SUBSAMPLING ACCURACY IN BEAM TRAWL CATCHES
advertisement

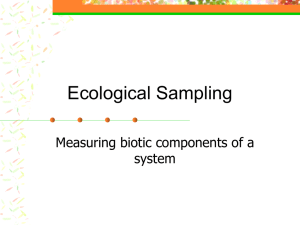
SUBSAMPLING ACCURACY IN BEAM TRAWL CATCHES Samuel Van de Walle 1, 2, Jochen Depestele1,2 Hans Polet1, Kris Van Craeynest1 & Magda Vincx2 1Institute for Agricultural and Fisheries Research, Animal Sciences – Fisheries ILVO, Biological Environmental Research, Ankerstraat 1, 8400 Oostende, Belgium 2Ghent University, Biology Research Group, Marine Biology Section Krijgslaan 281 (Campus Sterre – S8), B-9000 Gent, Belgium N° 58 Introduction 57 In commercial beam trawling data on fish catches is routinely available, while composition of invertebrate discards is ignored. However, from an ecosystem perspective such data is equally important yet is highly labor intensive to acquire. Subsampling the discards, and accepting the error this inherently imposes, is the only option. Here we investigate these error rates, which will allow for more accurate data or estimates in discard composition. 56 55 54 53 52 51 N° 50 -4 W° -2 Figure 1: Catch Sorting in Progress A S (4) = Rare Common 40 Abundant Dominant 30 20 10 0 p= proportion of catch analyzed a= Species abundance in subsample T= Species abundance in haul W= Total weight of haul 20 15 N1 (122,0kg) N3 (19,0kg) N5 (53,0kg) 10 (1) Categories of Abundance Index (n) 0 % Asterias rubens 24068 47,19 Echinidea sp. 21324 41,81 Buccinum undatum 901 1,77 Pagurus bernhardus 675 1,32 Liocarcinus depurator 636 1,25 Liocarcinus holsatus 608 1,19 Aphrodita aculeata 416 0,82 Palaemoninae sp. 318 0,62 Necora puber 247 0,48 236 0,46 … … … Totaal 51001 Table 1: Top Abundant Species Discussion Trawl N1 400 20 40 60 Sampling Error (S) (%) Rare Common Rare 95% CI Common 95% CI 300 200 150 100 50 0 0 20 40 60 80 100 Proportion of Catch Analysed (%) Sampling error (S) (%) 40 35 Abundant Dominant 30 25 Abundant 95% CI Dominant 95% CI 20 15 10 5 0 0 10 20 30 40 50 60 70 80 100 25 20 15 F1 (48,6kg) F3 (57,7kg) F5 (28,8kg) 10 80 90 100 Proportion of Catch Analysed (%) Corresponding author: +32(0)472 31 94 88 samuel.vandewalle@outlook.com F2 (86,3kg) F4 (28,0kg) A1 (110,0kg) 5 0 20 40 60 80 100 30 25 20 15 N1 (122,0kg) 95% CI 5 0 250 N2 (154,0kg) N4 (65,0kg) N6 (125,0kg) 5 Individuals Ophiura ophiura Mean Sampling error for 3/8 of haul analyzed 350 10 E° 25 10 C 8 30 Species 50 S (3) S 6 B Number of Species Recorded Number of Cases (Species x Trawl) (2) S 4 30 60 S 2 Figure 2: Trawling Location Materials & Methods Twelve trawls were performed, in February 2009(F1-F3), February 2010(F4&F5), April (A1) and November2011 (N1-N6) at the location in Fig. 2. Every species x trawl combination was assigned to an abundance category(1) based on an abundance index (n). Hauls were subdivided into 10L buckets(2) and individuals were identified. Different numbers of buckets(from 1 to all) from a trawl were used to simulate different subsample sizes. For a large number of random recombination's of buckets, a sampling error (S) was calculated(3) and from all these a mean error and a confidence interval was derived(4). A similar approach was then used to calculate the mean number of species in a subsample of a certain size. S 0 20 40 60 80 100 (A) Species Abundances Proportion of Catch Analysed (%) All hauls were highly dominated by starfish and sea urchins. A majority of species was found to be rare. (B) Species Richness An average of 23 species per trawl was found. Despite large differences in catch weight there was comparatively little variation in number of species. Most trawls follow a similarly shaped curve regardless of their weight or number of species. For N1 the confidence interval showed a constant range over al subsample sizes. (C) Sampling Error Results for N1 are given as example, other trawls revealed similar patterns. Average error decreased with increasing subsample size. Rare species showed the highest error rates while common species had intermediate error rates. For abundant & dominant species errors were up to 10 times lower and surprisingly similar to each other. This indicates that after a certain abundance threshold is reached(around 5ind./10kg), error rate is unlikely to drop much further, even at highest abundances. Future Prospects Currently most results are represent all trawls individually. In the next step, we aim to produce overall results for all trawls combined. Interpolation and/or a general linear mixed model approach should allow for this, while also accounting for the complex interdependence of all data points.