Document 10785065
advertisement

Stat 511 HW#7 Spring 2003 (revised)
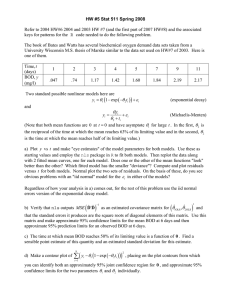
1. The classic textbook Statistics for Experimenters by Box, Hunter and Hunter contains a small
data set taken originally from the paper "Evaluation of methods for estimating biochemical oxygen
demand parameters" by Marske and Polkowski (1977, J. Water Pollut. Control Fed., 44. No. 10,
1987). It is reproduced below. y is BOD (biochemical oxygen demand) in mg/liter, a measure of
pollution. A small amount of domestic or industrial waste is mixed with pure water, sealed, and
incubated for a number of days, x . The data represent values obtained from n = 6 different bottles.
x
y
1
109
2
149
3
149
5
191
7
213
10
224
Apparently the original investigators considered the model
yi = β1 (1 − exp ( − β 2 xi ) ) + ε i
(for iid ε i ) to be potentially useful here. Note that in this model, β1 is a limiting BOD and β 2 is a
rate constant that controls how fast this limiting BOD is approached. For example, when β 2 x = 1 ,
the model says that mean BOD is at a fraction (1 − exp ( −1) ) = .63 of its limiting value.
Use R to help you do the following. Read in 6 × 1 vectors x and y.
a) Plot y vs x and make "eye-estimates" of the parameters based on your plot and the interpretations
of the parameters offered above. (Your eye-estimate of β1 is what looks like a plausible limiting
value for y and your eye-estimate of β 2 is the reciprocal of a value of x at which y has achieved 63%
of its maximum value.)
b) Add the nls package to your R environment. Then issue the command
> BOD.fm<-nls(formula=y~b1*(1-exp(-b2*x)),start=c(b1=##,b2=###),trace=T)
where in place of ## and ### you enter your eye-estimates from a). This will fit the nonlinear
model via least squares. What are the least squares estimate of the parameter vector and the
"deviance" (error sum of squares)
6
2
b
b OLS = 1 and SSE = ∑ yi − b1 (1 − exp ( − β 2 xi ) )
i =1
b2
(
(
))
c) Re-plot the original data with a superimposed plot of the fitted equation. To do this, you may
type
>
>
>
>
>
time<-seq(0,11,.1)
biochemical<-coef(BOD.fm)[1]*(1-exp(-coef(BOD.fm)[2]*time))
plot(c(0,11),c(0,250),type="n",xlab="Time (min)",ylab="BOD (mg/l)")
points(x,y)
lines(time,biochemical)
(The first two commands set up vectors of points on the fitted curve. The third creates an empty plot
with axes appropriately labeled. The fourth plots the original data and the fifth plots line segments
between points on the fitted curve.)
1
d) Get more complete information on the fit by typing
> summary(BOD.fm)
> vcov(BOD.fm)
(
ˆ ′D
ˆ
Verify that the output of this last call is MSE D
)
−1
and that the standard errors produced by the first
are square roots of diagonal elements of this matrix.
e) The time, say t100 , at which mean BOD is 100 mg/l is a function of β1 and β 2 . Find a sensible point
estimate of t100 and a standard error (estimated standard deviation) for your estimate.
f) As a means of visualizing what function the R routine nls minimized in order to find the least
squares coefficients, do the following. First create a sum of squares function
>
+
+
+
+
+
+
+
+
ss<-function (b1,b2)
{
(109-b1*(1-exp(-b2*1)))^2+
(149-b1*(1-exp(-b2*2)))^2+
(149-b1*(1-exp(-b2*3)))^2+
(191-b1*(1-exp(-b2*5)))^2+
(213-b1*(1-exp(-b2*7)))^2+
(224-b1*(1-exp(-b2*10)))^2
}
(I tried using vector calculations here, but ran into many hours worth of problems that I don't understand
trying to evaluate the sum of squares over a grid of parameter values. The above will work. You are
welcome to try something slicker if you like, but don't ask me to tell you why you get error messages, if
you do!)
As a check to see that you have it programmed correctly, evaluate this function at b OLS for the data in
hand and verify that you get the error sum of squares. Then type
>
>
>
>
beta1<-150:300
beta2<-seq(.2,1.4,.01)
SumofSquares<-outer(beta1,beta2,FUN=ss)
contour(beta1,beta2,SumofSquares,levels=seq(1000,10000,500))
This gives a "contour plot" or "topographic map" of the sum of squares surface. Identify on your plot the
Beale 90% approximate confidence region for the parameter vector ( β1 , β 2 ) (This is the set of parameter
vectors with corresponding residual sum of squares below some value). What does this region indicate
about how well this data set allows one to identify the limiting BOD ( β1 )? (By the way, as far as I can
tell, the Figure 14.9 of Box, Hunter and Hunter is wrong on this point. Their supposed Beale region
appears to be far too optimistic.)
g) To make the last part of part f) more concrete, do this. Make an approximately 90% confidence
interval for β1 by identifying on your plot those β1 for which there is a β 2 with
6
∑ ( y − β (1 − exp ( − β x ) ) )
i =1
i
1
2 i
2
1
≤ SSE 1 + F1,4
4
2
1
(If you find the contour on the plot corresponding to SSE 1 + F1,4 , this is the range of β1 values
4
covered by the region enclosed by that contour.)
h) Use the standard error for the first coefficient produced by the routine nls and make a 90% t
interval for β1 . How much different is it from your interval in g)?
Notice that the sample size in this problem is very small and reliance on any version of large sample
theory to support inferences is shaky at best. I would take any of the inferences above as very
approximate. We will later discuss the possibility of using "bootstrap" calculations to provide reliable
small sample inferences.
3