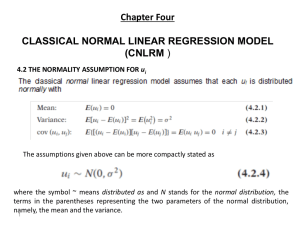
Assessing Normality – The Univariate Case
advertisement

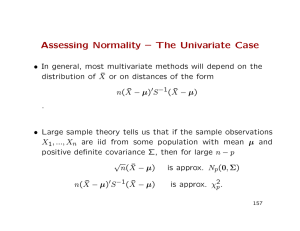
Assessing Normality – The Univariate Case • In general, most multivariate methods will depend on the distribution of X̄ or on distances of the form n(X̄ − µ)0S −1(X̄ − µ) . • Large sample theory tells us that if the sample observations X1, ..., Xn are iid from some population with mean µ and positive definite covariance Σ, then for large n − p √ n(X̄ − µ) is approx. Np(0, Σ) n(X̄ − µ)0S −1(X̄ − µ) is approx. χ2 p. 157 Assessing Normality (cont’d) • This holds regardless of the form of the distribution of the observations. • In making inferences about mean vectors, it is not crucial to begin with MVN observations if samples are large enough. • For small samples, we will need to check if observations were sampled from a multivariate normal population 158 Assessing Normality (cont’d) • Assessing multivariate normality is difficult in high dimensions. • We first focus on the univariate marginals, the bivariate marginals, and the behavior of other sample quantities. In particular: 1. Do the marginals appear to be normal? 2. Do scatter plots of pairs of observations have an elliptical shape? 3. Are there ’wild’observations? 4. Do the ellipsoids ’contain’ something close to the expected number of observations? 159 Assessing Normality (cont’d) • One other approach to check normality is to investigate the behavior of conditional means and variances. If X1, X2 are jointly normal, then 1. The conditional means E(X1|X2) and E(X2|X1) are linear functions of the conditioning variable. 2. The conditional variances do not depend on the conditioning variables. • Even if all the answers to questions appear to suggest univariate or bivariate normality, we cannot conclude that the sample arose from a MVN normal distribution. 160 Assessing Normality (cont’d) • If X ∼ M V N , all the marginals are normal, but the converse is not necessarily true. Further, if X ∼ M V N , then the conditionals are also normal, but the converse does not necessarily follow. • In general, then, we will be checking whether necessary but not sufficient conditions for multivariate normality hold or not. • Most investigations in the book use univariate normality, but we are also going to present some practical and recent work on assessing multivariate normality. 161 Univariate Normal Distribution • If X ∼ N (µ, σ 2), we know that q Probability for interval (µ − σ 2, µ + q q q q q σ 2) ≈ 0.68 Probability for interval (µ − 2 σ 2, µ + 2 σ 2) ≈ 0.95 Probability for interval (µ − 3 σ 2, µ + 3 σ 2) ≈ 0.99. • In moderately large samples, we can count the proportion of observations that appear to fall in the corresponding intervals where sample means and variances have been plugged in place of the population parameters. • We can implement this simple approach for each of our p variables. 162 Normal Q-Q plots • Quantile-quantile plots can also be constructed for each of the p variables. • In a Q-Q plot, we plot the sample quantiles against the quantiles that would be expected if the sample came from a standard normal distribution. • If the hypothesis of normality holds, the points in the plot will fall along a straight line. 163 Normal Q-Q plots • The slope of the estimated line is an estimate of the population standard deviation. • The intercept of the estimated line is an estimate of the population mean. • The sample quantiles are just the sample order statistics. For a sample x1, x2, ..., xn, quantiles are obtained by ordering sample observations x(1) ≤ x(2) ≤ ... ≤ x(n), where x(j) is the jth smallest sample observation or the jth sample order statistic. 164 Normal Q-Q plots (cont’d) • When the sample quantiles are distinct (as can be expected from a continuous variable), exactly j observations will be smaller than or equal to x(j). • The proportion of observations to the left of x(j) is often approximated by (j − 0.5)/n. Other approximations have also been suggested. • We need to obtain the quantiles that we would expect to observe if the sample observations were sampled from a normal distribution. For a standard normal random variable, quantiles are computed as j−1 1 1 2 2 =p . √ Pr(Z ≤ q(j)) = exp(− z )dz = (j) 2 n −∞ 2π Z q (j) 165 Normal Q-Q plots (cont’d) • For example, if p(j) = 0.5, then q(j) = 0 (or the median), and if p(j) = 0.95, then q(j) = 1.645. • Given the sample size n, we can compute the expected standard normal quantile (q(j)) for each ordered observation using p(j) = (j − 0.5)/n. SAS uses the Blom approximation with p(j) = (j − 0.375)/(n + 0.25). • If the plot of the pairs (x(j), q(j)) shows a straight line, we do not reject the hypothesis of normality. • If observations are tied, the associated quantile is the average of the quantiles that would have corresponded to slightly different values. 166 Example Ordered Observations x(j) Probability Level (j − 0.5)/n Standard Normal Quantiles q(j) -1.00 -0.10 0.16 0.41 0.62 0.80 1.26 1.54 1.71 2.30 0.05 0.15 0.25 0.35 0.45 0.55 0.65 0.75 0.85 0.95 -1.645 -1.036 -0.674 -0.385 -0.125 0.125 0.385 0.674 1.036 1.645 167 Example (cont’d) 168 Example (cont’d) • The normal quantiles can be computed with SAS using the probit function or the RANK procedure • Note that j − 0.5 j − 0.5 ) = probit( ) n n with Φ(a) the standard normal cumulative distribution function evaluated at a. q(j) = Φ−1( • SAS uses a different (Blom) approximation to the probability levels when the ”normal” option is executed in the Rank procedure: 3 j − 8 ). q(j) = Φ−1( n+1 4 169 Microwave ovens: Example 4.10 • Microwave ovens are required by the federal government to emit less than a certain amount of radiation when the doors are closed. • Manufacturers regularly monitor compliance with the regulation by estimating the probability that a randomly chosen sample of ovens from the production line exceed the tolerance level. • Is the assumption of normality adequate when estimating the probability? • A sample of n = 42 ovens were obtained (see Table 4.1, page 180). To assess whether the assumption of normality is plausible, a Q-Q plot was constructed. 170 Microwaves (cont’d) 171 Goodness of Fit Tests Correlation Test • In addition to visual inspection, we can compute the correlation between the x(j) and the q(j): Pn i=1 (x(i) − x̄)(q(i) − q̄) qP rQ = qP . n (x n 2 2 i=1 (i) − x̄) i=1 (q(i) − q̄) • We expect values of rQ close to one if the sample arises from a normal population. • Note that q̄ = 0, so the above expression simplifies. 172 Correlation Test (cont’d) • The sampling distribution of rQ has been derived (see Looney and Gulledge, The American Statistician 39:75-79) and percentiles of its distribution have been tabulated (see Table 4.2 in book). • Using the tabled values, we can test the hypothesis of normality and for a sample of size n can reject it at level α if rQ falls below the corresponding table value. • The critical values for rQ depend on both n and α. 173 Correlation Test (cont’d) • For the earlier example with n = 10, we have rQ = 0.994. • The critical value from Table 4.2 in book for α = 0.05 and n = 10 is 0.9198. • Since rQ > 0.9198 we fail to reject the hypothesis that the sample was obtained from a normal distribution. 174 Shapiro-Wilks’ Test • A weighted correlation between the x(j) and the q(j): Pn i=1 aj (x(i) − x̄)(q(i) − q̄) qP . W = qP n n 2 2 2 i=1 aj (x(i) − x̄) i=1 (q(i) − q̄) • We expect values of W close to one if the sample arises from a normal population. • SAS has stored values of aj for n < 2000 175 Empirical Distribution Function (EDF) Tests • Compare the EDF Number of observations ≤ x n to an estimate of the hypothesized distribution. Fn(x) = • For the hypothesized family of normal distributions, compare with x − x̄ F (x; µ̂ σ̂ 2) = Φ s 176 EDF Tests: Anderson-Darling Test • Order the observation from samllest to largest: x(1) ≤ x(2) ≤ · · · ≤ x(n) • Anderson-Darling Statistic A2 n = n Z ∞ h Fn(x) − F (x, θ̂) h i2 −∞ F (x, θ̂) 1 − F (x, θ̂) i dF (x, θ̂) n h i 1 X (2i − 1) ln(pi) + ln(1 − pn+1−i) = −n − n i=1 where pi = Φ x(i) −x̄ s 177 EDF Tests: Kolmogorov-Smirnov Test • Order the observation from samllest to largest: x(1) ≤ x(2) ≤ · · · ≤ x(n) • Kolmogorov-Smirnov Statistic − + Dn = max D , D + i−1 i − where Dn = max1≤i≤n pi − n and Dn = max1≤i≤n pi − n x(i) −x̄ and pi = Φ s 178 EDF Tests • Reject normality for large values of A2 n or Dn • Approximate upper percentiles for Dn are " Dn,0.05 = 0.895 " Dn,0.01 = 1.035 √ √ 0.85 n − 0.01 + √ n 0.85 n − 0.01 + √ n #−1 #−1 • Approximate upper percentiles for A2 n are 0.795 0.89 A2 − 2 n,0.05 = 0.7514 1 − n n 1.013 0.93 A2 = 1.0348 1 − − 2 n,0.01 n n 179 Assessing bivariate and multivariate normality • If sample observations X1, ..., Xn come from a Np(µ, Σ), we know that δi2 = (xi − µ)0Σ−1(xi − µ) ∼ χ2 p. • By substituting x̄, S for the population mean vector, we can estimate the sample squared distances d2 j . For large n − p (at least 25), the d2 j should behave approximately like independent χ2 p random variables. • A χ2 plot is similar to a Q-Q plot and can be computed for the sample squared distances. 180 Chi Square Plot • First order the squared distances from smallest to largest to get the d2 . (i) • Next compute the probability levels as before: p̂ = (i−0.5)/n. • Then compute the n chi-square quantiles qc,p(p̂i) for the χ2 p distribution. • Finally plot the pairs (d2 , q (p̂ )) and check if they approx(i) c,p i imately fall on a straight line. • For a χ2 distribution with ν degrees of freedom, the function to find the chi-square quantiles in SAS is qi,ν = cinv(p̂i, ν) 181 Stiffness of boards: Example 4.14 • Four measures of stiffness on each of n = 30 boards were obtained. • Data are shown on Table 4.3 in book, including the 30 sample squared distances. • The 30 probability levels are computed as 1 − 0.5 2 − 0.5 30 − 0.5 = 0.017, = 0.05, ...., = 0.983. 30 30 30 • Quantiles can be calculated using the cinv function in SAS, for p = 4 degrees of freedom and the 30 probability levels computed above. 182 Example (cont’d) 183 Formal Tests for Multivariate Normality - I • For any X, we have seen that X ∼ M V N iff a0X is univariate normally distributed. • How about taking a large (but random) number of projections and evaluating each projection for univariate normality? • Let us try with N (large, random) independent random projections on the unit vector. We will test each projection for univariate normality using the Shapiro-Wilks’ test. 184 Formal Tests for Multivariate Normality - I (contd.) • Note that we will have a large number of tests to evaluate, and this means that we will have to account for multiple hypothesis tests that have to be carried out. So we convert all the p-values into so-called q-values and if all the q-values are greater than the desired False Discovery Rate (FDR), then we accept the null hypothesis that X is multivariate normally distributed. • Note the large number of calculations needed: imperative to write R code efficiently. – code provided in testnormality.R. 185 The Energy Test for Multivariate Normality • Let X1, X2, . . . , Xn be a sample from some p-variate distribution. Then, consider the following “energy” test statistic: n n X X 2 1 ∗ 0 E = n IE k Xi − Z k −IE k Z − Z k − 2 k X∗i − X∗j k , n i=1 n i=1 where X∗i , i = 1, 2, . . . , n is the standardized sample and Z, and Z0 are independent identically distributed p-variate standard normal random vectors, and k · k denotes Euclidean norm. • The critical region is obtain by parametric bootstrap. • Implemented in R by the mvnorm.etest() function in the energy package also provided by authors Székely and Rizzo (2005). 186 Detecting Outliers • An outlier is a measurement that appears to be much different from neighboring observations. • In the univariate case with adequate sample sizes, and assuming that normality holds, an outlier can be detected by: 1. Standardizing the n measurements so that they are approximately N (0.1). 2. Flagging observations with standardized values below or above 3.5 or thereabouts. • In p dimensions, detecting outliers is not so easy. A sample unit which may not appear to be an outlier in each of the marginal distributions, can still be an outlier relative to the multivariate distribution. 187 Detecting outliers (cont’d) 188 Steps for detecting outliers 1. Investigate all univariate marginal distributions, visually by √ constructing the standardized values zij = (xij − x̄)/ σjj for the i-th sample unit and j-th variable. 2. If p is moderate, construct all bivariate scatter plots. There are p(p − 1)/2 of them. 3. For each sample unit, calculate the squared distance 0 −1 (x − x̄), where x is the p × 1 vector of d2 i i i = (xi − x̄) S measurements on the i-th sample unit. 2 are 4. To decide if d2 is ’extreme’, recall that the d i i 2 approximately χp . For example, if n = 100, we would expect to observe about 5 squared distances larger than the 0.95 percentile of the χ2 p distribution. 189 Example: Stiffness of lumber boards • Recall that four measurements were taken on each of 30 boards. • Data are shown in Table 4.4 along with the four columns of standardized values and the 30 squared distances. • Note that boards 9 and 16 have unusually large d2, of 12.26 and 16.85, respectively. The 0.975 and 0.995 percentiles of a χ2 4 distribution are 11.14 and 14.86. • In a sample of size 30, we would expect less than 0.75 obs. with d2 > 11.14 and less than 0.015 obs. with d2 > 14.86. • Unit 16, with d2 = 16.85 is not flagged as an outlier when we only consider the univariate standardized measurements. 190 Detecting if Outliers are Present • Mardia’s (1970, 1974, 1975) multivariate sample kurtosis measure b2,p(X) = n−1 n X (Xi − X̄)0S−1(Xi − X̄). (1) i=1 • Schwager and Margolin (1982) have shown that the presence of multivariate outliers can be ascertained, if b2,p(X) is greater than some cut-off. 191 Deciding Critical Region for Outlier Detection • Sample Z from Np(0, I), compute b2,p(Z) for each sample. • Obtain the reference distribution to obtain the estimated pvalue of b2,p(X). • Easily programmed in R. • Does not tell us the outliers: can look at individual values of (Xi − X̄)0S−1(Xi − X̄) 192 Transformations to near normality • If observations show gross departures from normality, it might be necessary to transform some of the variables to near normality. • The following are some suggestions which stabilize variances, but some people use them to transform to near-normality. Original scale Right skewed data x are counts Transformed scale log(x) √ x x are proportions p̂ logit(p̂) = 1/2 log[(p̂)/(1 − p̂)] x are correlations r Fisher’s z(r) = 1/2 log[(1 + r)/(1 − r)] 193 The Box-Cox transformation • Proposed by Box and Cox in a 1964 JRSS(B) article. • The Box-Cox transformation is a member of the family of power transformations: xλ − 1 , λ λ 6= 0, = log(x), λ = 0. x(λ) = • To estimate λ we do the following: 1. Assume that xλ ∼ N (µ, σ 2) for some unknown λ. 2. Do a change of variables to obtain the likelihood with respect to x. 3. Maximize the resulting log likelihood for λ. 194 Box-Cox Transformation (cont’d) • If xλ ∼ N (µ, σ 2), then with respect to the untransformed observations the resulting log-likelihood is n n X X n 1 (λ) L(µ, σ 2, λ) = − log(2πσ 2)− 2 (xi −µ)2+(λ−1) log(xi), 2 2σ i=1 i=1 where the last term is the logarithm of the Jacobian of the transformation |dxλ i /dxi |. • Substituting MLEs for µ and σ 2 we get, as a function of λ alone: X n 1 X (λ) 2 ¯ (λ) l(λ) = − [ (xi − x ) ] + (λ − 1) log(xi). 2 n i i • The best power transformation λ̂ is the one that maximizes the expression above. 195 Computing the Box-Cox transformation in practice • One way to find the MLE (or an approximation to the MLE) of λ is to simply plug into the log likelihood in the previous transparencies a sequence of discrete values of λ ∈ (−3, 3) or in some other range. • For each λ, we compute the log-likelihood and then we pick the λ for which the log-likelihood is maximized. • SAS will also compute the MLE of λ and so will R. 196 Radiation in microwave ovens example • Recall example 4.10. Radiation measurements on 42 ovens were obtained. • The log-likelihood was evaluated for 26 values of λ ∈ (−1, 1.5) in steps of 0.10. 197 Microwave example (cont’d) 198 The Multivariate Normal Distribution Microwave example (cont’d) 199 Another way to find a transformation • If data are normally distributed, ordered observations plotted against the quantiles under the assumption of normality will fall on a straight line. • Consider a sequence of values of λ: λ1, λ2, ..., λk and for λj fit the regression λ j = β0 + β1q(i),j + ei, x(i) λ j are the transformed and ordered sample values, where x(i) q(i),j are the quantiles corresponding to each transformed, ordered observation under the assumption of normality, and β0, β1 are the usual regression coefficients. 200 Alternative estimation method (cont’d) • For each of the k regressions (one for each λj ), compute the MSE. • The best fitting model will have lowest MSE. • Therefore, the λ which minimizes the MSE of the regression of sample quantiles on normal quantiles will be ’best’. • It can be shown that this is also the λ that maximizes the log-likelihood function shown earlier. 201 Transforming multivariate observations • By transforming each of the p measurements individually, we approximate normality of the marginals but not necessarily of the joint p-dimensional distribution. • It is possible to proceed as in the Box-Cox approach and try to maximize the log-likelihood jointly for all p lambdas. This is called the multivariate Box-Cox transform. 202 Bivariate transformation in microwave example • The bivariate log-likelihood was maximized for λ1, λ2. Contours are shown below. Optimum near (0.16, 0.16) 203