MITLibraries
advertisement

MITLibraries
Document Services
Room 14-0551
77 Massachusetts Avenue
Cambridge, MA 02139
Ph: 617.253.5668 Fax: 617.253.1690
Email: docs@mit.edu
http://libraries.mit.edu/docs
DISCLAIMER OF QUALITY
Due to the condition of the original material, there are unavoidable
flaws in this reproduction.
We have made every effort possible to
provide you with the best copy available. If you are dissatisfied with
this product and find it unusable, please contact Document Services as
soon as possible.
Thank you.
Antibody-Antigen Assay Design for Comnbinled
Optical Tweezers and Fluorescence
by
Jeniny Ta
Sitinnitted to the Departlment of Mechanical Engii ierin,
il p)artil fulfillmnent of thle requirernents
for the (leree of
Bachelor of Science i Mechanical Engineering
at the
MASSACHUSETTS INSTITUTE OF TECHNOLOGY
June 2004
(®)Massachusetts Institute of Technology 2004. All rights reserved.
~~~~~
Autho:r. A uthw
.
...................
'4 ..............................
. . . . . . . . . .;,
/.....
Department of Mechanical Engineering
Certifiedby.......
May 7, 2004
. ...
. .
.
-
-.--
h
Matthew
. .
.
-a
J. Laltlg
ig
Assistant Professor of Mechanical Engineering and Biological
Engineering
Thesis Supervisor
Accepted
by..........
....... ........-
......................
Ernest
G. Cra-vallho
Chlairmtn, Undergraduate Thesis Connnittee
MASSACHUSETTS INSTITUTE
OF TECHNOLOGY
OCT 2 8 2004
L B AR..i ,
. I
C
-6 I'$.
,1.AR,00F
Anttibody-Antigeti
Assay De,,,ign for Conlbined Optical
Twezers ad Fluorescence
})I
Submitted to lie Depart nwt I Mlechanic;al Etlgineering
on l\Iay 7. '2004, i prtial fulfillment of the
re(liiiremen-ts fo tle degree of
Bachelor 4t'Science in M[ecllanical Eng(ineering
Abstract
The re:ent development in combined optical trapping and fluorescence technology
lpomises to enable unbinding force st cli's
f receptor-liganld interactions, whose
specificity play a crucial role in the function of many biological systems. This thesis
tocuses on the development of assay designs for the study of antibody-anltigen binding
interactions llsing combined optical trappillg and single mnoleculefluorescence. The
assavs ,:reate the necessary linkage geometry between the antibody-anitigen system
un(der study to an optically trapped b)eadl.enabling force probing of the antibodyantigen b-inding interaction. In particular. two tether materials and fluorophores
were stl(ied: polyethylene glvc(-ol(PEG) with Cy-2. and (lsDNA with fluorescein.
\We (deilnolistratetether formtion in the dsDNA-fluorescein anitibody-antigen linkage
systeinmwith preliminary optical trapping Ldata.
Thesis Supervisor: Matthew .J. Lang
Title: Assistant Professor of Mechanical
l
Engineering and Biological Engineering
Acknowledgments
I would ike t(, hanik PrOfessm, Matthew J. Lang fr his supervision thr(llghout
p)roject a( i(
assays.
itrdoBra
f
training inc ill the miuch needed skills to I)epl)are my
I xoul( also like to thank all the other
throlghout
this
neibl)ers of the lab for guiding me
my work on this p)r ).ect.
In addit ion. I would( like t
tank
Mom and(lDad for their unwavering support for
my education. I would not have gotten this far without them.
Contents
1
Introduction
1.1
Ligand-Recelp)tol Interaction
..............
1.2
Optical Trapping and Fluorescence
1.3 Linkage Design ....................
.
.....
11
. . . . . . . . .
. . . . . . . . .
. . . . . . . . .
2 Materials and Methods
2.1
Polyethlyene
11
12
13
19
Glycol Linkage ..............
. . . . . . . . .
19
. . . . . . . . . . . . . . . .. . . . . . . . .
19
. . . . . . . . . . . . . . . .
20)
2.1.1
Bead Preparation
2.1.2
Bead-PEG
2.1.3
PEG-Biotin to Streptavidin-Cy2 Conjugation
Conjugation
Reaction
. . . . . . . . .
20)
2.2
DNA Tethers
. .. ... .. ... . ... .. .. .... . . . . . . . .
21
2.3
U V vis . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
23
3 Results
25
3.1
BSA Blocking Agent
3.2
DNA tethers
. . ..
4 Discussion
4.1
BSA concentration
.
4.2
Tether Length . . . .
4.3
Conclusion . . . . . .
...........................
...........................
...........................
......... ........... I.....
...........................
A DNA Tether Protocol
A.1
Maiterials
......
725
27
31
31
:31
32
35
..........................
I
35.
A.2
I'twedun . . . . . . . .
. . . . . . . . . . . . . . . . .
()
List of Figures
1-1 Th( experimental setup .........................
..........
.
13
1-2 Setup of optics for combined optical trapl)ilng and fluorescence detecti(ol[1] ....................................
14
1-3 A schematic of the variable components of the linkage system
1-4
Experimental
flow chart
.
....
.
15
. . . . . . . . . . . . . . . . . . . . . . .
17
2-1 Schematic of Cy2-PEG-bead complex .........................
.
19
2-2 Diagram of a, flow cell contructed out of a standard microscope slide,
2-3
etched coverslip, and double-sided sticky tape ..............
21
Schemnatic of DNA linkage system .....................
22
2-4 Gel of PCR p)roduclllts
used to make DNA-bead complexes.......
.
23
.
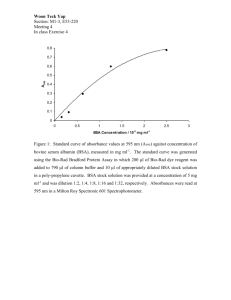
24
2-5 Plot of avidin bead (0.52imndiameter) con(centlrationand percent transmittance at 500 nnm
......................................
2-6 Plot of absorbance as a function of avidin bead (0.52pmndiameter)
concentration
as dletermined by the Beer-Lambert
Law .........
24
3-1 A plot of bead position response in voltage, as the stage is translated
in the x-axis across the center of the bead ................
29
3-2 A plot of bead position response in voltage, as the stage is translated
in the y-axis across the center of the bead ......................
.
29
3-3 Exampt)leof data profile collected from a bea(l stuck to the surface of
the slide...................................
1-1 Diagram of a tethered bead. (Not (lrawnllto scale.) ...........
30
32
8
List of Tables
3.1
Study of bead binding to suilface of (()verslil) with varying (oncentlations of BSA coverage ill the flow channel and no BSA pre-incubated
with the bead ...........................................
3.2
(i
Bead binding study with varying conlcelltration of BSA incubated with
beads and a fixed 2 mg/ml BSA inl flow channel ..
. . . . . . . . . . .
'
3.3 Bead binding study with fixed BSA-bead incubation of 2 mg/mL BSA
and varying concentration in flow cell.................
()
.
2.......
27
[()
Chapter
1
Introduction
This thesis project aims to develop preliminary assays that create the necessary geometry needed to study antibody-antigen binding interaction in combined optical
trapping and fluorescence studies. Type types of linkages, DNA and polyethylene
glycol (PEG), were tested along with two types of fluorophores, Cy2 and fluoresceill.
1.1 Ligand-Receptor Interaction
Recept(:)or-ligandinteractions an(l their specificity play a crucial role in tlie finin(tio(l
of many biological systems.
For example, information
transfer in a cell is aclhieve(1
by the binding of receptors to ligands. There are a wide variety of receptor-ligan(
interactiolns sch
as antil)bO(dyV-antigenbinding and hormnone-membrane p)rotein )ind-
ing. Growth factors utilize receptor-ligand interactions to regulate (ell reproduc(tion
and differentiation. Somnetypes of viruses begin attacking cells by binding to carbohydrates on the host cell's surface. The ability to study the miechlanicsof single
receptor-ligand bonds opens the door to understanding the nature of' aln hiological
systems. To this (late, however, mIost optical tweezers studies on bin(lding iteract
ions
have fo,:used on nicleotides and miuch remains to be developed for protein iternitions.
11
1.2
Optical Trapping anl Fluorescence
ltsedl opt i
\\,
ti t(,frei',
i
lI tral)pinig (also knmwll as )
i(al tw (/,zers ,L'laser taI))ing)
ivolved ill certain atil)()(l-an ti'i
pressll
( from a tightly
focusedt I ser 1
(I a l)eacidwith a (liameter between
iite'aeti(ms. Optic-al tilaipsuse radlian t.
Itap
i
) rt
icles. The las(r i fo(utse (I
).5 to '2tII. alol is Lse t() Il'asilt'e
I.1 to l()OpNrange with one nanoineter
t(1 res{olve
force' i
the
1('s,,ltioii.
The ultimate goal of the assay design is t() create ainex)erilnental
setiup suitable for
-olnll)inedoptical trapping and single-miolecile fluorescence studies of ligand-receptor
interactions. I the past, the two methodls 4f monitoring protein (confornmationusing
fluorescence and probing the protein withl an external force have )eell separated
in time andl space. However, recent advances in instrunientation have developed
a miethod of combining single-molecule fluorescence and optical trapping.
In single-fluorophore detection, a molecule is usually illuminated by a laser, and
the resulting emitted fluorescence is collected by an objective lens. The objective
lens is of a high numerical
aperture
and is coupled to (letectors such as a cooled
charge-coupled device (CCD), an image-intensified camera, avalanche p)hotodiode, or
a I)llphotoilllultiI)lier
tube. It has been d(emolstrated b)yLang et al.[1] that with careful
{)tical
esign and an appropriate choice of fluorophores. combillel otical trapping
aniidsingle-molecule
fluorescence can be used to measure forces andclsinmtltaneously
monitorthe molecular conformation. Combined single-molecule force and fluorescence
ineasurenments have been used to study the force required to split, a short length of'
DNA alid has also been used to observe the turnover of ATP by niyosinl[2]. This
technology has much potential applied to the study of ligand-receptor interactions.
This thesis aims to take a first step at develo)pingassays for studying ligand-receptors
at the single molecule level with a optical trap.
The main instrumentation used to test the tethers was a Nikon. Eclipse TE2000-U
micrIoscope modified with a three-axis piezo stage that could be controlled bv Labview
to nmove
at nlailonieter scale increments and a dete(tion branch with a position sensing
detector. Trap)ping
of the inicrospliere was (lone usilng a l()64ln
1)
I
(liode laser at 9
.\Il)s
(,I G6n111\V
. la(l d(
if,'tecti )11lasel.
(it
; u icriosIph'JI'
I
( ilgulne1- l)1
:,lation
Mlld beId movemienit was (let('(tl
sinigle lloleculc flluescen-
(
(Al,
1asCliStal('c
(ct
it, iv
xx
IIl' m ,sinig
7)14i11I I
t ; il,"i, , ';1u xvs (lone using A( ) ).
i'l;tit ye to Il(
(letectioll laser. \Vhil'
t) . as illusti t{[ iii the' opt lit, i Figure 1-2 was
lot
ar t >)(al)le to create linkag(s
i;>nvc•
t('sted ill this plm)j('ct,the IltiIInt,' goal of te
t hat canllbe use(l in SIF.
M.1rcury
Lamp
PSD
dt-ction
branch
pi ezo
stage
Figure 1-1: The experiieltal
1.3
setup.
Linkage Design
The goal of this )project is to develop a generalized linkage systeni as part of a coM)inecd (o)ptical tweezers a(l
fluorescence
assay to
13)
fl'ilitate
the stud(ly of antil)o(ldy-
bid
U
qkr
.
,
I
F-
a
.,
-...
..
I...I11-1 -
.
I
'kI
I :I;:S,>
1
, '
.
C_
i i
I
Zxty
y
:x / a_=se
i
:*J'
':
1,
,tl1
)1
p
,
..
...
H
..
...........
Iaser
....
.
..
...
.
....
.
...
..
Figure 1-2: Setup of optics for combined optical trapping and fluorescence detec-
tionil].
antigen binding.
While optical tweezers provide the technology needed to study
receptor-ligand mechanics at the single molecule level. further development of the
actual chemistry
that links the teclhnology to te
l)il(dilg sstem
itself is a.
lmajor
hurdle that must be surmountecd. This particular I)roject is aimed at (leveloping an
assay that links an antigen-a ntibody system to an optically trapped microsphere for
force probing. Most antibody-antigen interactions are amenable to study using such
a generalized linkage systeni because the structures coIlnmon to all antibodies can
l)e exploited in constructing the ssteml. The composition and length of te tether
linking the antigen to the ead will )beientical
for each of the (lifferent antigens.
The only conmp)onentthat must be clha.ngedis the chemistry that links te antigen to
11
tlie ttlIer to m((oiiiii(lflt(
1-3).
t
lifierenIt stlI( t lI's vari ouMs
Iate
tl alntil)((lv is i(lvpl
i
ohili ( l I V lloI
corresponding
leI
)e( 'ifiC ),iti ng td,
-s sIufI
I ( (.
ianltig'('niS atta(-ched to a mnlicrosphere by a tether.
tion is r)lobedlv atpl))vilg a '()l e using the ol)ticall
fluoro)ph()resserve( a.s
cence. Visualization
lv single
i I(e
a.!
Hlie bindinlg ilt
I c, 1. vwhile
tlli, (lill
.
tapt(l
ialkers tOl)visualizing the ssteni
xlli llile
hive
is
si,
Sirl(
'- c-
.ILt
mle('ulle l,I
uinoleculefluorescence serves as la )od ceck
linkage component of the systcim has been disrupted after a u)inding
f
xvIi( h
exler(nlil{,t.
-' \"m icrosphere
Force
\ anticen
i/
tether
.'
antibody \
\-
optical trap
-
__fluorophores
?
........
surface-modified glass coverslip
?
Figure 1-3: A schemnatic of the variable components of the linkage system.
The design criteria for this assay include a strong linkage suital)le for applying fotrce
and enablling the high-throughlput visualization of liganld-receptor binding with sinle
molecule fluorescence
(SMIF). which will allow its to see which interaction
has
>been
severed. During the force prolbing experiment, it is crucial that the receptor-ligandc
bond is the attachment.
whi(ch I)reaks when force is applied. The other linkages it I lhe
systenm must therefore b)e robst.
These linkages mnust also be straightforwamdl
n1d
easy to construct, thereby streamlining the experimental process for rapid probing of
molecular biomechanics. Relevant issues that miust be addressed include determoinlation of the suitable material, size a(d surface chem-istry of the lnicrospheres. x)loration of the tether imaterial an( length. and tpe
materials
used in this study
amId l)iocompatible
of fluorophores used. The tether
are p)olyetlhylenleglycol (PEG). which is water slIlile
and (IsDN.\.
Different fhllorophores were consiclered and chl (i.s(i
15
1,,
I()I Il tlheir
o l,';
lIt
1l (~,1vail;ibilitN-
\V'ious
I'
t
[,)i
1t
l
1)()s ',
)t
develop)inlg
Stoke's
straigltfi
s,
il.
11(1
ltil)
';- l(t aI11(deli;
(I,,con jlal-
protoco((l.
l(
ratios of ead( tr(, tetlher were tested to find the (,tftimal sult.(face(overage of
lbe.als
e
for the lkati
tl
w'Il11acteristics,
11ililg
ssteim.
and the timing of indli, i(llal stel)s il tihe linking
c(-edhi'c
were (m(liltni1 e(1. A v;ariety of miethods will 1 .
il)od-antigell
sst,'i
pxl)loredfr attlhing
the
(1 glass cover slips and subsequ ittlv linking t 1heantigen to
microsphere that can 1e op)tically trapped.
,(opient
Upon complhtii g (d'ev
iteractions
of the linkage system, tlt a tibodv-antigen binding
can he teste I b)ypulling on the microsphere aitad mneasuring
the force re-
(quiredto break the iteraition
with a procedure as outline l iII Figure 1-4. The linkage
fluorescence technology
assay developed is a crucial step in using optical tweezers land
to study ligand-receptor unbinding forces. Once the linkages are constructed, multiple
exl)eriments can )bec(ilctecd
lby repeating the experiment on the samneglass slide.
In order to make accurate force and position measurements, the position and force
sensing of the optical tweezers mnustbe calibrated. Since this technology allows for
performing
multiple unbinlding experiments
on one glass slide covered with multiple
tethered beads, a rapi(l ivestigation of these questions ciin be accomplished.
While this project f(c ses primarily on antibody-antigen systems, it can be further
developed to explore oth'er types of receptor-ligand intera(ctions. The variable nature
of this experimnenta!
phenomena.
syste'l will allow us to explore a plethora of interesting biological
The investigation
teums will elucidlate the pysical
of the forces involved in sch
mechanisms
lphenomenla.
1G
underlying
antibody-anltigen
ana.ny important
sys-
biological
Experiment
K
Aessay
ale;
5 5a '?|er
P:oter"i
LiiXa!;e
=-.
:'
Fitnd Tether
Sy stem
..
I
..
N
N-
I. I
·
.
......
-K Calibrate "
Position
Sensing
X
Center our
detector
-
o
r
v.........
--
-
.........
_
Ftuorescerce on
Apply Force
I
Record Breakagej
-T .
-
- _.
...
I Calibrate stiff ness I
Fr
E p. r .
t.
f w
Figure 1-4: Experiment a] flow chart.
I-
II.
Find n
new tether ))
Fn
18
Chapter 2
Materials and Methods
Polyethlyene Glycol Linkage
2.1
A preliminary assay for a PEG linkage was developed using Cy2 conjllgate(d Streptavidin (Rockland), amine-surface mod(lified0.5pn polystyrene bead (Bangs). and
heterofinctionalized Biotin-PEG-NHS 500() (Nektar Therapeutics)(Figure 2-1).
I
Cy2 Streptavidin
Amine modified
PS bead
Biotin-PEG-NHS
fr+'
*," f ' 11i
a
/.
0
w
Figure 2-1: Schematic of Cy2-PEG-beacl
compI)lex.
The following protocol was ldevelope(lto (create the cy2-biotin-PEG-bead
coin-
plexes.
Bead Stock Concentration:
2.1.1
2.1 e-9M1 (Bangs, 0).52/tin diameter, 0.5g,
10 e(
solids)
Bead Preparation
1. Alake 1/30 dilutions of beals
(.45e-11I\I)
ddH20 (1/1)0 dilution), then take 3/,p
1!)
)bydilution
10pL stock with 901/iL
o (1/10 dilution) andl (lilute with 70p/L
{(IH-:'i).
2. \\asli 1)eads. S)ill into pellet witl centrifuge f 0
app~o'.ximzately 41~/1of supernaltant
1llffer (Boralte !)Hil'r. 1.8.5).
2.1.2
minutes
t
R)I.Reinove
alnd replace with an equal vo)uiti, of reaction
Repeat 2 more times.
Bead-PEG Conjugation Reaction
1. Prepare Biotin-PEG-NHS. Stock concentration (2e-4M at 10p.L. 20/L, 501iL
aliquots). Add 90/iL ddH20 to 10/tL aliquot gives 2e-5M Biotin-PEG-NHS.
2. Add lO100Lof above 2e-5Nl Biotin-PEG-NHS to equal volume of prepared
beacls(wvith reaction buffer).
3. Allow to react overnight. (couple of hours may be ok)
4. Quench reaction. Add 100,L reaction sample to 100,L PBS (pH.7.4) and 20,uL
BSA (5mg/mL).
5. Spin
lown at 10K for 6in.
Remove supernatant, replace with PBS and
5mg/mL BSA. Repeat again to wash out excess Biotin-PEG-NHS in super-
natant.
2.1.3
PEG-Biotin to Streptavidin-Cy2 Conjugation
1. Incubate tethered
b)eads in BSA. Spin down, remove 80,uL supernatant,
70/IL PBS and 1L
aidd
5img/mL BSA. (incubated overnight, but an hour miay
just be neede(l)
2. Cy2 aliquots in freezer. 10/tL aliquot addcl90uL ddH20
3. Add 50uL tethcrel
4.
(1.85e-6M Cy2).
beals to t(),uLprepared (lye abl)ove. Incubate for 30 minutes.
aslh by) spinning at 10()Kfor Giii,
rel)lacing sllupernatant with PBS.
20
I
)ie( (5
II(I\'
(
( s we'l
(.()1st
ln'te(l
r
sta)Il( I l( lass llli('-I (,s o'IeTsli( eI \it IlItw o
Žl1(
( ,liII'-si(le( sticky ta)e se)arate(l h)Na)pr()xillatelv
)
lunIs(i(l t) l)(mnlld
thel ('(I,('s of te
flow channel. A glass cove(rsli) is then adh(ered on tol) of the flow
channe l (Fil'ie
2-2 ).
etched
-
coverslip -
flow chtaulel
I
t
\V /__
I
glass slide
tape
Figure 2-2: Diagram of a flow cell contructed out of a stalndard microscope slide,
etched (:oversli), and double-sided sticky tape.
Glass coverslips were etched by immersing in KOH and sonicating for 5 minutes.
The etching p)rocess remnovesany chemicals on the surface of the coverslip that may
have ben left' as residue during manufacturing and provides a clean surface with
low bha(kgroumndfluorescence.
The glass coverslips were then rinsed with ethainol and
milli)oem water and(l(ried in an oven for at least, 15 mninutes.
2.2
DNA Tethers
An anti--floies(ein
antibody-antigen
system was chosen for this tether b)ecallse of' the
availal)ility o(f'DNA primers conjugated to fluorescein. In this assay, tethers linking
dsDNA to an anti-fluorescein antibody and a 0.5/pm avidin-mnodifiedpolystyrene head
were created. (Figure 2-3)See Appendix A for protocol details.
The DNA was nmade 1)y PCR(Stratagene
Robocycler
Gradient
96): Taq DNA
polyilerase kit (Invitrogen, 10342020). M13mnp8l plasmicl (Bayou Biola)s. P-105).
an( frward
and reverse priners conjiugated to fluorescein ad
(NIX-G Biote(clh).The first primer had forescein
21
conjugated
biotin rest)e(tively
to the
' end(lwithl the
~:treptaviJ Jiitllu
t), ,;'d
,.
Fc.orce
DNA liker
f~~~~itw
anti-fluorescei n
antibody .
.,
biotirn
...f
(.((
surface-modified
a'Hff glass coverslip
Figure 2-3: Sclhenmatic of DNA linkage
s t i m.
following sequence:
5'-TTG AAA TAC CGA CCG TGT GA-:3'
The second primer had biotin conjugated to the 5' end and lhadthe following sequence:
5'-TAT TGC GTT TCC TCG GTT TC-3'
The block temperatures
48 degrees: elongate,
set were: supermelt,
94 degrees: melt. 94 (legrees: anneal,
72 degrees; final extension.
72 degrees.
PCtI p)roducts were
pllrified with a QiaQuick Purifcation Kit (Qiagen, 27104). P('R products were run
on an agarose gel and viewed with a UV viewer(FluorChenl
900( - Alpha inotech)
to check the approximate length of DNA. (Figure 2-4) The DNA used for the tether
is 1010 base pairs, approximately
300nm. All DNA (lilutionlls were( lnmad(le
with TE
Buffer (pH 8.5) to perserve the fluorescence of fluorescein.
This protocol was adapted fromnKeir Nelnian's protocol on linking anti-DIG antil)o(ldy to dsDNA. Bead-DNA complexes were mnade by icul)atillg
1001l of 20pM
DNA withl 10()0il of GOpMl)eads for at least four hours.
Tbe tethers were then niade bv flowing 25,uL of 2(hng,/iiil
oclonal atibody
(lolecular
Probes. ati-fiuorescein/Oieg)}i
'22
anti-fllores(cein mnon-
Green, mouse lgG2a,
Fi,;vlr 2-4: Gel of PCR
lII(nLo( [onai
t)ciatur'
!i.,(edto malakeDNA-beacl (ol1)lexes.
plod(-t>
4-4-20) into the flow (cllanitl anld allowing it, to i(ulbate
h) more than 4 minutes.
the'i ri]s('t out by flowing 500OOLof
`xa(e'ssantibodY not a(lllere(l to the glass is
nd 2()(}IL of 2.()mg/mL BSA
2lig//mL BSA
is flowed in to coat any exposed surflces.
The
it room temn-
The BSA is incbate(d
for 20 minutes.
ec{(.ad-DNA
complexes are thet flo)we(linto the chanber (0/iL) and allowed to
incull)ite for 20 minutes. Afterwards, aiv unbound bead(lsare flushe(l out with 400,uL
of ()0.2nlg/mL BSA. A final flow of 200t/L of 2.0mg/mL
BSA is flowed in if deemed
lle'essar'y.
2.3
UV vis
UV' visil)le spectroscopy was used to (leterinine the concentration
pr(odutct. It was also used to dletermine the concentration
Thle relationship between the o)ti(cal asorbance
of DNA in the PCR.
of avidin b)eads.
rea(ding and the (oncentration
of l)beads i solution was calculated by measuring a series of l)ead cncentrations
and
using the Beer-Lambert equation.
.4
whe:e A is absorbance,
Ebc
(2.1)
is the wavelength-dependenlt molar alsorpltivity coeffi-
cient. 1) is )pathlength, and c is aalyNte concentratiolln.
This Beer-Lamibert relationship w-asused to determine the concenltration of beads
afte twv were modified to have Streptavidin conjugated to the suif'ace.
2)
f i-
.1 -
I LU. LI
100.0
0.0
60.0
'%..
-
40.0
-t~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
%.,
II
20.0
111
0.0
10
100
1000
Bead Concentration [M1
Figure 2-5: Plot of avidin bead (0.52mli diameter) concentration and percent transinittance at 500 nmn.
I
I
--
U
-0.1
-0.2
-0.3
-0.4
8 -0.6
-0.68
-0.9
-1
0
100
200
300
400
500
600
700
800
900
1000
Bead Concenbtraion
Figure 2-6: Plot of absorbance as a function of avidin bead (0.52irn diameter) concentration as determined )y the Beer-Lambl)ertLaw.
Chapter 3
Results
The l)olyethylene glycol linkage assay came across difficulties 1)ecause the beoa(l-PEG
complexes frequently adhered to the sides of the plastic containers, preventing the
beads from being spun down i
the centrifilge and washed.
WVhileaddition
of BSA
blocking agents were able to decrease the amount of beads adhered to the sides of the
container, it was deemed necessary to mlove on with work on the DNA tetheis.
\\Whilethe DNA tethering assay also came across the issue of sticky b)cals, the
addition of BSA seemed to alleviate the problem, but its optimal conicentration had
to be dleterniiiiled. The tethlers were spotted by viewing the flow cell in the microscope
ald watching for jiggling motion. This type of motion indicated that te
bedl was
attachedl to the surface, but still able to freely float in solution within a certain radius.
indicating possible formation of a DNA tether.
illovemellIt,
Based on the calculated ra(lins of
it was possible to visnallv pick ouit possible tethers and test the tethers
using a Labviewv progranm.
3.1
BSA Blocking Agent
OUr initial observations of the beads under the microscope revealed that many beads
were actually stuck onIto the surface of the coverslip instead of freely floatilg
tethere(l.
,Ve hyll)othesized that te
coll(centration of blocking agent i te
)bovineseruli albumnin(BSA), nee(led to )e optimized.
an(d
pr tocol.
B
i,
ill'1111
,a
its teWltIf -( t ) ilii-ie-
5
ii<'ll
)teilI
prpmII
Ing/inlI (21)() /I. fto\-(,(l imlt the channel
't
' tl e
of BSA
't 1f 'is.
as
,i locking
oi ,sefI
bhlckilng :,it
a nd incul)atd
ilmajority of t l
m
nicroslelre,
,
Vi:g foulil tl,'
\\'
)ilcl I(, ilrfa('ti'.
proto(t I.;.\}SlA I ( 1 t giniilly b)eelnusedl
lse(d tttl
(4,ol
at te
;
,
w w((falise
l
itilal
c(om l('ttratiol
;Iss;lvx'
of 3
1,,: 2()mnimites). I (,wcver, w ltl
- t ill stiuck. I'
was iin optiilliZd(lthroulgh exlperilments to del ,t nine the
( )llc('ntrati( ll
coinLt(',ittrattil
that
b)est prcve('ts; the tleats 'fromsticking.
ofvariables il
ott of the assay to sinplif v the 1_nnibt('of'
First DNA wPastken
the system.
Flow cells were constructed
three li-f(lrent cone(iitrations
as shown i
Figire 2-2 and incilb)ated with
of BSA fr 20 minuttes. 1i{/tL of Streptvidinl-co at(d
microsplheres were then flowed into the channel and inc.iu)ated for 20 minttes.
the flow chlmel
was flowed with 400 ,L of 0.2 mg/il
Finally,
BSA and another 200 /L of the
variable BSA (oncientcation. The slides were then viewe(l under the mi('croscope'and
12 field of views w-ere observed, counting the number of stuck beads in each. (Table
3-1)
BSA[mg/ml]
Stuck Beads
0.2
2
20
95
72
20
Table 3.1: Stuly of bead(lbindillg to surface of covers]iip with varying (oll(celntratiolis
of BSA coverage in the flow channel and no BSA pre-incnbated with the bead.
While it was foun( inl this study that a concentration
of 20 mg/nl
fewest nulmber of 1,ie(ls stuck onl the sturface, it was noted that tlhat
was much higher thatn mentioned in the literature[4].
BSA ha(d the
oncentratioll
It was hypotlhesize(l that te
20 mg/mlL BSA hald f'ewer number of beads stuck because there was a large enoughl
concentration of BSA to fully (coat the surface of the glass slide and also coat the
surface of the beads. decreasing the bead surface avwilable to nonspe(ifically bind
to the glass su'fiee.
For this reason, the next study used a, fixed flow cell BSA
concentration of 2.() nig/nli and varied the col(entrait ion of BSA incubated with the
nlicrosp)lheres p)ritr to tflowing into the flow channel. (Tabl)ie 3-2)
BSA[mg/ml]
Sttuck Beads
0.2
295:;
2
41(
20
83
i
liditlg
Neail
stu(ly with varying ci-nceutl ation
Tabtle ,.2:
beads :Inl i tixed 2 m /nlmlBSA in flow chainel.
Th
nation
of 2.0 mg/minL BSA on the mnicrospheres i comn)i-
on(eltration
i(llmted
f BSA iicnul)at (d witli
ithl the 2.) ing/nml BSA flowed into the flow- channel resuilted in te
numbelr of heads stuck.
least
of B3SA
\Ve then did a final stldy fixing the concentration
arying the concentration o)f1SA
incubate!l (1, the microspheres at 2.0 mg/mL and
flowed ont(o the surface. (Table 3-3)
BSA[mg/ml]
Stuck Beads
0.2
52
2
41
20
16
Table 3.3: Bead binding study with fixed BSA-bead incubation of 2 ing/mL BSA and
varying c(enltration
in flow cell.
While the al)ove results show that the 20 mg/mnLof BSA flowed ito te (chlanel
resulte
in t e fewest number of beads stuck to the surface, it is a ten-fold manitude
higher thll
Not,:
he concentration
BA
prior to is(.
specified in the Keir Neiuman protocol.
,eeds to be prepared fresh on, a weeAly basis and it needs to bt filtered
If tle BA
is to be uised wnth a fluorescent
be dega.,,s(d tulth o.dditions,, of beta-mercaptolethanol
molecule,
t also
eeds to
to help protect the flnorecs:enr1,(e
of
the /.nooph'orh . 0iygqi i the solution is highly 'reactiveand may damaye florescent
molecules.
3.2
DNA tethers
After varyiuY the c(tcentration
DNA complexes.
of BSA incutlmated (on the coverslip and on tl, hea(d-
it was (leterminedl that
conventration
27
aroundl 2mg/ml
ilnmue the
h(''(I lest
I)N:\
St i( 1\- (1
a(d
the
t
e'
,l wer'
Sll do
Fl()w cell,
('.
v'itW'\\-( itiler
\\ re
(l'
the i(IOt
1li
-
'1,,.
II
( d lCeltIlt
io)ll
of
'Vs ' t'If pinIl))Oilt'd 1)v
c'\v I)yviewing the Inicfrosph('res ()i te surface f the (over:-] ) 11:(l)i king out b)ea(ds
titi were
sl-1
that
ppaentlv
movin d
it st'Ved i
Olne l('atiOI.
t
thermalll llotioll I)lt li 1lt'v'(l
A\labview pro?,raii was 1s( to test out te p)ossille t('e
bebhived like a microsphere
tethered
to the ghlass surface.
t'l
'
(Itto the surfatce
t,( S(
'
if they indee(l
i thle Lahview prograin,
thl(' iezo stage scans a distance across the nhicr spherlt ald i
[>ohfi('(it the movement
of te bead relative to the detection laser is recorded. (Figues :s3-1.3-2) As the stage
morved far enolugh away such that the DNA tether is xteo{e'(I to its full length, the
bead will begin to
e dragged in the direction of the stage mnovemelt. As the stage
lmoves under the center of the inicrosphere,
the DNA linkage is dillowe(lto freely miove
and the microsphere can be -visualized as jiggling cldueto th(lrnlal motion. Notice the
tethered bead has a portion of no motion of the bead(l whichl is seen as a flat region
in the plot.
In the case of a bead that is adhered to the surface of the (overslilp, the scanning
of the piezo stage creates a data. profile with no flat region i the enter because the
bead moves with the piezo stage throughout the scan. (Figirie 3-3)
28
. 0-I
>
4 .0 3.0
0
to
c
3. 0 2}.0
·1.0 -
+
I'X
0.0 0
0
-1 .0 -2.0
-
._
--3.0
'Cl -4.0
-
'n
mt
(D
s
-5.0
-6.0
-
36500
:37000
37500-
3800-
38500 39000
39500 40000 4500
Stage Posifon [nm]
Figure 3-1: A 1)1()tof bead position response ill voltage, as the stage is tranlslated il
the x-axis across te center ()oftle b)ead.
3 A....
.....
.
..............................................................
I
E 2.01.0
0 1C
9 0.0-
lftp
-1.0 -
0
-2.90 -
.
-3.0 -
j
i
i
i
'i -4-5.0
;-;
i
I
I
I
Ii
i
,I
,
9000
1000
00FJ
I,
'1
1000
1000
0 1:-; 00
Stage Position [nm]
Figure 3-2: A plot of bead position response in voltage, as the stage is translated il
the y-axis cross te center of tlte bead.
,1 I--_
4.
-
E
...
s
^
c
S
..
- m
i
a- 1.0
1].i
1.0
0.0
- a.0
; -.,. U M
-3.0 46500
l
I
I
I
I
47000
47500
48000
46500
i
49000
Stage Position [nm]
Figure 3-3: Example of (lata pr()file(collected from a bead stuck to the surface of the
slide.
3()
Chapter 4
Discussion
There emains much work to be done to optimize the assay (lesigns. ()ther tether
materials can be studied, and other types of blocking agents such as casein. gelatin,
silane agents, or hydrogels can be used.
4.1
BSA concentration
It was ound fom this project that te
concentration of BSA needed to keep the
microsphleres from nonspecifieally adhering to the surface of the coverslip changes
with the components
under st udy.
A BSA concentration that is too low allows uncovered glass surface to be exp)osed,
allowing the microspheres to adhere to the glass. On the otherlhand, too mluch BSA
mav cause BSA protein to lver within the flow cell interfering with the tether formation aind also increases stickiness of the mnicrosphere. An optimal concentrationI
needs t be found by testing the syste
4.2
using different concenitrations of BSA.
Tether Length
The DNA lusedl to construct
the DNA tethers is 1010 bp long, which apl)proxiLnatelv
equlls 300()nm. The diameter of the polstree
fo)rmtion,
b)eadlsis 00nn.
Knowing this in-
it is p)ssil)le to a))pproximate the (distance a bead (an freely travel when
:31
t ,I II, I O
(II,I,(,I 1" t
I [!
x
I
c1V2
-/2
Figlnc -1-1: Diagrami of actethered bead. (Not drawn to scale.)
The length c i the al)ove figure is the distance the bead is allowed to reelv move.
r is the radius of the bead, x is the length of the tether material and
is the angle
between the surface of the coverslip and the tether. Approximating the length of the
DNA tether as 3Wnm and assuming the bead is touching the surface of the coverslip,
we c(an let r = 250nm, x -- 300nm and calculate a value for c.
This gives cv
63
degrees and d -- 980nnt.
The length d. which the microsphere is allowed to freely move within is extracted
from the tether testing (data l)y measuring the peak to peak distance o the curve.
From the data is Figure 3-1, the peak to peak distance is 2,500nrn. This is over a
factor of two times longer than expected. It may indicate that the DNA is eing
stretched during the experiment.
4.3
Conclusion
It is shown in the project that tethers were formed for an fluorescein antibody-antigen
systen with a DNA tether linked to a polystyrene bead through a biotin-streptavi(lin
bond. The preliminary data has shown that the length of the tethers can be extractedl.
The assay
as not
et b)een ol)tinize(d to increase the aount
of tethers per flow
cell. A goo(l point for future stud(ly is to stludy the antilbody coverage on the surfaice
and its effect on the
minmler of tethers
formeed.
32
lWhile
simle molecule finoresemece
t'
,l (all
)l1, ib'
; goo1
) lt ]o(,1 . t
;ilt l-hlorcs(ceil IaIlti)ody indi( lt'(
hv .)()
itpO
atit'l
>( lwtsl to filnl
binding.
thr
SOuilce
t I
, Ia .
s-
t iat
t( :11t
t }1)il
TlliS I llilx )1\(
(,
intl)(l(hieS
t ),
I ;ll Ilf( tI e
S ill{
'tII tit
iS knownl to qillncl
a. problem
t llit () not quench
for SMF.
the
l
tle
fliloleseell(e
so it would
fluorescene.
34
Appendix A
DNA Tether Protocol
Protocol is adapte(l fron iKeir Nemian's p)rotoc( fr making flow cells for fluores'(ence
unlbindi ng ex)perinmelnt s.
A.1
Materials
* Flow Cell (with etched coverslil)s)
* 2'0pM DNA comp)lexes(made fromlfluoiescein and biotin conjugated priiners(MWG
Biotech))
* 60pM avi(lin-oated
50()niln polystyrene
* PBT* (l()0OnMIPhosphate
hl)(ids (Bangs)
Buffer, pH 7.5, ().1% Tween) 1860/,uL 1IM NaH 9 Po4;
81401 L 1Ii Na2HPo4: ()OliL (lH2(): 1)0)/IltTween; Filter with 0.2mli filter
* 20mg/rnL Anti-fluorescein(imake fresh dilution daily) (lolecular Probes: antifluorescein/Oregonl
anti-fluorescein
Green.
mouse IgG, monoclonal
4-4-20) 20/,IL 20011ng/mnL
in PBS (frozen aliqu(lllots):180/tL PBT*
* 2.0ing/mnL BSA(niake fresh weekly) Filter with ().2pm filter
* 0.2mg/miL BSA Filter witll (}.2tm filt(er
* 2 O)ni
L PBS2A(
\ ('c.i-((t)(ha
2,li1lte(l il (dE12,)
A.2
jl''
aet'1viug
;)(iI,
'3I
flurescel('(')
I
t-1uicapto ethan
WS:-\
Procedure
1. ILcubat,
beads
.1d
D)NA (oi)lexes
for 3 hours at 4 degrees Celsius.
cltough for severa.l sli(les. using 40/tL of milix for ech
Make
Sli(le.
Bead:DNA Mix
100/z 2pM DNA complexes
100/J,L 6OpMNbeads
2. Spin down beads at 10K for 6 minutes. Remove supernatant and replace with
2.0mg/nmL BSA. Allow to incubate for at least 20 minutes.
A 200,L pipette tip attached to tubing, a filter flask, and vacuum line is used
to assist flow through of solution for more consistent flow rates.
3. Flow in 25,uL of anti-florescein
into each flow cell and incubate
for one hour
at roonmtemperature.
4. Flow in 500L
of ().1lmg/nmL BSA.
5. Flow in 200tiL of 2.0mng/mL BSA. Incubate for 20 minutes at room temperature.
6. Sonicate any bead--DNA coniplexes that have been sitting around for a few days.
(add ice to cup sonicator for sonicate for 2 milnuttesat 40%)
7. Flow in 400/iL of bead-DNA
complexes. Incubate for 20 minutes.
8. Flow in 400/iL of 0.2mng/mlI BSA.
9. Flow in 200/t of 2.Omg/mnL (degassedBSA. Add small pools of BSA on the sides
of the flow cell to prevent the flow cells from drying up and place in huniidity
chamber (I used ol p1j)i)ette
(containerswith a little water in them and placed
slides on a raised plftfornl insi(dle(ontainer.)
3(i
Tethers will last one to two dlays.
1
Bibliography
[1] Matthew J. Lang Cowtu)led optical trajpl/.qy]and
J. Biol.. vol. 2, p.
[21 Ni. I. Wallace
6
siqhl -molecule fluorescel(e
(2003).
Combimncd single-molcculec forc( ad
fluorescence
measure-
ments for biology.. Biol., vol. 2, p. 4 (2003).
[3] A. Ishijima Simultancous observation of im(ivi(lual ATPase and mnechanical
events by a single myosin molecule duringqinteraction with actin Cell, vol.92,
pp.161-171 (1998).
[4 Bernard P. Chan Effect of Streptavidin RGD Mutant on the Adhesion of
Endothelial Cells Biotechnol. Prog., 20, 566-575)(2004).
37