BCB 444/544 - F07 Study Guide Final For Final Exam (Dec 10)
advertisement

BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 1 of 13
BCB 444/544 - F07
Study Guide Final - KEY
For Final Exam (Dec 10)
Answers will be discussed in Review Session on Thurs Dec 6
FINAL EXAM Mon Dec 10 9:45 – 11:45 AM in MBB 1420 & 1340
NOTE CHANGE: ENTIRE FINAL EXAM WILL BE OPEN BOOK/NOTES!!
General Comments:
Final Exam will include 100 pts and contain 2 sections:
1) a 50-minute written Exam, open-book, open-notes, open-computer
20 pts In Class: Comprehensive
40 pts In Class: New material (since Exam 2)
All topics covered in class, lab and assigned readings:
Lectures 27-38, HW5-6, Chps 9-11 & 17-19, Labs 9-11
2) a 50-minute lab practical Exam, open-book, open-notes, using
computers
40 pts In Lab: Practical (Comprehensive)
Some questions will involve computation; bring your calculators if you like.
All required formulae or tables will be provided.
Some questions will require short essay-like answers that demonstrate
your understanding of key concepts covered in the course.
How to study:
Review all topics, problems & correct answers included in:
o Study Guides 1 & 2 & 3/Final (this document, see below)
o Exams 1 & 2
o Graded HW Assignments
Review topics on which you missed points or need review in:
o Lecture PPTs (1 - 38) (39 will not be covered)
Spend time reviewing procedures and answers for:
o Lab Exercises (1-11) (Lab 12 will not be covered)
BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 2 of 13
Hints:
For “Comprehensive” part of Final Exam, focus will be on key vocabulary,
concepts and problems covered on Exams 1 & 2 (Lectures 1-26) and
skills covered in Lab Exercises 1-8.
For example, everyone should know:
differences between eukaryotic and prokaryotic cells
differences between replication, transcription, translation
how to read a genetic code table
how to fill in a simple dynamic programming matrix
differences between PAM and BLOSUM scoring matrices
how to retrieve sequences and structures from online databases
how to visualize and manipulate protein structures with PyMol
how to predict genes in a given DNA sequence
how to predict protein function from sequence
Strong hints:
transcription/translation, dynamic programming & HMM
problems similar to those on Exams 1 & 2 are almost
guaranteed to appear!!
For the "New Material" part of Final Exam, focus will be on material
covered in:
Lectures 27 - 38 (not Lecture 39)
Lab Exercises 9 – 11 (not Lab 12)
Final Exam WILL include question(s) based on BCB
544 Project Presentations
Strong hints:
You should understand basic principles of:
Phylogenetic analysis
Machine learning algorithms
Microarray and proteomics techniques & analysis
You should solve practice problems included in this Study
Guide: New Material (Part IB – New Material) - below
BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 3 of 13
Practice Problems for Part IA - Comprehensive Section (20 pts)
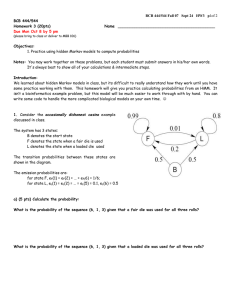
IA-1. Using this hidden Markov model and assuming that you start in state 1
calculate the most probable path for sequence AGAT.
The most probable path is: :
1–1–1-1
For complete credit, show your work by completing this probability table:
A
1
2
3
G
A
T
0.25*max{0.25*0.5 0.25*max{0.03125*0.5
0*0.2
0.03*0.2
0*0}
0.005*0}
= 0.03125
= 0.00391
0.25*max{0.00391*0.5
0.0018*0.2
0.0025*0}
= 0.00049
0
0.4*max{0.25*0.3
0*0.6
0*1}
= 0.03
0.1*max{0.03125*0.3
0.03*0.6
0.005*1}
= 0.0018
0.1*max{0.00391*0.3
0.0018*0.6
0.0025*1}
= 0.00025
0
0.1*max{0.25*0.2
0*0.2
0*0}
= 0.005
0.4*max{0.03125*0.2
0.03*0.2
0.005*0}
= 0.0025
0.4*max{0.00390*0.2
0.0018*0.2
0.0025*0}
= 0.00031
0.25
BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 4 of 13
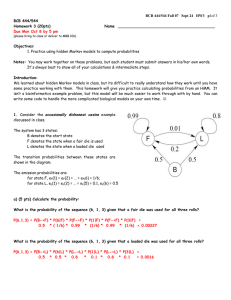
IA-2.1 Fill out the dynamic programming matrix for determining an optimal
global alignment between the sequences TCG and TCCAG.
Scoring: +3 for matches; -2 for mismatches and spaces.
T
C
C
A
G
0
-2
-4
-6
-8
-10
T
-2
3
1
-2
-4
-6
C
-4
1
6
4
2
0
G
-6
-2
4
4
2
5
2.2
What is the score(s) of the optimal alignment(s)? 5
matrix)
(Circle in DP
2.3.
There are 2 optimal alignments. For full credit, draw both of them
below & show your traceback arrows in the DP matrix above.
T
C
C
A
G
T
C
C
A
G
T
-
C
-
G
T
C
-
-
G
+3 -2 +3 -2 +3 = 5
+3 +3 -2 -2 +3 = 5
Practice Problems & Review Questions for Part IB – New Material (40 pts)
BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 5 of 13
IB-1. Phylogenetic Analysis
Find the parsimony score for the tree below using Fitch’s algorithm. For full
credit, show your work.
The parsimony score for this tree is 9
Reminder re: how Fitch’s algorithm works:
v in a tree has a set X v .
If v is a leaf, X v is the nucleotide (or amino acid for protein based trees) observed
at v .
If v is a node with descendants u and w :
o Let Y X u X w
o If Y make X v Y
o If Y make X v X u X w and count one evolutionary step.
Each node
Basically, we examine the characters at children of each internal node. If they have some
character(s) in common, we write down the common character(s). If there is no overlap, we
write down all of the characters and count an evolutionary step (noted with a *). At the end,
we count up all of the *’s to get the total number of evolutionary steps.
C
A
G T
{A,G}*
G
C
{G,T}*
T
A
{C,T}*
T
T
{A,T}*
A
G
C
{C,G}*
{A,C,G}*
{A,C,G}*
{A,C,G,T}*
{A,T}
{T}
{T}
{A,C,G,T}*
BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 6 of 13
IB-2. Microarray Analysis – Clustering
2.1 Using hierarchical clustering, how would you clusters genes (A,B,C,D)?
A
1.00
0.95
0.50
0.25
A
B
C
D
B
0.95
1
0.70
0.50
C
0.50
0.70
1
0.65
D
0.25
0.50
0.65
1
You may find it helpful to use the following table to calculate your clusters:
Iteration
1
2
3
Clusters = [AB]
Object 1
A
C
[AB]
&
Object 2
B
D
[CD]
Correlation
0.95
0.65
0.49
New Object
[AB]
[CD]
[ABCD]
[CD]
2.2 Draw a simple tree that illustrates this grouping of the genes.
A
C
B
D
BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 7 of 13
IB-3. Vocabulary & Short Answers Questions
What is/are:
A perceptron?
A single layer neural network that takes a set of (weighted) inputs and
maps them to an output value by applying a function; the value of the
function is compared with a threshold to determine whether the
perceptron “fires” (output=1) or not (output =0).
A kernel function?
o A function that map inputs into a higher dimensional “feature space” in
which the members of different classes are (hopefully) linearly
separable.
Parsimony?
o The parsimony principle means that the simplest explanation is the best.
Parsimony is often used in phylogenetics to select a tree that explains
the observed data with the smallest number of evolutionary steps.
Bootstrapping?
o A method for estimating how many positions in a multiple sequence
alignment support a particular branch point in a phylogenetic tree.
Basically, bootstrapping estimates how much of the input data supports
each branch point.
The Jukes-Cantor model?
o A simple model of evolution in which each position in a sequence has the
same probability of mutating, and when a mutation occurs, every possible
mutation is equally likely.
A new high-throughput “massively parallel” sequencing technique?
o 454 or Pyro sequencing
The HAPMAP project?
o Project to identify and catalog common genetic variants (e.g. SNPs) in
humans. HAPMAP stands for “haplotype map;” a haplotype is a set of
SNPs that are inherited together.
SAGE?
o Serial Analysis of Gene Expression - a high throughput method for
analyzing RNA expression; provides a “snapshot” of RNAs expressed in a
particular sample (similar to that provided by microarrays)
The two main microarray platforms?
o cDNA arrays - probes are cDNAs spotted onto glass slides
o Oligo arrays – probes are short synthetic oligonucleotides synthesized
directly on chips
BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 8 of 13
Two types of machine learning algorithms used to recognize patterns in
microarray data?
o Clustering algorithms (e.g., hierarchical, k-means)
o Classification algorithms (e.g., k-nearest neighbor, SVM)
What are the main differences between:
Brendel’s (ISU) GeneSeqer & Burge’s (MIT) GenScan programs for gene
prediction?
o GeneSeqer uses splice site prediction & spliced alignment; it does not
depend on level of sequence homology between query sequence and
known genes; GenScan relies on HMMs and works best for sequences
that share sequence similarity with known genes
Supervised vs unsupervised machine learning algorithms?
o Supervised algorithms require the data to have labels, for example
binding or non-binding, and learn from these labeled examples. Our
lab exercise on machine learning used only supervised algorithms
(Naïve Bayes, SVM, decision trees). Unsupervised algorithms work
with unlabeled data and attempt to discover correlations in the data
without the guidance of labels. Our lab exercise on microarray
analysis used examples of unsupervised algorithms (clustering).
Distance-based & parsimony-based phylogenetic tree-building algorithms
o The main difference is the input used by the programs. Distancebased methods use a distance matrix, which is very quick to compute
for any sequences. Parsimony-based methods use a character matrix
derived from a multiple sequence alignment. The distance matrix
condenses all of the information about differences between two
sequences into a single number, whereas the multiple sequence
alignment contains information about every specific difference.
Regulation of gene expression in prokaryotes vs eukaryotes?
o Everything is much more complicated in eukaryotes. In prokaryotes,
almost all regulation of gene expression occurs at the level of
transcription initiation. In eukaryotes, regulation occurs at multiple
levels, many of which occur after transcription. This includes
processes such as RNA processing, transport, and stability, & protein
processing, post-translational modification, transport, and stability.
BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 9 of 13
Hierarchical and k-means clustering?
o Hierarchical clustering proceeds by grouping the two most similar
objects and repeating until all objects are in a single cluster; thus the
final number of clusters is determined by choosing where to “cut” the
hierarchy. K-means clustering starts with k randomly chosen points
(centroids), computes the distance from each object to these k
centroids, and assigns each object to the closest centroid. New
centroids are then computed by averaging all points assigned to each
centroid. Then the distance from each object to each new centroid is
calculated, and each object is reassigned to the closest centroid. This
process is repeated until the centroids don’t move very much. The
number of clusters is determined by value of k chosen.
Promoters & enhancers?
o In eukaryotes, both promoters and enhancers are binding sites for
transcription factors. Promoters are essential for initiation of
transcription and are located relatively close to the transcription
start site. Enhancers are needed for regulated transcription and can
be very far from the transcription start site.
Cis- vs trans- acting regulatory factors/elements?
o Cis-acting regulatory elements are signals in DNA (or RNA)
sequences and include promoters, enhancers, and silencers. Transacting regulatory factors are “diffusible” and recognize such signals;
they usually are proteins, such as transcription factors or chromatin
remodeling complexes, but some small regulatory RNAs are also
trans-acting.
Briefly list & describe the major sequence signals used in gene prediction software.
o These include regulatory signals for:
Transcription: TF binding sites, promoters, initiation sites,
terminators, CpG islands, etc.
RNA Processing: Splice donor/acceptor sites, polyA signals
Translation: Start (AUG = Met) & stop (UGA,UUA, UAG) codons,
ORFs, codon usage
What are the two major types of distance-based methods for generating
phylogenetic trees – and what are the relative advantages and disadvantages of
each?
Clustering methods and optimality methods. Clustering methods attempt to
build a tree in which the most similar sequences are close together in the
tree. Advantage – very fast. Disadvantage - produce a single tree, without
BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 10 of 13
consideration of alternative tree topologies. Optimality methods compare all
possible trees and select the tree that best fits the distance matrix.
Advantage – consider multiple tree topologies. Disadvantage – computationally
slow.
Why are SNPs important?
SNPs are Single Nucleotide Polymorphisms or single-base variations in
genomic DNA sequences that are present in at least 1% of the population.
They are important because specific SNPs are associated with predisposition
for certain diseases or can be indicative an individual’s response to a
particular therapy.
Which student project presentation did you find most interesting? Why? (Be
specific)
What is the most important “new” thing you learned in this course? Explain.
This Genetic Code table will be provided for Part I of the Final.
BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 11 of 13
Study Suggestions for Part II – Lab Practical (40 pts)
You will be given an amino acid sequence for a “mystery” protein. You will be
required to use servers/programs used in lab (such as BLAST) and to gather and
analyze information to characterize this mystery protein.
Knowledge of which servers perform which function will also be tested.
There will not be sufficient time to perform any analysis of actual microarray
data as part of the practical exam. However, you should be prepared to discuss in
a paragraph or two some of the important issues in setting up a microarray
experiment, and how to normalize and filter your data prior to subsequent
analysis (e.g. clustering/machine learning etc.). This may be included as part of
the mystery protein portion of the practical, or as a standalone question or three.
For the practical, you will not be expected to discuss the clustering or
visualization portions of the lab, as there was less guidance about the best ways
to do this.
You will be expected to know how to retrieve structure files from PDB using
keyword search, direct retrieval by PDB id, or by BLAST query (either at PDB,
under Advanced Search, or using NCBI blastp, by specifying PDB as the database
to query). You will also be expected to use PyMOL to perform very basic
manipulations of how such a structure file is displayed.
The following list of links will be provided for Part II of the Final.
Entrez Web site (http://www.ncbi.nlm.nih.gov/entrez/)
BLAST at NCBI (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi)
OMIM (Online Mendelian Inheritance in MAN)
UniProt (http://www.pir.uniprot.org/)
READSEQ (http://thr.cit.nih.gov/molbio/readseq/)
SRS server (http://srs6.ebi.ac.uk/ )
Dothelix (www.genebee.msu.su/services/dhm/advanced.html)
LALIGN (www.ch.embnet.org/software/LALIGN_form.html)
BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 12 of 13
SSEARCH (http://pir.georgetown.edu/pirwww/search/pairwise.html)
PRSS (http://fasta.bioch.virginia.edu/fasta/prss.htm)
Biology Workbench
ClustalW (http://www.ebi.ac.uk/Tools/clustalw/)
TCoffee (http://igs-server.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi)
MultAlign (http://prodes.toulouse.inra.fr/multalin/multalin.html)
DCA server (http://bibiserv.techfak.uni-bielefeld.de/dca/submission.html)
PRRN server (http://prrn.ims.u-tokyo.ac.jp/)
ORF Finder at NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html)
Gene Seqer (http://www.plantgdb.org/)
GeneMark (http://opal.biology.gatech.edu/GeneMark/genemark24.cgi)
PROSITE (http://ca.expasy.org/prosite/)
Super Family (http://supfam.org/SUPERFAMILY/hmm.html)
JAFA meta server (http://jafa.burnham.org/)
PDB (http://pdb.org)
CDM
PSIPRED
Proteus
TMPRED
Phobius
SWISS-MODEL server
WHAT IF server
3-D Jury (BioInfoBank) META server
mfold Web server
BCB 444/544 Fall 07
Study Guide Final KEY – Dec 5
p 13 of 13
RNAfold server
RNAalifold
UCSC genome browser (http://genome.ucsc.edu/)
MEME/MAST (http://meme.sdsc.edu/meme/)
TESS
Gene Expression Pattern Analysis Suite (GEPAS, at http://gepas.bioinfo.cipf.es/)
Expression Profiler (http://www.ebi.ac.uk/microarray/index.html)