02
#10
«5E-02
36
0
N/A
#11
8
-2.25
2.E-06
O:0~Z3 26
#12
«5E-02
24
0.0369
6
5.n
#13
26
0.0400
12.51
«5E-02
3
#14
«5E-02
20
0.0308
4
7.22
#15
20
1
0.0308
28.86
«5E-02
#16
0.0200
«5E-02
13
3
6.25
#17
11
0.0169
0
N/A
«5E-02
Other
41
7
Totals
3192
938
Table 5-5. Quantitation of the 17 most common
CD44 alternatively spliced isoforms in
aggregated samples of (ALL + AML) versus aggregated samples of NM were obtained by
quantitative isoform profilingas described in Chapter 1 and Chapter 2. We define Cancer
ALL + AML. Isoform Signatures describe the known expressed variant exons where '6'
refers to '6b' and we include exons 18 and 19 which are the mutually exclusive tails.X
Ratio (where X = sample type) = ratiobetween counts of each exon out of total count of
exons. C.f.Ratio ratio between Cancer and NM (negative values indicate up-regulation
of NM). Statisticallysignificant up-regulation of Cancer is highlighted in light blue and upregulation of NM is high-lighted in lightgreen. We report the following novel isoforms for
NM (highlighted in yellow): #1 -#3, #5 - #9, #12 - #14, and #16. The p-values were
calculated by a standard large-sample test procedure for evaluating the difference
between proportions obtained in two different populations [Dev91].
---------------13 ------19
------------------14---19
------------12---------19
------10---------------19
------------1213------19
-----------12---14---19
-7 ---------------------19
--------------1314---19
-7 ------------13 ------19
---------11------------19
------------121314---19
6----------------------19
-----9-----------------19
------10------13------19
----------------------18----8-------------------19
---------111213------19
966
823
547
303
124
78
92
35
25
C.
Ratio
0.3026
1.2662
0.8415
0.4662
0.1908
0.1200
0.1415
0.0538
0.0385
0.0554
,,,
=
=
Regarding NM samples, we have identified(see Chapter 3.4) both exon 8 (V4)
and exon 9 (V5) in NM samples (one of which is human adult whole bone
marrow purified cellsand the other is human peripheral blood purified B-cells)
that were previously not found in normal bone marrow cells or in normal
peripheral blood cells [BenDD]. The isoform signatures that include them are #16
and #13, respectively. Here we report the following novel isoforms for NM: #1 #3, #5 - #9, #12 - #14, and #16.
The results of comparing Cancer to NM are particularlyinteresting in context of
the results of quantitative exon profilingin Chapter 3.4. We saw that in Cancer
no exons except for exon 11 (V7) were up-regulated with statistical
significance-and exon 11 (V7) was up-regulated (with statisticalsignificance)
only because itwas not found to be expressed in NM. In stark contrast, almost
allof the common isoforms are up-regulated (with statisticalsignificance)
compared to NM. Therefore, itis clear that looking only at variant exon inclusion
and exclusion is not enough-analysis
of the combinatorics of the exons in
transcripts is crucial.
Varma, Chris
45 of 65
Ph. D. Thesis
5.7. Conclusion
In this chapter we have provided the first comprehensive quantitative analysis of
isoforms performed on human cells, or almost any organism or cell type. The
developed methods of quantitative isoform profiling have made this possible.
We have found many statistically significant up-regulations and down-regulations
of particular isoforms as well as several cases, even among the common
isoforms, where isoforms were not present in a particular sample type-usually
NM. We have also reported the specific novel isoforms for each samples type.
Comparison of uncommon isoforms (i.e. by definition rare isoforms that may only
be expressed in a few samples) were not analyzed here. In the next chapter we
use the quantitative results obtained here to generate statistically significant
findings of isoforms that may serve as potential diagnostic markers or may be
considered for further analysis for therapeutic targets.
Varma, Chris
46 of 65
Ph.D. Thesis
Chapter 6: In Pursuit of Exclusive Converging-lsoforms
Note that in order to derive more robust and universally accepted results, we
compare the leukemias against a heterogeneous mix of normal samples
(containing independent samples of human peripheral blood purified B-cells,
human cord blood purified B-cells and, human adult whole bone marrow purified
cells) designated as NM.
6.1. Introduction
In this chapter we strive to develop a new paradigm that will enable the analysis
of isoforms (that we have identified in Chapter 5) in order to qualify them for
further evaluation as potential diagnostic markers, therapeutic targets, and
perhaps even therapeutics themselves.
6.2. Introducing Isoform Convergence
In order to robustly compare isoforms of AML, ALL, and NM-especially for a
small number of total samples-we will need to develop a unified framework that
allows us to capture the qualities that will most effectively and flexibly
differentiate between different sample types through the comparison of isoforms.
Those qualities include the presence of a particular isoform that is consistently
expressed in almost all of the samples of a sample type - but with the flexibility to
consider less consistently expressed isoforms, the consideration of and
prioritization of rare isoforms as a function of complex splicing, and the ability to
identify those isoforms that are present in one sample type but not in another
with a certain degree of confidence.
We begin by introducing the concept of Isoform Convergence. We first assume
that for each member of a population, it's unique isoforms (i.e. the different types
of isoform signatures expressed) have been characterized. In addition, we
require that each unique isoform of a member has an associated number of
occurrences--that is how many times that unique isoform was detected in the
member. This also enables us to then determine the total number of alternatively
spliced isoforms expressed in each member which is the sum of the unique
isoforms occurrences. Then, the Convergence Value of a population is
determined as follows. Members of a population are ordered according to the
following operator: a member with a larger number of unique isoforms is listed
before a member with a smaller number of unique isoforms. If two members
have equal number of unique isoforms, then we order them based on the total
number of alternatively spliced isoforms. If two members have equal numbers of
total alternatively spliced isoforms then we choose one based on coin toss. We
Varma, Chris
47 of 65
Ph.D. Thesis
then intersect the set of unique isoforms of each member in order starting by
intersecting the first member in the ordered list by itself (for appropriate indexing
of intersections). After each intersection, each remaining unique isoform is
associated with the least occurrence value (i.e. min(occurrence of member 1,
occurrence of member 2)). We continue to intersect members until we have
intersected at least 3/4 of the members and the number of unique isoforms
remaining Converges - that is, until an intersection (n) and its subsequent
intersection (n + 1) result in the same number of remaining unique isoformsadditionally, intersection n is now identified as the Converging Intersection and
the members of the population intersected in reaching the Converging
Intersection are known as the Converging Members. This list of unique isoforms
(including their occurrences) is assigned a Convergence Value of 1. Assume
that k intersections were required to reach a Convergence Value of 1, then a list
of unique isoforms selected from a previous intersection (k - 1) has a
Convergence Value that is equivalent to the ratio k -1 / k. Thus, each isoform of
a set of unique isoforms resulting from an intersection can be assigned a
Convergence Value. We then further define a unique isoform that has a
Convergence Value (derived from the process of Isoform Convergence) of
greater than 2/3 ( - 0.667) as a Converging Isoform of its population. A unique
isoform that has a Convergence Value of 1 is designated as Maximally
Convergent. Furthermore, we rank isoforms from most converging to least
converging, we prioritize first by Convergence Value and second by number of
occurrences.
Lastly, we introduce the following additional concepts as part of our unified
framework: Exclusive and Maximally Exclusive as applied to alternatively spliced
isoforms. A Converging Isoform, i of a population, m is said to be Exclusive
against another population, n if it does not occur in n's set of Converging
Isoforms. More stringently (assuming the same comparison population n), a
Converging Isoform, i of a population, m is said to be Maximally Exclusive
against another population, n if it does not occur in any member of n.
The presented unified framework is fundamentally based on the mathematical
operators of intersection, difference, and union - it applies these operators
through a flexible algorithm that enables rational identification of isoforms that are
consistently exclusive to a sample type. Thus, Converging (or Maximally
Converging) Isoforms that are Exclusive (or Maximally Exclusive) have the
following properties: 1) they occur in a majority of the samples of a sample type with the flexibility to range from a minor majority to a significant majority and
assigning a level of confidence based on preponderance of occurrence, 2) they
can be rare because the unified platform prioritizes the evaluation of samples
with more complex splicing, 3) they can be used to distinguish between different
sample types with a certain degree of consistency (without requiring absolute
consistency). Furthermore, the processes of Isoform Convergence guarantees
that an isoform present after an intersection was also present in all prior
intersections.
Varma, Chris
48 of 65
Ph.D. Thesis
We will now apply these concepts to our samples of AML, ALL, and NM.
6.3. Finding Convergence
We first apply the concept of Isoform Convergence to each of AML, ALL, and NM
(see Figure 1~ The Converging Intersection for each of AML, ALL, and NM is
th
rd
4 , 3 , and 4 ,respectively. Between the first intersection and the Converging
Intersection we lose 87.50/0 of unique isoforms for AML, 61.1 % of unique
isoforms for ALL, and 76.5% of unique isoforms for NM.
Convergence of Isoforms Upon Series of Intersections
30
25
en
E
.E
0
~AML
-
20
..0
15
~
-ALL
NM
0
~
Q)
E
:]
Z
10
5
•
o
1st
2nd
3rd
4th
5th
Number of Intersections
Figure 6-1. The process of Isoform Convergence is used to analyze each set of samples,
AML, ALL, and HM. Each set of samples has been found to reach Convergence.
Varma. Chris
49 of 65
Ph.D. Thesis
6.4. Converging-Isoforms
We determine the set of Converging Isoforms for each of AML, ALL, and NM by
applying the process of Isoform Convergence. Here we use all isoforms found
for each sample type - not just the common isoforms as used previously.
In AML, we have identified 7 Converging Isoforms (see Table 1). 3 of the 7
isoforms are Maximally Convergent.
Counts of Converging Isoforms for each pair
(intersections,Convergence Value)
Conver~dn2 Isoform (AML)
3,0.75
-7------------13 ------19
-7--------------------19
------10--------------19
--------11-----------19
-----------12---------19
--------------13------19
-----------------14---19
Totals
2
5, 1
0
0
9
0
0
57
19
85
4,1
0
0
10
0
0
77
115
202
10
10
1
49
77
115
264
Table 6-1. Converging Isoforms for AML as determined by the process of Isoform
Convergence on samples of AML. Convergence was reached after 4 intersections.
In ALL, we have also identified 7 Converging Isoforms (see Table 2). All 7 of 7
isoforms are Maximally Convergent. Note that since we only had 4 samples of
ALL, it does not make sense to report values for 2,1 since any new isoforms
identified would not be Converging Isoforms. This is because a Converging
Isoform must have a Convergence Value of greater than 2/3.
Counts of Converging Isoforms for each
pair (intersections,Convergence Value)
Converein2 Isoform (ALL)
6---------------------19
-7------------13 ------19
-7---------------------19
------10-------------19
-----------12---------19
--------------13------19
----------------14---19
Totals
3, 1
1
1
6
6
8
11
4, 1
1
1
1
3
5
5
8
24
32
65
Table 6-2. Converging Isoforms for ALL as determined by the process of Isoform
Convergence on samples of ALL. Convergence was reached after3 intersections. Note
that the values of 2,1 were not reported as these do not meet the definition of a
Converging Isoform and thus would not be valid.
Varma, Chris
50 of 65
Ph.D. Thesis
In NM, we have identified 6 Converging Isoforms (see Table 3). 5 of the 7
isoforms are Maximally Convergent.
Counts of Converging Isoforms for each pair
(intersections,Convergence
Value)
Conver2in2 Isoform (NM)
3,0.75
4,1
5, 1
-7------------13------19
------10-------------19
---------12---------19
-----------12---14---19
------------13------19
----------------14---19
1
8
19
0
8
18
0
26
0
8
2
0
11
24
24
7
39
30
Totals
104
76
45
Table 6-3. Converging Isoforms for NM as determined by the process of Isoform
Convergence on samples of NM. Convergence was reached after 4 intersections.
6.5. Exclusive
Converging-Isoforms
Here we determine and identify the Exclusive and Maximally Exclusive
alternatively-spliced, unique isoforms from each sample type's set of Converging
Isoforms (see Table 4) by comparing different sample types. Each type of
sample is found to have at least one Exclusive Converging Isoform. Both AML
and ALL have two Exclusive Converging Isoforms and for AML one of these is
Maximally Exclusive. Both of ALL's Exclusive Converging Isoforms are
Maximally Converging (see previous section).
Counts of Exclusive Isoforms for each sample type and pair
(intersections,Convergence
Value)
AML 3, 0.75
ALL 3,1
AML
N/A
None
NM 3,0.75
----------12---14---19
3,0.75
ALL
None
N/A
-----------12---14---19
3,1
NM3,
-7 ---------------------19
6----------------------19
N/A
0.75
Count: 10
Count: 1
NM4,1
NM
UNION
Count: 7
Count: 7
---------11-----------19
-7 ---------------------19
Count: 1
Count: 6
-7 ---------------------19
6----------------------19
Count: 10
Count: 1
---------11-----------19
-7 ---------------------19
Count: 1
Count: 6
---------11-----------19
None
N/A
N/A
Count: 1
Table 6-4. Exclusive Converging Isoforms and Maximally Converging Isoforms (ifany) of
each sample type determined based on the process of Isoform Convergence and subject
Varma, Chris
51 of 65
Ph.D. Thesis
to the concepts of Exlusive and Maximally Exclusive. Count indicates least occurrence of
the converging isoform in the set of Converging Members.
6.6. Identification
of Possible Candidate Targets
Finally, we compare our results of the identification of the Exclusive Converging
Isoforms with our previous results for isoform profiles (Chapter 5) and for exon
profiles (Chapter 4) to determine which of our Exclusive Converging Isoforms are
also up-regulated with statistical significance (see Table 5).
Exon Profile
Isoform Profile
Exclusive
Converging
Isoform
Occurr
-ence
Ratio
Up regulated
vs. NM
Pvalue
Occurr
-ence
Ratio
Up regulated
vs. NM
Pvalue
AML#l
.09
2.9%
No
N/A
-7---------------------19 2.5%
Yes
Yes
.03
Yes
.0002
0.4%
AML#2
1.1%
-------11-----------19
.02
.02
1.5%
Yes
ALL#l
6----------------------19 2.7%
Yes
Yes
.0006
Yes
.005
ALL#2
5.8%
-7--------------------19 4.5%
<.05
NM#l
-----------12---14---19 3.7%
Yes*
Table 6-5. Isoforms for both AML and ALL that were identified as Exclusive Converging
Isoforms were evaluated for statistically significant up-regulation Vs. NM (from Chapter 4).
In addition, the single variant exon present in each Exclusive Converging Isoform was
evaluated for statistically significant up-regulation vs. NM (from Chapter 3). *NM's
Exclusive Converging Isoform was compared to both AML and ALL separately and its upregulation was found to be statistically significant. The Occurrence Ratio refers to the ratio
of the occurrences of the Exclusive Converging Isoforms (or exons) to all alternativesplicing isoforms (or variant exons) identified for each sample type.
Therefore, we propose the following Exclusive Converging Isoforms as potential
candidates for further study as targets for diagnostic probing or therapeutic
intervention since the included variant exons are expressed on the extracellular
proximal domain of CD44 which is involved in ligand binding: AML#2, ALL#1, and
ALL#2. AML#2 may be particularly relevant as exon 11 has not been found to be
present in normal cells in our studies or in others [BenOO, Leg98, Ben04, Kha96].
Furthermore, inclusion of exon 11 (V7) enables direct binding to chondroitin
sulfate, heparin, and heparin sulfate in addition to HA [Sle97] which could confer
additional functionality that may be exploited in malignancy.
6.7. Conclusion
In this Chapter we proposed a process for Isoform Convergence as a method to
select robustly expressing isoforms from a population of all unique isoforms
expressed by a particular sample type. The method of Isoform Convergence first
Varma, Chris
52 of 65
Ph.D. Thesis
requires that Convergence be obtained with a population of samples. This allows
for the identification of Converging Isoforms. The processes of Isoform
Convergence guarantees that an isoform present after a particular intersection
was also present in all prior intersections. Finally, we determine exclusivity of a
Converging Isoform by comparing it against the Converging Isoforms of other
relevant populations.
Furthermore, we propose three Converging Isoforms (1 of AML, and 2 of ALL)
that are Exclusive and are up-regulated with statistical significance (compared to
NM). These isoforms may serve as potential diagnostic probes or therapeutic
targets, though significant studies need to be completed.
Varma, Chris
53 of 65
Ph.D. Thesis
Conclusion
We have presented
a new paradigm
by which to quantitatively
study the
alternative splicing of any molecule in any clinical sample through Polony
Technology [Mit99] and our methods of quantitative exon profiling and
quantitative isoform profiling. Furthermore, we extended this paradigm to include
Isoform Convergence-a process by which we can potentially qualify particular
isoforms as candidate diagnostic markers, potential therapeutic targets, and
perhaps even as precursor therapeutics themselves. We applied this paradigm
to quantitatively investigate the alternative splicing of CD44 in two leukemias acute myeloid leukemia and acute lymphocytic leukemia. To address some of the
controversy in the CD44 leukemia literature, we suggested several corrections to
previously made claims about the presence of specific CD44 exons and of
specific CD44 isoforms.
Furthermore, we provided not only the first
comprehensive characterization of CD44's (or any molecules) alternative exon
splicing in human cells, but also the resulting quantities of the exons and of the
exact isoforms present to a resolution of 1.E.+06 molecules (of CD44). Through
this process, we identify a plethora of novel isoforms of CD44 expressed in acute
myeloid leukemia and in acute lymphoblastic leukemia. Finally, we identify
specific isoforms in each leukemia that may serve as candidate markers or
possibly as therapeutic targets.
In future work, we hope to establish our paradigm as a new and rational method
for the identification of therapeutic targets that result in the generation of
successful therapies. In order to do this, we need to identify Exclusive
Converging Isoforms that are able to demonstrate therapeutic intervention
through intracellular techniques such as antisense oligonucleotides and smallinterfering RNAs (siRNAs) or through extracellular techniques such as
monoclonal antibodies-for example, chemoimmunotherapy. Intracellular
techniques would attempt to knock-down defective alternative splicing and
restore normal splicing patterns in targeted cells. Extracellular techniques would
target the Exclusive Converging Isoforms expressed on the targeted cell's
surface in order to induce apoptosis, to reduce cell viability, or to induce an
inflammatory response against the cell.
Varma, Chris
54 of 65
Ph.D. Thesis
Supplementary Results
S.1. TM-Exon Skipping
Note: A portion of the results of this section after our initial observation of TMExon Skipping, not including the quantitative comparisons, were completed in
collaboration with Jun Zhu, formerly a post-doc in the laboratory of George
Church at Harvard Medical School and now an Assistant Professor at Duke
Univeristy.
Initial Observation, Identification
& Background
During our initial studies of isoform profiling, we found that transmembrane exon
of CD44 (i.e. exon 17) was spliced out in some isoforms when compared to the
constant exon 5 (see Figure 1).
Cy3-dA
CyS-dU
Merge
Exon-17
Exon-5
Merge
Figure 1. Identification of TM-skipping in human CD44 on polony slides. Arrows point to
the TM-skipped isoforms in the merged image. Note also that the merged image shows
cases were the constant exon 5 is not present - this is due to cryptic splicing as
previously identified.
In order to determine if the identified TM-skipping polonies expressed a true
alternatively-spliced isoform, a polony with such expression was cut out of the
polony gel and sequenced (this was also done by gel electrophoresis for further
confirmation) (see Figure 2). The results of the shown sequence (Figure 2)
clearly demonstrate that exon 17 has been appropriately alternatively-spliced out
due to the absence of the entire TM exon. Furthermore, the flanking sequence
provides a U1 binding 5'ss signal (GGT) that is consistent with 80% of exonskipping [Ast04].
Varma, Chris
55 of 65
Ph.D. Thesis
. -R~<b ':\~~,o~
~~
~~
~"
~~
"'-v
~,,\A ~ ~,,\A.'"
Exon 16
I
AC C C C AA A T T C C A G G T G T G G G C A G A A G
,....,
..., ,..,
rnfl
1\
,.
~
I
,
~
~ij
,
Vv\.A
Figure 2. Confirmation
J'..
.AA
of TM-exon skipping and subsequent
.\
A
sequencing.
It is theorized that TM-skipping would cause the expressed TM- CD44 protein to
be secreted. CD44 is known to be present in soluble form in quantities of ug/ml
in human blood, however this was considered to be due to the process of CD44
shedding (see Figure 3) - caused by proteolytic cleavage of a post-translated
CD44 protein on its extracellular domain. This form of soluble CD44 is then
regulated at the post-translational level and does not express the tail region. In
stark contrast, our identified soluble CD44 is regulated at the pre-mRNA level
and does express the tail region - this may confer special properties upon this
form of soluble CD44 as well as represent a different highly regulated function.
NH2
NH2
>-- Anti-CD44ecto
CD44 full length
Ab
CD44 ectodomain
proteolytic cleavage
Extracellular
.. Alternative
splicing site
Soluble CD44
Alternative
splicing site
Membrane
Intracellular
>-- Anti-CD44cyto
Ab
CD44 cleavage
product
COOH
>-- Anti-CD44cyto
Ab
COOH
Figure 3. CD44 shedding occurs via proteolytic
which confers soluble CD44.
cleavage of a portion of the TM exon
In mice, soluble CD44 has been associated with several functions [Yu96].
Soluble CD44 has been shown to block endogenous CD44 from binding and
internalizing its primary ligand, HA - acting as a decoy. Soluble CD44 has been
shown to inhibit TA3 cell invasion of HA-producing cell monolayer and has been
shown to inhibit tumor formation when intravenously injected into mice.
Varma, Chris
56 of 65
Ph.D. Thesis
Furthermore, soluble CD44 has been found to induce apoptosis of invading
tumor cells.
Isoform Expression Profiles
Alternative-exon
splicing of exon 17 (TM-exon) was performed via quantitative
isoform profiling as described previously (Chapter 1 and 2). Counts of each
identified TM- isoform were obtained for AML vs. NM, AML vs. ALL, ALL vs. NM,
and Cancer (AML and ALL aggregated) vs. NM (see Tables 1 to 4). Ratios of
each type were obtained over total CD44 counts as provided in Table 2 of
Chapter 3. All samples expressed TM- CD44.
Isoform
AML
AML Ratio
NM
0.001137
0.000023
0.000023
333
0
1
NM
C.f.
Ratio
Ratio
0.005550
0.000000
0.000017
-4.88
N/A
1.40
<<5E-02
1.E-01
4.E-01
4.E-02
2.E-01
---------------------- ( ---- 19
--------------- 13--- ( )----19
------------------ 14( )----19
341
7
7
-------12---( )---- 19
------10------------ -----19
2
4
0.000007
0.000013
2
0
0.000033
0.000000
-5.00
N/A
--7-----------------( )----19
--------------- 1314( )----19
-----------1213---()
---- 19
0
1
0
0.000000
0.000003
0.000000
0
0
1
0.000000
0.000000
0.000017
N/A
N/A
N/A
P-value
N/A
3.E-01
1.E-02
Totals
300000
60000
Table 1. Quantitation of the 8 most common CD44 TM skipped isoforms in aggregated
samples of AML and NM were obtained by quantitative isoform profiling as described in
Chapter 1 and Chapter 2. AML Ratio and NM Ratio are calculated over the total number of
respective CD44 molecules as reported in Table 2 of Chapter 3. C.f. Ratio = ratio between
AML and NM (negative values indicate down-regulation in AML). Statistically significant
up-regulation of AML is highlighted in light blue and down-regulation is high-lighted in
light green. The p-values were calculated by a standard large-sample test procedure for
evaluating the difference between proportions obtained in two different populations
[Dev91].
Isoform
AML
AML Ratio
0.001137
436
ALL
Ratio
0.004360
C.f.
Ratio
-3.84
P-value
------------------------
19
---------------..
--()
19
7
0.000023
1
0.000010
2.33
2.E-01
--------------1.4( )----19
12---- ---- 19
---------------- 10------------- -- -19
7
2
4
0.000023
0.000007
0.000013
0
1
0
0.000000
0.000010
0.000000
N/A
-1.50
N/A
6.E-02
4.E-01
1.E-01
.. 19
( )----
0
--------------- 13 14( )----19
------------ 1213-)----19
Totals
1
0
300000
-7----------
341
ALL
0.000000
0.000003
0.000000
2
0
0
100000
<<5E-02
0.000020
N/A
7.E-03
0.000000
0.000000
N/A
N/A
3.E-01
N/A
Table 2. Quantitation of the 8 most common CD44 TM skipped isoforms in aggregated
samples of AML and ALL were obtained by quantitative isoform profiling as described in
Chapter 1 and Chapter 2. AML Ratio and ALL Ratio are calculated over the total number of
respective CD44 molecules as reported in Table 2 of Chapter 3. C.f. Ratio = ratio between
Varma, Chris
57 of 65
Ph.D. Thesis
AML and ALL (negative values indicate down-regulation in AML). Statistically significant
up-regulation of ALL is highlighted in light blue and down-regulation is high-lighted in
light green. The p-values were calculated by a standard large-sample test procedure for
evaluating the difference between proportions obtained in two different populations
[Dev91].
Isoform
--------------------- ( )---- 19
--------------- 13---( ---- 19
------------------ 14( )----19
------------ 12------ 19
------ 0------------ ()----19
7--------------)----19
.(
--------------1314()---- 19
------------ 1213---( )----19
ALL
ALL Ratio
436
1
0
1
0
2
0
0
NM
0.004360
0.000010
0.000000
0.000010
0.000000
0.000020
0.000000
0.000000
333
0
1
2
0
0
0
1
NM
Ratio
C.f.
Ratio
0.005550
0.000000
0.000017
0.000033
0.000000
0.000000
0.000000
0.000017
-1.27
N/A
N/A
-3.33
N/A
N/A
N/A
0.00
P-value
4.E-04
2.E-01
1.E-01
1.E-01
N/A
1.E-01
N/A
1.E-01
Totals
100000
60000
Table 3. Quantitation of the 8 most common CD44 TM skipped isoforms in aggregated
samples of ALL and NM were obtained by quantitative isoform profiling as described in
Chapter 1 and Chapter 2. ALL Ratio and NM Ratio are calculated over the total number of
respective CD44 molecules as reported in Table 2 of Chapter 3. C.f. Ratio = ratio between
ALL and NM (negative values indicate down-regulation in ALL). Statistically significant
up-regulation of ALL is highlighted in light blue and down-regulation is high-lighted in
light green. The p-values were calculated by a standard large-sample test procedure for
evaluating the difference between proportions obtained in two different populations
[Dev91].
Isoform
--------------------( )----19
--------------- 13..--() ---- 19
14(--------------)----19
------------ 12------( )----19
------ 0-----------( )--- 19
7--------------(
-)--19
--------------- 13:14( )---- 19
------------ 1213-..--( )----19
Cancer
777
8
7
3
4
2
1
0
Cancer
Ratio
0.001943
NM
0.000020
0.000018
0.000008
0.000010
0.000005
0.000003
0.000000
333
0
1
2
0
0
0
1
NM
Ratio
0.005550
C.f.
Ratio
-2.86
P-value
0.000000
0.000017
0.000033
0.000000
0.000000
0.000000
0.000017
N/A
1.05
-4.44
N/A
N/A
N/A
0.00
N/A
<<5E-02
5.E-01
4.E-02
N/A
N/A
N/A
5.E-03
Totals
400000
60000
Table 4. Quantitation of the 8 most common CD44 TM skipped isoforms in aggregated
samples of (AML + ALL) and aggregated samples of NM were obtained by quantitative
isoform profiling as described in Chapter 1 and Chapter 2. Cancer = AML + ALL. Cancer
Ratio and NM Ratio are calculated over the total number of respective CD44 molecules as
reported in Table 2 of Chapter 3. C.f. Ratio = ratio between Cancer and NM (negative
values indicate down-regulation in Cancer). Statistically significant up-regulation of
Cancer is highlighted in light blue and down-regulation is high-lighted in light green. The
p-values were calculated by a standard large-sample test procedure for evaluating the
difference between proportions obtained in two different populations [Dev91].
Varma, Chris
58 of 65
Ph.D. Thesis
Universal Expression
We additionally characterized the expression of TM- CD44 in other cells types
beyond leukemic and normal through a comprehensive tissue panel. TM- CD44
expression was found in all tissue and was most significantly expressed in the
tissues of the lung and salivary gland (Figure 4).
I::I.G.J
TM+
TM-
Figure 4. Expression of TM- CD44 in all tissues queried via a tissue panel.
5.2. Exon Expression Profiles of Cell Lines
Obtaining Samples
The following EBV-transformed human cell lines were purchased from The
Coriell Institute: 3638 (B-cell ALL) and 3797 (Normal B-celllymphoblast).
The
following EBV-transformed human cell line RNA was purchased from Ambion:
KG-1 (AML).
Varma, Chris
59 of 65
Ph.D. Thesis
Quantitative Comparisons
Exon
AML
EXON 6B
LN
0
0 .0000
EXON 7 (V3)
EXON 8 (V4)
EXON 9 (V5)
20
0
0
0.0070
0.0000
0.0000
EXON 10
(V2)
(V6)
AML Ratio
NM LN
NM Ratio
C.f.
P-value
0
0.0000
Ratio
N/A
N/A
97
0
2
0.0889
0.0000
0.0018
-12.65
N/A
N/A
1.E-04
N/A
3.E-01
1.E-02
10
0.0035
42
0.0385
-10.95
EXON 11 (V7)
0
0.0000
1
0.0009
N/A
3.E-01
EXON 12
EXON 13
EXON 14
153
180
229
0.0538
0.0633
0.0805
0
73
147
0.0000
0.0669
0.1347
N/A
-1.06
-1.67
1.E-02
5.E-01
5.E-02
(V8)
(V9)
(V10)
Table 1. NM LN = normal cell line. C.f. Ratio = ratio between AML_LN and NM_LN
(negative values indicate down-regulation in AML_LN). The p-values were calculated by a
standard large-sample test procedure for evaluating the difference between proportions
obtained in two different populations [Dev91].
Exon
AML LN
AML Ratio
ALL LN
ALL
Ratio
C.f.
Ratio
P-value
0
34
0.0000
0.0457
N/A
-6.50
N/A
N/A
N/A
8.E-03
N/A
2.E-01
EXON
6B
0
0.0000
EXON
7
(V3)
20
0.0070
EXON
8
(V4)
0
0.0000
0
0.0000
EXON
9
(V5)
0
0.0000
3
0.0040
0.0035
4
0.0054
0.0000
0
0.0000
(V2)
-1.53
4.E-01
N/A
N/A
EXON 12 (V8)
153
0.0538
1
0.0013
40.01
2.E-02
EXON 13 (V9)
180
0.0633
22
0.0296
2.14
1.E-01
EXON 14 (V:10) 229
0.0805
264
0.3548
-4.41
<<5.E-02
Table 2. C.f. Ratio = ratio between AML_LN and ALL_LN (negative values indicate downregulation in AML_LN). The p-values were calculated by a standard large-sample test
EXON
10
EXON 11
(V6)
(V7)
10
0
procedure for evaluating the difference between proportions obtained in two different
populations [Dev91].
Exon
(V2)
ALL
LN
ALL Ratio
NM LN
NM
Ratio
C.f.
Ratio
P-value
0
0.0000
34
0.0457
0
0.0000
97
0.0889
N/A
-1.95
N/A
N/A
7.E-02
N/A
EXON
6B
EXON
7
EXON
8
(V4,)
0
0.0000
0
0.0000
EXON
9
(V5)
3
0.0040
2
0.0018
2.20
4.E-01
EXON
10
(V6)
4
0.0054
42
EXON
11
(V7)
0
0.0000
1
EXON
12
(V8)
1
0.0013
0
0.0385
0.0009
0.0000
-7.16
N/A
N/A
4.E-02
4.E-01
3.E-01
EXON
13
(V9)
22
264
0.0296
0.3548
73
147
0.0669
-2.26
8.E-02
(V3)
0.1347
2.63
<<5.E-02
Table 3. NM LN = normal cell linae Cf_ Ratin = ratin bhtween ALL LN and NM LN
EXON
14
(V10)
(negative values indicate down-regulation in ALL_LN). The p-values were calculated by a
standard large-sample test procedure for evaluating the difference between proportions
obtained in two different populations [Dev91].
Varma, Chris
60 of 65
Ph.D. Thesis
Exon
Cancer
C. Ratio
NM LN
NM Ratio
LN
EXON 6B (V2)
EXON
7
(V3)
EXON 8 (V4)
EXON
9
EXON
10
EXON 11
EXON 12
EXON
13
(V5)
('6)
(V7)
(V8)
(V9)
0
54
0
3
14
0
0.0000
0.0150
0.0000
0.0008
0.0039
0.0000
0
97
0
2
42
1
0.0000
0.0889
0.0000
0.0018
0.0385
0.0009
154
0.0429
0
202
0.0563
73
C.f.
P-value
Ratio
N/A
N/A
-5.91
1.E-04
N/A
N/A
-2.19
-9.87
4.E-01
3.E-03
0.0000
N/A
N/A
3.E-01
2.E-02
0.0669
-1.19
3.E-01
493
0.1374
147
0.1347
1.02
5.E-01
Table 1. CancerLN = aggregation of AML_LN and ALL_LN exon counts. NM_LN = normal
EXON 14
(V'10)
cell line. C.f. Ratio = ratio between CancerLN and NM_LN(negative values indicate downregulation in CancerLN). The p-valueswere calculated by a standard large-sample test
procedure for evaluating the difference between proportions obtained in two different
populations [Dev91].
Varma, Chris
61 of 65
Ph.D. Thesis
References
[AacO4]
Aach, J and Church, GM: Mathematical models of diffusion-
constrained polymerase chain reactions: basis of high-throughput
nucleic acid assays and simple self-organizing systems. J. Theoret.
[Alb94]
Biol. 2004 May 7;228(1):31-46.
Abbas AK, Lichtman AH, Pober JS: Cellular and Molecular
Immunology, 4 th Ed. W.B. Saunders Company. 2000.
Aksk E, Bavbek S, Dalay N: CD44 variant exons in leukemia and
lymphoma. Path. Onc. Res. 2002; 8(1):36-40.
Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J: Molecular
[ApaO3]
Apaydin MS, Brutlag DL, Guestrin C, Hsu D, Latombe JC, Varma
[AbbOO]
[Aks02]
Biology of the Cell, 3rd Ed. Garland Publishing. 1994.
C: Stochastic Roadmap Simulation: An efficient Representation and
Algorithm for Analyzing Molecular Motion. J. of Comp. Biology.
2003:10;257-281.
[ArsOO]
Ars E, Serra E, Garcia J, Kruyer H, Gaona A, Lazaro C, Estivill X:
Mutations affecting mRNA splicing are the most common
molecular defects in patients with neurofibromatosis type 1. Hum
Mol Genet 2000, 9:237-247.
[AstO4]
Ast G: How did Splicing Evolve? Nat. Gen. Oct. 2004; 5:777.
[Bar97]
http://www.surqery.wustl.edu/bicmdl/mdl.htm
[BenOO]
Bendal LJ, Bradstock KF, Gottlieb DJ: Expression of CD44 variant
exons in acute myeloid leukemia is more common and more
complex than that observed in normal blood, bone marrow or
CD34+ cells. Leukemia. 2000. 14:1239-1246.
[BenO4]
Bendall L: Role of CD44 variant exon 6 in acute lymphoblastic
leukemia: association with altered bone marrow localisation and
increased tumor burden. Leukemia (correspondence). 2004. Online
Pub doi: 10. 1038/sj.leu.2403393.
[Bio98-1]
[Bio98-2]
[BleOO]
http://www.bioscience.org/1998/v3/d/lesley/2. htm
http://www.bioscience.org/1998/v3/d/bou rguig/4.htm
Blencowe BJ: Exonic splicing enhancers: mechanism of action,
diversity and role in human genetic diseases. Trends Biochem Sci
2000, 25:106-110.
[Bou97]
Bourguignon L, Zhu H, Chu A, lida N, Zhang L, Hung M: Interaction
between adhesion receptor, CD44, and the oncogene product,
p185-HER2, promotes human ovarian tumor cell activation. J. Bio.
Chem. 1997. 272:27913-27918.
[CarO3]
[Cot99]
[Dev91]
Varma, Chris
Cartegni L, Krainer AR.: Correction of disease-associated exon
skipping by synthetic exon-specific activators. Nat. Struct. Biol.
2003. 10(2):120-5.
Cotran RS, Kumar V, Collins T: Robbins Pathologic Basis of
Disease, 6 th Ed. W.B. Saunders Company. 1999.
Devore JL: Probability and Statistics for Engineering and the
Science, 3 rd Ed. Wadsworth Publishing, Inc. 1991.
62 of 65
Ph.D. Thesis
[EmeO4]
[Fer99]
[Fie99]
www.emedicine.com
Fersht, A: Structure and Mechanism In Protein Science. W.H.
Freeman & Company. 1999.
Friedman, K.J., J. Kole, J.A. Cohn, M.J. Knowles, M.J. Silverman
and R. Kole (1999) Correction of aberrant splicing of CFTR gene by
[Fin95]
antisense oligonucleotides. J. Biol. Chem. 27436193-36199.
Finke LH, Terpe HJ, Zorb C, Haensch W, Schlag PM: Colorectal
cancer prognosis and expression of exon-v6-containing CD44
proteins. Lancet 1995. 345:583.
[Foe99]
[Gha96]
Foekens JA, Dall P, Klijn JG, Skroch PA, Claassen CJ, Look MP,
Ponta H, van Putten WL, Herrlich P, Henzen-Logmans SC:
Prognostic value of CD44 variant expression in primary breast
cancer. Int. J. Cancer 1999. 84:209-215.
Ghaffari S, Dougherty GJ, Eaves AC, Eaves CJ: Altered patterns of
CD44 epitope expression in human chronic and acute myeloid
leukemia. Leukemia. 1996. 10:1773.
[Has01]
Hastings ML, Krainer AR: Pre-mRNA splicing in the new
millennium. Current Opinions in Cell Biology 2001, 13:302-309.
[HeaO2]
http://health.allrefer.com
[HeaO4]
http://www. healthcentral.com/mhc/top/000570.cfm
[Hei93]
Heider KH, Hofmann M, Horst E, Van Den Berg F, Ponta H,
Herrlich P, Pals ST: A human homologue of the rat metastasis-
associated variant of CD44 is expressed in colorectal carcinomas
[Hen96]
and adenomatous polyps. J. Cell Biol. 1993. 120:227-233.
Henke C, Bitterman P, Roongta U, Ingbar D, Polunovsky V:
Induction of fibroblast apoptosis by anti-CD44 antibody:
implications for the treatment of fibroproliferative lung disease. Am
[HerOO]
J Pathol. 1996 Nov;149(5):1639-50.
Herrlich P, Morrison H, Sleeman J, Rousseau VO, Konig H,
Remers SW, Ponta H: CD44 Acts Both as a Growth- and
Invasiveness-Promoting Molecule and as a Tumor-Suppressing
[Jan96]
Cofactor. Annals New York Academy of Sciences 2000, 106-120.
Janeway CA and Travers P: Immunobiology: The Immune System
[Kat99]
In Health and Disease, 2 nd Ed. Garland Publishing Inc. 1996.
Katagiri Y, Sleeman J, Fujii H, Herrlich P, Hotta H, Tanaka K,
Chikuma S, Yagita H, Okumura K, Murakami M, Saiki I, Chambers
A, Uede T: CD44 variants but not CD44s cooperate with B1-
containing integrins to permit cells to bind to osteopontin
independently of arginine-glycine-aspartic acid, therby stimulating
[Kau95]
[Kha96]
Varma, Chris
cell motility and chemotaxis. Cancer Res. 1999. 59"219-226.
Kaufmann M, Heider KH, Sinn HP, von Minckwitz G, Ponta H,
Herrlich P: CD44 variant exon epitopes in primary breast cancer
and length of survival. Lancet 1995. 345:615-619.
Khaldoyanidi S, Achtnich M, Hehlmann R, Zoller M: Expression of
CD44 variant isoforms in peripheral blood leukocytes in malignant
63 of 65
Ph.D. Thesis
lymphoma and leukemia-inverse correlation between expression
and tumor progression. Leukemia Research. 1996. 20:839-851.
[KhaO2]
Khan SA, Lopez-Chua CA, Zhang J, Fisher LW, Sorensen ES,
Denhardt DT: Soluble osteopontin inhibits apoptosis of adherent
endothelial cells deprived of growth factors. J Cell Biochem.
2002;85(4):728-36.
[Kin99]
Kincade PW: Blasting away leukemia. Nature Med. 1999; 5(6):619-
620.
[Kra97]
[LacOO]
Krainer AR: Eukaryotic mRNA ProcessinQ. Oxford University Press.
1997.
Lacerra G, Sierakowska H, Carestia C, Fucharoen S, Summerton
J, Weller D, Kole R: Restoration of hemoglobin A synthesis in
erythroid cells from peripheral blood of thalassemic patients. Proc
Natl Acad Sci USA 2000, 97:9591-9596.
[Leg98]
Legras S, Gunthert U, Stauder R, Curt F, Oliferenko S, KluinNelemans HC, Marie JP, Proctor S, Jasmin C, Smadja-Joffe F: A
strong expression of CD44-v6 correlates with shorter survival of
patients with acute myeloid leukemia. Blood. 1998; 91(9):34013413.
[LeuO4]
[Liu01]
[MagOl]
[MarOO]
http://www.leukemia-lymphoma.org
Liu H-X, Cartegni L, Zhang MQ, Krainer AR: A mechanism for exon
skipping caused by nonsense or missense mutations in BRCA1
and other genes. Nat Genet 2001, 27:55-58.
Magyarosy E, Sebestyen A, Timar J: Expression of metastasis
associated proteins, CD44v6 and NM23-H1, in pediatric acute
lymphoblastic leukemia. Anticancer Res. 2001. 21:819-823.
Maroney PA, et al: Functional recognition of the 5' splice site by
U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early
spliceosome assembly. Mol Cell 2000, 6:317-328.
[MedO2]
[Mit99]
http://www.nlm.nih.gov/medlineplus/ency/article/000570.htm
R. D. Mitra, G. M. Church: In situ localized amplification and contact
replication of many individual DNA molecules. Nucleic Acids Res.
[MitO3]
27, e34 (1999).
R. D. Mitra et al.: Digital Genotyping and Haplotyping with
Polymerase Colonies Proc. Natl. Acad. Sci. U.S.A. 100, 5926
[Mu194]
Mulder JWR, Kruyt PM, Sewnath M, Oosting J, Seldenrijk CA,
(2003).
Weidema WF, Offerhaus GJA, Pals ST: Colorectal cancer
prognosis and expression of exon-v6-containing CD44 proteins.
Lancet 1994. 344:1470-1472.
[Yu96]
Yu Q, Toole B: A new alternatively spliced exon between v9 an v10
provides a molecular basis for synthesis of soluble CD44. JEM.
[RozOO]
1997. 18:1985.
S. Rozen and H. J. Skaletsky: Primer3 on the WWW for general
users and for biologist programmers. In: Krawetz S, Misener S
Varma, Chris
64 of 65
Ph.D. Thesis
[Sie99]
[SieOO]
(eds) Bioinformatics Methods and Protocols: Methods in Molecular
Biology. 2000. Humana Press, Totowa, NJ, pp 365-386
Sierakowska, H., M.J. Sambade and R.Kole: Sensitivity of splice
sites to antisense oligonucleotides. RNA. 1999. 5369-377
Sierakowska, S. Agrawal and R. Kole (2000) Antisense
oligonucleotides as modulators of pre-mRNA splicing. Methods Mol
Biol. 133223-33.
[SkoO3]
Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F.:
Bifunctional antisense oligonucleotides provide a trans-acting
splicing enhancer that stimulates SMN2 gene expression in patient
fibroblasts. Proc Natl Acad Sci U S A. 2003. 100(7):4114-9.
[Sle96]
Sleeman J, Rudy W, Hofmann M, Moll J, Herrlich P, and Ponta H:
Regulated clustering of variant CD44 proteins increases their
hyaluronate binding capacity. J. Cell Biol.1996. 135:1139-1150.
[Sle97]
Sleeman J, Kondo K, Moll J, Ponta H, Herrlich P: Variant exons v6
and v7 together expand the repertoire of glycosaminoglycans
[Str95]
[Suw02]
bound to CD44. J. of Bio. Chem. 1997. 272:31837-31844.
Stryer, L: Biochemistry, 4 th Ed. W.H. Freeman & Company. 1995.
Suwanmanee, T., H. Sierakowska, S. Fucharoen, and R. Kole.
(2002) Restoration of human beta-globin gene expression in murine
and human IVS2-654 thalassemic erythroid cells by free uptake of
antisense oligonucleotides. Mol Pharmacol. 2002 Sep;62(3):545[Tan94]
53.
Tanabe KK & Saya H: Crit. Rev. Oncog. 1994, 5:201.
[Uku01]
http://www.uku.fi/laitokset/anat/PG/ha_funct. htm
[Van93]
van Weering DHJ et al: PCR Methods Appl 1993, 3:100.
[WauO3]
http://www. neuro.wustl.edu/neuromuscular/pathol/spliceosome.htm
[Wil99]
Wilton, S.W., F. Lloyd, K. Carville, S.Fletcher, K. Honeyman, S.
Agrawal and R.Kole (1999) Specific removal of nonsense mutation
from the mdx dystrophin mRNA using antisense oligonucleotides.
[Yu99]
[Zhu03]
Neuromuscular Disorders 9330-338.
Yu Q, Stamenkovic I: Localization of matrix metalloproteinase 9 to
the cell surface provides a mechanism for CD44-mediated tumor
invasion. Genes Dev. 1999. 13:35-48.
Zhu J, Shendure J, Mitra RD, Church GM: Single Molecule Profiling
of Alternative Pre-mRNA Splicing. Science. 2003. 301:836-838.
Varma, Chris
65 of 65
Ph.D. Thesis
Room 14-0551
77 Massachusetts Avenue
MITLibraries
Document Services
Cambridge, MA 02139
Ph: 617.253.5668 Fax: 617.253.1690
Email: docs@mit.edu
http: Illibraries. mit. edu/docs
DISCLAIMER OF QUALITY
Due to the condition of the original material, there are unavoidable
flaws in this reproduction. We have made every effort possible to
provide you with the best copy available. If you are dissatisfied with
this product and find it unusable, please contact Document Services as
soon as possible.
Thank you.
Some pages in the original document contain color
pictures or graphics that will not scan or reproduce well.