BCB 444/544 - F07 Study Guide Final For Final Exam (Dec 10)
advertisement

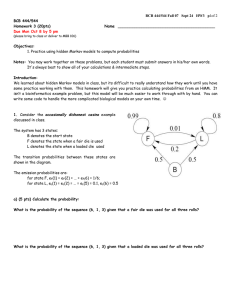
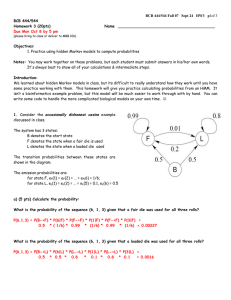
BCB 444/544 Fall 07 Study Guide Final – Dec 3 p 1 of 8 BCB 444/544 - F07 Study Guide Final For Final Exam (Dec 10) Answers will be discussed in Review Session on Thurs Dec 6 FINAL EXAM Mon Dec 10 9:45 – 11:45 AM in MBB 1420 & 1340 NOTE CHANGE: ENTIRE FINAL EXAM WILL BE OPEN BOOK/NOTES!! General Comments: Final Exam will include 100 pts and contain 2 sections: 1) a 50-minute written Exam, open-book, open-notes, open-computer 20 pts In Class: Comprehensive 40 pts In Class: New material (since Exam 2) All topics covered in class, lab and assigned readings: Lectures 27-38, HW5-6, Chps 9-11 & 17-19, Labs 9-11 2) a 50-minute lab practical Exam, open-book, open-notes, using computers 40 pts In Lab: Practical (Comprehensive) • • • Some questions will involve computation; bring your calculators if you like. All required formulae or tables will be provided. Some questions will require short essay-like answers that demonstrate your understanding of key concepts covered in the course. How to study: • • • Review all topics, problems & correct answers included in: o Study Guides 1 & 2 & 3/Final (this document, see below) o Exams 1 & 2 o Graded HW Assignments Review topics on which you missed points or need review in: o Lecture PPTs (1 - 38) (39 will not be covered) Spend time reviewing procedures and answers for: o Lab Exercises (1-11) (Lab 12 will not be covered) BCB 444/544 Fall 07 Study Guide Final – Dec 3 p 2 of 8 Hints: • For comprehensive part of Final Exam, focus will be on key vocabulary, concepts and problems covered on Exams 1 & 2 (Lectures 1-26) and skills covered in Lab Exercises 1-8. For example, everyone should know: differences between eukaryotic and prokaryotic cells differences between replication, transcription, translation how to read a genetic code table how to fill in a simple dynamic programming matrix differences between PAM and BLOSUM scoring matrices how to retrieve sequences and structures from online databases how to visualize and manipulate protein structures with PyMol how to predict genes in a given DNA sequence how to predict protein function from sequence Strong hints: • transcription/translation, dynamic programming & HMM problems similar to those on Exams 1 & 2 are almost guaranteed to appear!! For the "New Material" part of Final Exam, focus will be on material covered in: Lectures 27 - 38 (not Lecture 39) Lab Exercises 9 – 11 (not Lab 12) Final Exam WILL include question(s) based on BCB 544 Project Presentations Strong hints: You should understand basic principles of: Phylogenetic analysis Machine learning algorithms Microarray and proteomics techniques & analysis You should solve practice problems included in this Study Guide: New Material (Part IB – New Material) - below BCB 444/544 Fall 07 Study Guide Final – Dec 3 p 3 of 8 Practice Problems for Part IA - Comprehensive Section (20 pts) IA-1. Using this hidden Markov model and assuming that you start in state 1 calculate the most probable path for sequence AGAT. The most probable path is: For complete credit, show your work by completing this probability table: A 1 0.25 2 0 3 0 G A T BCB 444/544 Fall 07 Study Guide Final – Dec 3 p 4 of 8 Fill out the dynamic programming matrix for determining an optimal global alignment between the sequences TCG and TCCAG. Scoring: +3 for matches; -2 for IA-2 mismatches and spaces. λ T C C A G λ 0 -2 -4 -6 -8 -10 T -2 C -4 G -6 2.2 What is the score(s) of the optimal alignment(s) ? (Circle in the DP matrix) 2.3. There are 2 optimal alignments. For full credit, draw both of them below & show your traceback arrows in the DP matrix above. BCB 444/544 Fall 07 Study Guide Final – Dec 3 p 5 of 8 Practice Problems & Review Questions for Part IB – New Material (40 pts) IB-1. Phylogenetic Analysis Find the parsimony score for the tree below using Fitch’s algorithm. Show your work. C A G T G C T A T T A G C BCB 444/544 Fall 07 Study Guide Final – Dec 3 p 6 of 8 IB-2. Microarray Analysis – Clustering Answer the following questions based on the provided table of correlation coefficients for each pair of genes, calculated from microarray expression levels. A B C D A 1.00 0.95 0.50 0.25 B 0.95 1 0.70 0.50 C 0.50 0.70 1 0.65 D 0.25 0.50 0.65 1 2.1 Using hierarchical clustering, how would you clusters genes (A,B,C,D)? You may find it helpful to use the following table to calculate your clusters: Iteration 1 2 3 Object 1 Object 2 Correlation New Object 2.2 Draw a simple tree that illustrates this grouping of the genes. In this Study Guide, a framework is provided – all you need to do is label the leaves: BCB 444/544 Fall 07 Study Guide Final – Dec 3 p 7 of 8 IB-3. Vocabulary & Short Answers Questions What is/are: • • • • • • • • • • A perceptron? A kernel function? Parsimony? Bootstrapping? The Jukes-Cantor model? A new high-throughput “massively parallel” sequencing technique? The HAPMAP project? SAGE? The two main microarray platforms? Two types of machine learning algorithms used to recognize patterns in microarray data? What are the main differences between: • • • • • • • Brendel’s (ISU) GeneSeqer & Burge’s (MIT) GenScan programs for gene prediction? Supervised vs unsupervised machine learning algorithms? Distance-based & parsimony-based phylogenetic tree-building algorithms Regulation of gene expression in prokaryotes vs eukaryotes? Hierarchical and k-means clustering? Promoters & enhancers? Cis- vs trans- acting regulatory factors? Briefly list & describe the major sequence signals used in gene prediction software. What are the two major types of distance-based methods for generating phylogenetic trees – and what are the relative advantages and disadvantages of each? Why are SNPs important? Which student project presentation did you find most interesting? Why? (Be specific) What is the most important “new” thing you learned in this course? Explain.. BCB 444/544 Fall 07 Study Guide Final – Dec 3 p 8 of 8 Study Suggestions for Part II – Lab Practical (40 pts) You will be given an amino acid sequence to a mystery protein. You will be required to use servers/programs used in lab (such as BLAST) and to gather and analyze information to characterize this mystery protein. Knowledge of which servers perform which function will also be tested. There will not be sufficient time to perform any analysis of actual microarray data as part of the final practical. However, you should be prepared to discuss in a paragraph or two some of the important issues in setting up a microarray experiment, and how to normalize and filter your data prior to subsequent analysis (e.g. clustering/machine learning etc.). This may be included as part of the mystery protein portion of the practical, or as a standalone question or three. For the practical, you will not be expected to discuss the clustering or visualization portions of the lab, as there was less guidance about the best ways to do this. You will be expected to know how to retrieve structure files from PDB using keyword search, direct retrieval by PDB id, or by BLAST query (either at PDB, under Advanced Search, or using NCBI blastp, by specifying PDB as the database to query). You will also be expected to use PyMOL to perform very basic manipulations of how such a structure file is displayed.