Important for specification of identity along the A-P axis. Hox Genes
advertisement

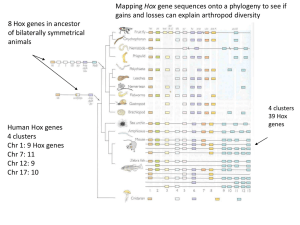
Hox Genes Important for specification of identity along the A-P axis. Insects Segment identity controlled by segment identity (aka homeotic, aka selector) genes. Discovered through homeotic mutations. This is a mutation that causes the transformation of one structure to another homologous structure. (Homologs have evolutionarily related ancestry—both derived from a common ancestor structure). Eg. Antennapedia causes the transformation of antennae to legs; ultrabithorax causes the transformation of halteres to wings (T3 to T2). Drosophila homeotic genes encode transcription factors. These belong to a family with related DNA binding domains called homeodomains. The DNA sequence that codes for the homeodomain is called a homeobox. Genes that encode homeodomain proteins are knicknamed Hox genes. **It is important to note that homeotic and Hox are NOT synonomous. Homeotic refers to a mutant phenotype and Hox refers to a sequence motif. Not all Hox genes give homeotic mutant phenotypes and not all genes (especially in other organisms) that give homeotic mutant phenotypes encode homeodomain proteins. The segment identity genes are chromosomally located in 2 gene complexes (in Drosophila). ! The Antennapedia complex controls head and thorax segment identities ! The Bithorax complex controls abdominal segment identities Most insects (and other segmental animals) contain all the segment identity genes in one large complex, HOM-C. It appears that the complex was split during Drosophila evolution. Thus, Antennapedia + Bithorax = Hom-C. Each gene within the complex is expressed in a discrete set of segments to control identity of that region. There is a correlation between the pattern of gene positions within the HOM-C complex and the pattern of expression in the embryo. Genes located toward the 5’ end of the complex tend to be expressed in more anterior segments. Segment identity gene expression is initially regulated by gap and pair-rule genes. Expression patterns are refined and maintained by interactions with other pair-rule genes. General rule is that genes expressed more posteriorly repress genes expressed more anteriorly genes. Eg. The normal Antennapedia gene is expressed in T2 and specifies the identity of T2. The Antennapedia mutant is a dominant gain-of-function mutant where the Antp gene is ectopically expressed in a head segment. This represses the normal Hox gene in that (more anterior) head segment and converts the identity to (more posterior) T2, causing the homeotic transformation of antennae to legs. Eg. Ultrabithorax is normally expressed in T3, where it represses the expression of Antp. In the ubx loss-of-function mutant, the without Ubx repression, Antp expression expands posteriorly into T3, transforming it to T2 identity. Thus the fly has 2 T2 segments and the halteres in T3 are transformed to wings like T2. Segment identity genes control the expression of realizator genes—the genes that actually direct morphogenesis. An example is the distal-less gene that promotes leg formation. Vertebrates Insects: Antennapedia complex + bithorax complex = HOM-C. Mammals: 4 copies of HOM-C = HoxA – HoxD Order of genes in complex similar to flies Expression pattern similar to flies Order of genes on the chromosome is the same as the order of expression along the A-P axis Some are so similar to fly homeotic genes that they can be swapped Mammalian Hox genes expressed along dorsal axis in neural tube, neural crest, paraxial mesoderm and surface ectoderm and in derivatives of these tissues. Experimental evidence that Hox genes specify identity along A-P axis: 1. Targeted gene disruptions result in homeotic transformations. Generally get anterior transformations just like flies, indicating that posterior genes inhibit anterior. Good direct evidence. ! Eg. Knockout mouse Hoxc-8 converts a lumbar vertebra to a thoracic vertebra#get extra rib 2. Retinoic Acid teratogenesis. [RA] is normally high in Hensen’s node and probably plays a role in axis formation. Treat gestating mice with RA and get babies with homeotic transformations. Also see corresponding change in homeotic gene expression pattern, and some transformations can be mimicked by ectopic Hox gene expression. For example, ectopic expression of Hoxa-7 generates cranio-facial defects similar to RA treatment. 3. Comparative anatomy. Mammals and chicks have different numbers of cervical, thoracic and lumbar vertebrae. The constellation of Hox gene expression predicts the type of vertebra. For example, mice have 7 cervical and 13 thoracic vertebrae while chicks have 14 cervical and 7 thoracic. In both cases, the Hox-5 gene is always expressed in the last cervical vertebra while Hox-6 is in the first thoracic. Evolution Hox genes appear to have played important roles in morphological evolution. Many differences in morphology between species/phyla appear due to evolutionary changes related to Hox genes. There are 4 major ways this could happen: 1. Changes in the response of realizator genes to Hox genes. Eg. Fly vs. butterfly. Fly Ubx --| genes for wing formation (get haltere in T3) Butterfly Ubx does not block wing genes (get 2nd wing in T3) Therefore, the Hox genes did not change but the response to them did. 2. Changes in Hox gene expression within a body segment. Ubx --| distalless # limb formation Drosophila, have Ubx expression throughout abdominal segments, thus no legs Butterflies, get late downregulation of Ubx expression in small regions of abdominal segments. This allows distalless expression and the formation of prolegs. 3. Changes in Hox gene expression in different segments. Eg. The different Hox gene expression patterns in mammals and birds, discussed above, produces altered body plans with different numbers of cervical, thoracic and lumbar vertebrae. Eg. Snakes. Most vertebrates form forelimbs anterior to anteriormost expression domain of Hoxc-6. Snakes express Hoxc-6 and Hoxc-8 throughout# specifies many thoracic vertebrae and no forelimb. 4. Changes in Hox gene number. There is a correlation between the complexity of the organism and the number of copies of the HOM-C comples. All invertebrates have a single Hox complex simplest = sponges (1 or 2 Hox genes) more complex = insects Hom-C Amphioxis – primitive invertebrate chordate (1 HOM-C) Jawless fishes (earliest vertebrates) – 4 HOM-C ; represents a major leap in complexity from amphioxis