Analysis of Covariance with Spatially Correlated Secondary Variables
advertisement

Revista Colombiana de Estadística
Junio 2008, volumen 31, no. 1, pp. 95 a 109
Analysis of Covariance with Spatially Correlated
Secondary Variables
Análisis de covarianzas con variables secundarias correlacionadas
espacialmente
Tisha Hooks1,a , David Marx2,b , Stephen Kachman2,c ,
Jeffrey Pedersen3,d , Roger Eigenberg3,e
1 Department
of Mathematics and Statistics, Winona State University, Winona,
United States
2 Department
3 Department
of Statistics, University of Nebraska, Lincoln, United States
of Agronomy and Horticulture, USDA-ARS Research, University of
Nebraska, Lincoln, United States
4 USDA-ARS
U.S. Meat Animal Research Center, Clay Center, United States
Abstract
Advances in precision agriculture allow researchers to capture data more
frequently and in more detail. For example, it is typical to collect “on-the-go”
data such as soil electrical conductivity readings. This creates the opportunity to use these measurements as covariates for the primary response
variable to possibly increase experimental precision. Moreover, these measurements are also spatially referenced to one another, creating the need for
methods in which spatial locations play an explicit role in the analysis of
the data. Data sets which contain measurements on a spatially referenced
response and covariate are analyzed using either cokriging or spatial analysis of covariance. While cokriging accounts for the correlation structure of
the covariate, it is purely a predictive tool. Alternatively, spatial analysis
of covariance allows for parameter estimation yet disregards the correlation
structure of the covariate. A method is proposed which both accounts for
the correlation in and between the response and covariate and allows for
the estimation of model parameters; also, this method allows for analysis of
covariance when the response and covariate are not colocated.
Key words: Covariance Analysis, Spatial Analysis, Cokriging, Covariate.
a Assistant
Professor. E-mail: THooks@winona.edu
E-mail: DMarx1@unl.edu
c Professors. E-mail: SKachman1@unl.edu
d Geneticist and Professor. E-mail: JPedersen1@unl.edu
e Researcher. E-mail: REigenberg2@unl.edu
b Professors.
95
96
Tisha Hooks, et al.
Resumen
Los avances en agricultura de precisión permiten a los investigadores
obtener datos con más frecuencia y en detalle. Por ejemplo, es común colectar “en el transcurso” datos como lecturas de electro-conductividad del suelo.
Esto crea la oportunidad de usar estas medidas como covariables para incrementar la precisión experimental de la variable de respuesta. Aún más,
estas medidas están espacialmente relacionadas entre sí, creando la necesidad
de métodos en los cuales la ubicación espacial representa un papel explícito
en el análisis de los datos. Se analizan conjuntos de datos que contienen
variables de respuesta y covariables espacialmente relacionadas, usando el
método cokriging o el análisis espacial de covarianza. Aunque el método cokriging usa la estructura de correlación de la covariable, es una herramienta
puramente predictiva. Alternativamente, el análisis espacial de covarianza
permite la estimación de parámetros pero sin tener en cuenta la estructura
de correlación de la covariable. El presente artículo propone un método que
tiene en cuenta la correlación en la covariable, así como la correlación entre
la covariable y la variable de respuesta, permitiendo la estimación de los
parámetros del modelo. De la misma manera, este método permite el análisis espacial de covarianza cuando la variable de respuesta y la covariable no
están colocalizadas.
Palabras clave: análisis de covarianzas, covarianza espacial, cokriging, covarianza.
1. Introduction
With recent advances in precision agriculture, researchers are now able to capture data more frequently and in more detail. For example, it is typical to collect
“on-the-go" data such as soil electrical conductivity readings. This creates the
opportunity to use these measurements as covariates for the primary response variable to possibly increase experimental precision. Moreover, these measurements
are also spatially referenced to one another, creating the need for methods in which
spatial locations play an explicit role in the analysis of the data. These covariates
are usually measured before the treatment is applied and hence the assumption
that the covariate is not affected by the treatment is met.
A standard framework for the analysis of spatial data considers a response
variable, Y (s), which is in principle obtainable at any location, s, within a twodimensional spatial region R. Let these spatial locations be indexed by site si .
Data values ysi = yi are obtained from locations si , i = 1, 2, . . . , n, and are
assumed to follow the model
Y (si ) = µ(si ) + e(si ),
i = 1, 2, . . . , n
where Y (si ) = Yi is the response variable, µ(si ) is the deterministic trend, and
e(si ) is a stochastic error term with some spatial covariance structure (Dubin
1988). Applications of this model fall into two broad categories: spatial prediction
problems and estimation problems. In spatial prediction problems, the objective
is to predict the value of the response variable at some arbitrary location, s0 ,
Revista Colombiana de Estadística 31 (2008) 95–109
Analysis of Covariance with Spatially Correlated Secondary Variables
97
given the data y = (y1 , y2 , . . . , yn ). In estimation problems, interest centers on
estimating model parameters such as treatment effects. A thorough discussion of
methods applicable to these types of problems can be found in texts such as Journel
& Huijbregts (1978), Isaaks & Srivastava (1989), Cressie (1993), and Goovaerts
(1997).
Often, data collected on the response variable are supplemented by additional
information collected on the covariates. The model given above can be extended to
include the covariates. In spatial prediction problems, the joint data are analyzed
using the cokriging approach (Goovaerts 1997). In this case, the model can be
expressed as
Y1 (s1i ) = µ1 (s1i ) + e1 (s1i ),
i = 1, 2, . . . , n1
Y2 (s2i ) = µ2 (s2i ) + e2 (s2i ),
i = 1, 2, . . . , n2
where Y1 (s1i ) is the response variable, Y2 (s2i ) is the covariate, µ1 (s1i ) and µ2 (s2i )
represent deterministic trend, and e1 (s1i ) and e2 (s2i ) are random error terms with
some spatial covariance structure. This approach is most advantageous when Y2
is more densely sampled than the response variable. Cokriging explicitly accounts
for spatial cross-correlation between the response and secondary variable in that
e1 (s1i ) and e2 (s2i ) have a spatial correlation structure and a cross-correlation between them. Incorporated into the procedure is the usual restriction that the covariance matrix associated with the cross-covariogram be positive definite (Isaaks
& Srivastava 1989). Ordinary cokriging then predicts Y1 (s0 ) using information
from the data Y1 (s1i ) and the covariate Y2 (s2i ). Universal cokriging allows for a
trend or large scale structure in the prediction equations under the assumption of
coregionalization. However, in order for the covariate to have any influence on the
estimation of the large scale structure parameters, a restriction on the parameter space that jointly involve parameters in the response and covariate variables
must be made (Helterbrand & Cressie 1994). Making such a restriction, such as
assuming the mean of the response variable and covariate are equal, it is often too
restrictive to make universal cokriging of any practical use for estimation. Universal cokriging can also be used as a regression procedure (Stein & Corsten 1991).
While the cokriging approach is an extremely useful geostatistical tool, it is used
only in prediction problems and does not easily allow for the estimation of treatment effects.
In spatial estimation problems where interest centers on estimating model
parameters or testing for treatment differences, data consisting of measurements
on a response variable and a covariate are usually analyzed using a spatial analysis
of covariance. Analysis of covariance methods use information about the response
variable that is contained in another variable in order to improve estimation, and
a detailed description of this tool can be found in Searle (1971) and Milliken &
Johnson (2002). The basic analysis of covariance model is as follows:
yij = αi + βi xij + eij
where i = 1, 2, . . . , t, j = 1, 2, . . . , ni , the mean of yi for a given value of X is αi +
βi X and eij ∼ i.i.d. N (0, σ 2 ). Note that this model has t intercepts and t slopes
Revista Colombiana de Estadística 31 (2008) 95–109
98
Tisha Hooks, et al.
and thus represents a collection of simple linear regression models with a different
model for each level of the treatment. If equal slopes are assumed, the model used
to describe the mean of y as a function of the covariate is
yij = αi + βxij + eij
where eij ∼ i.i.d. N (0, σ 2 ) (Milliken & Johnson 2002).
If the assumption on the error term is relaxed and eij is alternatively assumed
to have some spatial covariance structure, the model becomes a spatial analysis
of covariance model (Cressie 1993, Zimmerman & Harville 1991). In this case,
the analysis uses information from both the response variable and the covariate
in order to obtain more accurate parameter estimates. Dubin (1988) proposed
this approach for spatially autocorrelated error terms. However, this method accounts for only the spatial correlation that exists in the response variable. If the
covariate has a spatial correlation structure and a cross-correlation with the response variable, the analysis does not take these spatial correlation structures into
account.
Consider the case where the primary attribute of interest (response variable)
and a secondary variable (covariate) possess some spatial structure, and assume interest lies in estimating treatment effects. For example, consider the situation and
an agronomic trial is conducted where the field fertility contains spatial structure.
The experimental design of the study can be any appropriate classical design, but
the appropriate analysis will include a spatial component (Marx & Stroup 1993).
If soil testing of the area has been conducted recently, then the soil fertility can
be more accurately estimated using these measurements. There will generally be
fewer soil sample locations than plots and these will not be colocated with the
centers of the plots. A hypothetical schematic is given in Figure 1, where there
is a 6 × 6 arrangement of plots with 20 gridded soil sample locations throughout
the field. In another example, a recent study conducted at the U.S. Meat Animal
Research Center in Clay Center, Nebraska, compared a cover crop treatment and
a no-cover crop treatment for values of ammonia nitrogen. Also, soil electrical
conductivity readings were used as a covariate. In this situation, the response
variable and a covariate possessed a spatial structure. These spatial correlations
and the cross-correlation structure that exists between them need to be included
in the analysis as it is in the cokriging approach, but parameter effects need to
be estimated as in spatial analysis of covariance. The objective of this work is
to develop a model that accounts for the correlation in and between the response
and covariate and allows for the estimation of model parameters. In the following pages, the proposed model and methodology are described, the analysis of a
simulated data set in which the response and covariate are colocated is presented
(colocated data occur when the response variable and the covariate are measured
at the same geographic coordinates), the methodology is applied to a simulated
data set in which the data are not colocated; and the methodology is used to
analyze a soils data set obtained from the U.S. Meat Animal Research Center.
Revista Colombiana de Estadística 31 (2008) 95–109
Analysis of Covariance with Spatially Correlated Secondary Variables
99
Figure 1: Hypothetical agronomic field trial where additional information is available
through soil test samples.
2. Model and Methods
For colocated data, the model can be expressed as
y = Xτ + βu + ey
(1)
u = 1µ + eu
where y is an n × 1 vector containing the measurements of the observed y(si )’s at
sites in Syu , u is an n × 1 vector containing the covariate observations u(si )’s at
sites in Syu , X is an n × p design matrix for treatment effects, τ is a p × 1 vector
of treatment effects, β is the regression coefficient for the covariate, 1 is an n × 1
vector of 1’s, and µ represents the mean of the covariate. Also, the model assumes
ey ∼ N (0, σy2 R)
and
eu ∼ N (0, σu2 R)
where R represents a spatial correlation structure (e.g., spherical, gaussian, or
exponential)(Cressie 1993). It can be seen that
E(y) = E(Xτ + βu + ey ) = Xτ + βE(u) = Xτ + β1µ
E(u) = E(1µ + eu ) = E(1µ) = 1µ
Also, since the term βu in equation (1) can contain any correlation between y
and u, it can be assumed that Cov(ey , eu ) = 0. Then,
V ar(y) = V ar(Xτ + βu + ey ) = β 2 V ar(u) + V ar(ey ) = β 2 V ar(eu ) + V ar(ey )
V ar(u) = V ar(1µ + eu ) = V ar(eu )
Cov(y, u) = Cov(Xτ + βu + ey , 1µ + eu ) = Cov(Xτ + β(1µ + eu ) + ey , 1µ + eu )
= Cov(βeu + ey , eu ) = Cov(βeu , eu ) + Cov(ey , eu ) = βV ar(eu )
Revista Colombiana de Estadística 31 (2008) 95–109
100
Tisha Hooks, et al.
Finally, the model assumptions can be written as
y
∼N
u
Σ
Xτ + β1µ
, yy
Σuy
1µ
Σyu
Σuu
!
where
Σyy = β 2 Σuu + Σyy = (β 2 σu2 + σy2 )R
Σuu = V ar(eu ) = σu2 R
Σyu = βΣuu = βσu2 R
thus,
y
∼N
u
2 2
(β σu + σy2 )R
Xτ + β1µ
,
βσu2 R
1µ
!
βσu2 R
σu2 R
(2)
For the univariate case, the variance of the response variable is defined as
κ2y = V ar(yi ) = β 2 σu2 + σy2
(3)
and the variance for the covariate is defined as
κ2u = V ar(ui ) = σu2
(4)
κyu = Cov(yi , ui ) = βσu2
(5)
Also,
Let ρ represent the correlation between the covariate and response variable.
The relationship between ρ and β can be described as follows:
κyu
κyu
= 2
σu2
κu
Cov(yi , ui )
κyu
=⇒ κyu = ρκy κu
ρ= p
=
κy κu
V ar(yi )V ar(ui )
κyu = βσu2 =⇒ β =
thus,
ρκy κu
ρκy
=
2
κu
κu
ρ̂κˆy
β̂ =
κˆu
β=
(6)
Note that κy is also the square root of the sill for the response and κu is the
square root of the sill for the covariate. The covariance matrix in (2) can easily be
reparameterized to have just one error term. Since, in our situation, we have two
variables, we chose to have two error terms rather than just one and the other error
term then being a proportionality constant times the first error term. This is in
agreement with Oliver (2003) (equations (3), (4), and (5)). Additionally note that
Revista Colombiana de Estadística 31 (2008) 95–109
Analysis of Covariance with Spatially Correlated Secondary Variables
101
Oliver constructs the cosimulation by using independent Gaussian random variables which again implies that the assumption of zero covariance is made without
loss of generality.
This model notation can be extended to non-colocated data (Banerjee
et al.
S
2004). Let y be the vector of observed y(si )’s at S
the sites in Syu Sy and u be
the vector of observed u(sj )’s at the sites in Syu Su . Let y ′ denote the vector
of missing y observations in Su and u′ denote the vector of missing u observations
in Sy . Then, the vectors for the response and covariate observations from the
previous discussion can be replaced with the augmented vectors yaug = (y, y ′ ) and
uaug = (u, u′ ). After permutation to line up the y’s and u’s, they can be collected
y
yaug
of equation (2).
which is analogous to the vector
into a vector
uaug
u
A program which models the covariance structure in equation (2) was written in
R (R Development Core Team 2004). The program uses generalized least squares
and yields restricted maximum likelihood (REML) estimates of the range, the
sills for the response variable and for the covariate, treatment effects, and the
correlation between the response and the covariate. Also, the results give the F value for testing for an overall difference in treatment effects, and an approximate
z-test is constructed using the asymptotic variance of ρ to test whether or not ρ
(and equivalently β) is significantly different from zero. The denominator degrees
of freedom for the F -test can be found by subtracting the number of fixed effects
from the number of observations on the response variable. Finally, an estimate for
β can be found using the relationship between β and ρ given in equation (6). The
program, simulated data sets, and soils data set may all be obtained from the first
author.
3. Example Using Simulated Colocated Data
The simulated data set consisted of 20 replications of five treatments. The
treatments were laid out in a completely randomized design on a 10 × 10 arrangement of plots. For each of the 100 points, both a spatial floor (Y ) and a
spatial covariate (X) were generated using the method of gaussian cosimulation
(Oliver 2003).
In this example, the spherical covariance function was used for the construction
of both variables. The function is as follows:
(
σ 2 {1 − 32 ( ha ) + 12 ( ha )3 }, if 0 ≤ h ≤ a;
C(h) =
0,
if h > a.
where h is the distance between observations, a is the range of the corresponding
spherical semivariogram, and σ 2 is the sill of the semivariogram. The response
variable was simulated with a range of 8 and a sill of 10, and the covariate was
simulated with a range of 8 and a sill of 1. Two different correlations of ρ = 0.3
and 0.8 were simulated when modeling the cross-covariance between the spatial
floor and the covariate. Finally, the mean of both the response variable and the
Revista Colombiana de Estadística 31 (2008) 95–109
102
Tisha Hooks, et al.
covariate was simulated to be 10, and treatment effects were generated with the
treatment vector τ = (−0.4, −0.2, 0, 0.2, 0.4).
The data were analyzed in three ways: using a nonspatial analysis of covariance, a spatial analysis of covariance, and the proposed analysis which accounts
for the spatial structure of both the response and the covariate. The results are
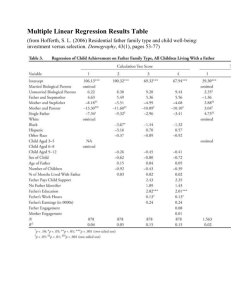
summarized in Tables 1-3. The spatial analyses yield more accurate estimates
than the analysis which disregards the location of the observations. As displayed
in Tables 1 and 2, the estimate for β is much closer to the simulated value and
the average standard error of treatment differences is much smaller for the spatial
analyses. Also, the estimates for σy2 and for the regression coefficient for the
covariate are very close for the proposed method and spatial analysis of covariance. While the proposed method provides the most accurate representation of the
treatment effects as seen in Table 2, it is observed that the average standard error
of treatment differences is very close to that obtained from a spatial analysis of
covariance. Table 3 shows the results of hypothesis tests for treatment differences
and significance of the covariate for all three analyses. Overall, the results from the
proposed method are very close to the results obtained from the spatial analysis
of covariance, and it appears that little is gained when accounting for the spatial
structure of the covariate when the response and covariate are colocated. The same
results occur for ρ = 0.3 as seen in Table 4. In addition to the proposed method
giving a slightly smaller average standard error of treatment differences, the least
square means were slightly closer to the simulated data means (not shown). Thus,
one may choose to use a simple spatial analysis of covariance to test for treatment
differences since the precision that is gained via use of the proposed method may
not be worth the extra effort that is required.
Table 1: Parameter estimates from the three analyses conducted in the example using
simulated colocated data with ρ = 0.8.
Parameter of
Interest
Range
sill for response
sill for covariate
β
Analysis of
Covariance
–
–
–
3.37
Spatial Analysis
of Covariance
8.74
4.31∗
–
2.33
Proposed
Method
8.61
9.73
1.02
2.33
Simulated
Data
8.00
10.00
1.00
2.53
∗ Note that 4.31 actually represents σ̂ 2 , whereas the sill for the response is κ2 .
y
y
For the proposed method, σ̂y2 is calculated to be 4.20 using equation (3). The
simulated σy2 is 3.60.
However, this is not to imply that the proposed method is not extremely useful.
The true strength of this analysis is more than simply accounting for the spatial
structure of the covariate. Its power lies in the fact that this model allows for
covariate measurements that are not colocated with measurements of the primary
attribute of interest. An example which considers a data set in which the covariate
is measured at locations different from the response variable is presented in the
next section.
Revista Colombiana de Estadística 31 (2008) 95–109
103
Analysis of Covariance with Spatially Correlated Secondary Variables
Table 2: Least squares means for treatments and average standard errors of treatment
differences from the three analyses conducted in the example using simulated
colocated data with ρ = 0.8.
LSMEAN for
treatment 1
LSMEAN for
treatment 2
LSMEAN for
treatment 3
LSMEAN for
treatment 4
LSMEAN for
treatment 5
Average standard
error of treatment
differences
Analysis of
Covariance
Spatial Analysis
of Covariance
Proposed
Method
Simulated
Data
10.860
10.150
9.980
9.6
10.670
10.290
10.110
9.8
10.930
10.460
10.280
10.0
11.580
10.780
10.610
10.2
11.560
10.720
10.540
10.4
0.478
0.260
0.255
–
Table 3: Results of hypothesis tests for treatment differences and significance of the
covariate from the three analyses conducted in the example using simulated
colocated data with ρ = 0.8.
test for treatment
differences
test for significance
of covariate
Analysis of
Covariance
F = 1.54
(p -value = 0.1976)
F = 327.90
(p -value < 0.0001)
Spatial Analysis
of Covariance
F = 1.75
(p -value = 0.1448)
F = 119.57
(p -value < 0.0001)
Proposed
Method
F = 1.20
(p -value = 0.3174)
z = 3.668
(p -value < 0.0001)
Table 4: Parameter estimates from the three analyses conducted in the example using
simulated colocated data with ρ = 0.3.
Parameter of
Interest
Range
sill for response
sill for covariate
β
Average standard
error of treatment
differences
Analysis of
Covariance
–
–
–
1.220
0.778
Spatial Analysis
of Covariance
7.220
6.320∗
–
0.800
0.333
Proposed
Method
10.05
9.40
0.90
0.83
0.328
Simulated
Data
8.00
10.00
1.00
0.95
–
∗ Note that 6.32 actually represents σ
by2 whereas the sill for the response is κ2y .
4. Example Using Simulated Non-Colocated Data
The simulated experiment consisted of twenty replications of five treatments.
The treatments were laid out in a completely randomized design on a 10 × 10 arrangement of plots. Within each plot, another 3×3 grid of points was constructed.
For each of the 900 points, a spatial floor (Y ) and a spatial covariate (X) were
generated using the method of gaussian cosimulation (Oliver 2003).
Revista Colombiana de Estadística 31 (2008) 95–109
104
Tisha Hooks, et al.
The spherical covariance function was used for the construction of both variables. The response variable was simulated with a range of 25 and a sill of 5,
and the covariate was simulated with a range of 25 and a sill of 1. Correlations
of ρ = 0.8 and 0.3 were simulated when modeling the cross-covariance between
the spatial floor and the covariate. Finally, the mean of the response variable
and the covariate was simulated to be 10, and treatment effects were generated
with the treatment vector τ = (−0.5, −0.25, 0, 0.25, 0.5). These treatment effects
correspond to treatments A, B, C, D, and E, respectively.
The final data set was constructed as follows. First, the center observation
from each of the 100 plots was used as the response variable. Then, 25 of the 100
remaining responses were randomly chosen and deleted from the data set. Finally,
90% of the 900 covariate observations were randomly selected and deleted. A
representation of this simulated data set is given in Figure 2. For each plot, the
treatment (A, B, C, D, or E) is identified, and the black squares represent the
location of the sampled covariates. Clearly, the response and the covariate are
not colocated in this example, and there are more covariate observations than
measurements on the response.
Figure 2: Simulated data set in which the response and covariate are not colocated.
Treatments A-E for each plot and the locations of the sampled covariates are
identified.
The data were first analyzed using the proposed analysis which accounts for the
spatial structure of both the response and the covariate. In order to compare this
to a spatial analysis of covariance, a data set in which the response and covariate
were colocated was constructed by using the covariate measurement which was
closest to the central observation of each plot as the covariate for that plot. If
two covariate measurements were equally distant from the center of the plot, the
covariate was calculated as their average. A spatial analysis of covariance was then
conducted using this data set. The results from both analyses are summarized in
Tables 5-7.
Revista Colombiana de Estadística 31 (2008) 95–109
Analysis of Covariance with Spatially Correlated Secondary Variables
105
Table 5: Parameter estimates from the three analyses conducted in the example using
simulated colocated data with ρ = 0.8.
Parameter of
Interest
Range
sill for response
sill for covariate
β
Spatial Analysis
of Covariance
27.43
4.75∗
–
0.83
Proposed
Method
27.35
5.16
1.02
2.02
Simulated
Data
25.00
5.00
1.00
1.79
∗ Note that 4.75 actually represents σ
by2 , whereas the sill for the res-
ponse is κ2y . The simulated σy2 is calculated to be 1.80 using
equation (3).
Table 6: Least squares means for treatments and average standard errors of treatment
differences from the three analyses conducted in the example using simulated
colocated data with ρ = 0.8.
LSMEAN for
treatment 1
LSMEAN for
treatment 2
LSMEAN for
treatment 3
LSMEAN for
treatment 4
LSMEAN for
treatment 5
Average standard
error of treatment
differences
Spatial Analysis
of Covariance
Proposed
Method
Simulated
Data
10.000
9.540
9.50
9.930
9.360
9.75
10.460
10.200
10.00
10.560
10.340
10.25
10.680
10.270
10.50
0.304
0.241
–
Table 7: Results of hypothesis tests for treatment differences and significance of the
covariate from the three analyses conducted in the example using simulated
colocated data with ρ = 0.8.
Test for treatment
differences
Test for significance
of covariate
Spatial Analysis
of Covariance
F = 2.10
(p -value = 0.0909)
F = 10.21
(p -value = 0.0021)
Proposed
Method
F = 6.06
(p -value = 0.0003)
z = 1.9580
(p -value = 0.0502)
As seen in Table 5, the proposed analysis yields a much more accurate estimate
of β than the spatial analysis of covariance. Moreover, the estimate of the sill
for the response variable (κ2y ) from the proposed analysis is very close to the
simulated value, but the estimate of σy2 from the spatial analysis of covariance is
considerably different from the simulated value. Also, as illustrated in Table 6, the
proposed method provides more accurate representations of most of the treatment
effects, and the average standard error of treatment differences provides a 20%
improvement over the average standard error of treatment differences from the
Revista Colombiana de Estadística 31 (2008) 95–109
106
Tisha Hooks, et al.
Table 8: Parameter estimates from the three analyses conducted in the example using
simulated colocated data with ρ = 0.3.
Parameter of
Interest
Range
sill for response
sill for covariate
β
Average standard
error of treatment
differences
Spatial Analysis
of Covariance
20.520
4.200∗
–
0.100
Proposed
Method
20.330
5.120
0.760
0.520
0.335
0.329
Simulated
Data
25.00
5.00
1.00
0.67
–
∗ Note that 4.20 actually represents σ
by2 whereas the sill for the
response is κ2y .
spatial analysis of covariance. The F -statistic is larger for the proposed analysis
(Table 7), but there is no reason to expect that the F -statistics be comparable
since the proposed method uses all covariate points and the spatial analysis of
covariance uses a subset of the covariates. Moreover, some covariate points are used
twice in the spatial analysis of covariance. A relatively large correlation between
the response variable and covariate was chosen so that the analysis of covariance
would be very effective. To see if the results held for a smaller correlation, a
correlation of ρ = 0.3 was simulated using the same parameters (range, sill for
response and covariate, treatment means) as were used for the ρ = 0.8 simulation.
These results, shown in Table 8, indicate that the same trend as seen for ρ = 0.8
continue, but with a lesser effect. The average standard error of the treatment
differences was reduced slightly using the proposed method compared to spatial
analysis of covariance and the least square means were slightly closer to the simulated means using the proposed method (data not shown). Our conclusion is
that the stronger the association between the response variable and covariate the
greater the improvement by using the proposed method. The true strength of the
proposed method is that even the spatial analysis of covariance should not be run
except when the data are colocated. Our proposed method is the only procedure
which allows for non-colocated data and tests of treatment effects.
5. Example Using Actual Field Data
These data were obtained from a study site located at the U.S. Meat Animal
Research Center located in Clay Center, Nebraska. The site was treated with four
replications of a winter wheat cover crop versus four replications of a no-cover
crop. The treatments were laid out in a randomized complete block design with
subsampling. The response variable consisted of ammonia nitrogen levels obtained
from soil cores. Also, soil electrical conductivity measurements taken prior to
treatment application were used as a covariate, and the geographical coordinates
(northing and easting) were recorded at each measured location. A representation
of this data set is given in Figure 3.
Revista Colombiana de Estadística 31 (2008) 95–109
Analysis of Covariance with Spatially Correlated Secondary Variables
107
Figure 3: Soils data set in which the response and covariate are not colocated. Locations where soil cores were taken for the response are marked by diamonds,
and locations of the sampled covariates are also identified.
The proposed method was used for the analysis since the response variable and
covariate are not colocated. The data were analyzed as a randomized complete
block design with subsampling and fixed block effects. A spherical covariance
structure was assumed, and the results are summarized in Tables 8-10.
All parameter estimates in Table 9 are reasonable. Table 10 shows the least
squares means of the two treatments and the standard error of their difference.
Finally, as shown in Table 11, there does not appear to be a difference in values
of ammonia nitrogen for cover crop treatment versus a no-cover crop treatment.
Moreover, the covariate (soil electrical conductivity) is not significant.
Table 9: Parameter estimates from the soils data example.
Parameter of
Interest
Range
sill for response
sill for covariate
β
Estimate from
Proposed Method
12.0600
0.9800
43.5100
−0.0350
Revista Colombiana de Estadística 31 (2008) 95–109
108
Tisha Hooks, et al.
Table 10: Least squares means for treatments and average standard errors of treatment
differences from the analysis of the soils data.
Estimate from
Proposed Method
LSMEAN for
treatment 1
LSMEAN for
treatment 2
Standard error of
treatment difference
3.9900
3.8800
0.2032
Table 11: Results of hypothesis tests for treatment differences and significance of the
covariate from the analysis conducted in the soils data example.
Test for treatment
differences
Test for significance
of covariate
F = 0.55
(p -value = 0.5125)
z = −0.75
(p -value = 0.4532)
6. Discussion and Conclusion
First of all, it should be noted that the proposed method and analysis make
the usual intrinsic coregionalization assumption of equal ranges for the response
variable and the covariate. The model could be altered to allow for this assumption
to be relaxed, but the current framework assumes the correlation matrices are
proportional to each other.
Also, the results from the proposed method and the spatial analysis of covariance are very similar when the data are colocated. It appears that little is gained
by accounting for the correlation structure of the covariate, and if the response
variable and the covariate are colocated, the authors recommend analyzing the
data using a simple spatial analysis of covariance. In fact, when data are colocated,
the results are a generalized form of the error-in-variables problem. However, the
true power of this analysis lies in the fact that the model allows for analysis of
covariance when data are not colocated. Using this method, it is not essential that
the covariate and response be measured at the same location or to have the same
number of observations for the response and covariate. Although a data set in
which the response and covariate are not colocated can be manipulated so that a
spatial analysis of covariance is appropriate, more than one choice for a covariate
exists. For example, the covariate could have been constructed by averaging all
covariate measurements within a plot, had the data allowed for this. The construction of the covariate is arbitrary and has not statistical validation associated with
it; thus, using the proposed method is superior to the alternative. The spatial
locations of the covariates play a role in the proposed analysis, and this leads
to more precision in testing for treatment differences. Lastly, and perhaps most
importantly, experiments where the covariate is available at fewer locations than
Revista Colombiana de Estadística 31 (2008) 95–109
109
Analysis of Covariance with Spatially Correlated Secondary Variables
the primary variable still allows this supplementary information to be incorporated
into the analysis.
Recibido: noviembre de 2007 — Aceptado: mayo de 2008
ˆ
˜
References
Banerjee, S., Carlin, B. P. & Gelfand, A. E. (2004), Hierarchical Modeling and
Analysis for Spatial Data, Chapman and Hall/CRC Press.
Cressie, N. A. C. (1993), Statistics for Spatial Data, John Wiley and Sons, Inc.,
New York, United States.
Dubin, R. (1988), ‘Spatial Autocorrelation’, Review of Economics and Statistics
70(466-474).
Goovaerts, P. (1997), Geostatistics for Natural Resources Evaluation, Oxford University Press, New York, United States.
Helterbrand, J. D. & Cressie, N. (1994), ‘Universal Cokriging Under Intrinsic
Coregionalization’, Mathematical Geology 26(2), 205–226.
Isaaks, E. H. & Srivastava, R. M. (1989), An Introduction to Applied Geostatistics,
Oxford University Press, New York, United States.
Journel, A. G. & Huijbregts, C. J. (1978), Mining Geostatistics, Academic Press,
New York, United States.
Marx, D. & Stroup, W. (1993), Analysis of Spatial Variability using PROC
MIXED, in ‘Proceedings of the 1993 Kansas State University Conference of
Applied Statistics in Agriculture’, pp. 40–59.
Milliken, G. A. & Johnson, D. E. (2002), Analysis of Messy Data, Volume III:
Analysis of Covariance, Chapman and Hall/CRC Press.
Oliver, D. S. (2003), ‘Gaussian Cosimulation: Modeling of the Cross-Covariance’,
Mathematical Geology 35, 681–698.
R Development Core Team (2004), R: A Language and Environment for Statistical
Computing, R Foundation for Statistical Computing, Vienna, Austria. ISBN
3-900051-07-0.
*http://www.R-project.org
Searle, S. R. (1971), Linear Models, John Wiley and Sons, Inc., New York, United
States.
Stein, A. & Corsten, I. C. A. (1991), ‘Universal Kriging and Cokriging as a Regression Procedure’, Biometrics 47, 575–587.
Zimmerman, D. L. & Harville, D. A. (1991), ‘A Random Field Approach to the
Analysis of Field-Plot Experiments and other Spatial Experiments’, Biometrics 47, 223–239.
Revista Colombiana de Estadística 31 (2008) 95–109