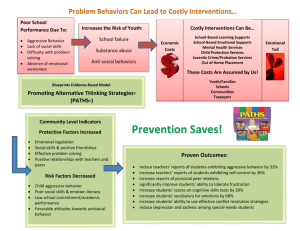
Minimal Path Enumeration to Predict Gene Knockout Effects in Microbial Communities
advertisement

Minimal Path Enumeration to Predict Gene Knockout Effects in Microbial Communities Steven 1School 1 Pastor , Yemin 1 Lan , Gail L 2 Rosen of Biomedical Engineering, Science, and Health Systems, Drexel University, 3141 Chestnut St Philadelphia, PA 19104, USA 2Department of Electrical and Computer Engineering, Drexel University, 3141 Chestnut St Philadelphia, PA 19104, USA Introduction Supraorganism Topological Model In complex microbial communities, it is a challenge to trace activities performed by certain organisms. Applied the path finding pipeline to the metabolic interactions of 3 fungi: 1. Trichoderma reesei (Tr), 2. Phanerochaete chrysosporium (Pc) 3. Rhodotorula graminis (Rg). To address this, we simulate effects of gene knockouts in a community using a supraorganism topological model. 140 Trichoderma reesei Focused Model (Wild Type) 127 T. reesei individual biosynthesis of Trehalose Boolean Satisfiability Over the years, SAT solvers have been used for many applications and are renown for their speed in solving complex formulas. Consumed/produced by Tr Trehalose 59 3 1 3 2 3 0 4 5 6 7 8 9 10 11 12 Gene Knockout Effects on Minimal Paths in Trehalose Biosynthesis 10 Pc and Rg 8 Minimal Paths 7 Procedure Trichoderma reesei Focused Model, Knockout Example - 2.3.1.Condition Encode CNF T. reesei individual biosynthesis of Trehalose Legend Not consumed by Tr Consumed by Tr & produced by: Tr+Pc+Rg, Tr+Pc, Tr+Rg, Tr Pc and Rg http://en.wikipedia.org/wiki/Boolean_satisfiability_problem SAT Solving 63 Condition Number Consumed/produced by Tr Condition Number Total Solutions Minimal Solutions Tr Wild Type 1 3 3 Supraorganism Wild Type 2 127 7 KO: 2.3.1.- 3 3 3 KO: 3.2.1.28 (Tr) 4 63 4 KO: lpqX 5 127 7 KO: 3.1.3.12 6 123 4 KO: 3.6.3.- 7 123 4 KO: 2.3.1.122 8 120 6 KO: 2.4.1.15 (Tr) 9 126 6 KO: 2.3.1.- & 3.2.1.28 (Tr) 10 0 0 KO: lpqX & 3.2.1.28 (Tr) 11 63 4 KO: 3.1.3.12 & 3.2.1.28 (Tr) 12 59 4 Trehalose Color-Coded Rows Correspond to Model Color-Codes and Denote Knockout Conditions 7 6 6 5 4 3 Minimal Paths 120 35 For this reason, and the short-comings exhibited by other metabolic models, it is of interest to encode a complex metabolic network into a propositional formula to determine the minimal routes from a source metabolite to target metabolite. Automatically encode the metabolic pathway into boolean logical formula1,2 126 63 Consumed by Tr & produced by: To determine satisfiable solutions, one must encode their system of interest into a boolean propositional logic formula. KEGG, MetaCyc, Unipathway: Substrates Products Enzymes 123 Not consumed by Tr Tr+Pc+Rg, Tr+Pc, Tr+Rg, Tr Databases 123 70 0 Legend 127 105 Total Paths In the presence of the other 2 organisms, trehalose production is both complemented and augmented, expanding metabolism to 7 minimal paths. Previous iterations of this technique only addressed single organism/single pathway systems. SAT solvers automatically calculate the satisfiability of a propositional logic formula3. Gene Knockout Effects on Total Paths in Trehalose Biosynthesis By itself, TR synthesizes trehalose via 3 minimal paths. Using this model and aided by a boolean satisfiability (SAT) solver, we can enumerate minimal paths through metabolic networks. Our model allows for insights into how a knockout perturbs metabolic networks within a community. Community-Level Knockout Effects 0 3 4 4 4 4 3 0 1 2 3 4 5 6 7 8 Condition Number 9 10 11 12 Conclusions Expanded upon single organism methods to create an automated pipeline that enumerates the total and minimal paths in microbial community metabolism. This multi-organism approach serves as a hypothesis-generating tool for more granular experimental microbial ecological analyses. Applying the minimization process allowed for pinpointing of essential genes required for key metabolic functions. Future iterations of this process will extend the minimization via stoichiometric coefficient matrices. - - - - - Denotes a Double Knockout Automated process to find shortest route from source metabolite(s) to target metabolite(s). FInds Total Paths: Source Metabolite(s) Target Metabolite(s) References 1. Soh T, et al. Evaluation of the prediction of gene knockout effects by minimal pathway enumeration. International Journal On Advances in Life Sciences, 2012. 4,3 and 4,154-165. 2. Peres S, Morterol M, and Simon L. SAT-Based Metabolics Pathways Analysis Without Compilation. 12th Conference on Computational Methods in Systems Biology (CMSB'14), 2014. 20-31. 3. Ohrimenko O, et al. Propagation = Lazy Clause Generation, Principles and Practice of Constraint Programming. Lecture Notes in Computer Science, 2007. 4741, pp. 544–558