Linear Models Stat 430

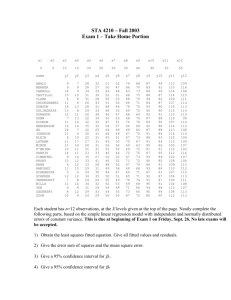
Linear Models
Stat 430
Outline
•
Project
•
Normal Model
•
F test
•
Model Comparison and Selection
Flights Project
•
Accuracy is measured as
1
�
( y i
− y ˆ i
)
2 n
(lower is better) i m i +1 ,j +1 m ij log θ ij
= log = ...
= β i +1
− β i m i +1 ,j m i,j +1
•
What YOU should do: divide data set into 90%
Training Set and 10% Test Data Set
= ...
= β
•
Fit model on training set, compute accuracy of your model on test set.
( n
1
( s 2
1
−
+ s 2
2
1) + s 4
2
) 2
/ ( n
2
− 1)
•
Compute accuracy on test data.
s 2
1
/n
1
¯
+
2 s
−
2
2 d
/n
2
¯ s/
− µ n
0
�
ˆ
ˆ
(1
−
− p
0 p ) /n
�
ˆ
1
(1 − p
1 p ˆ
1
− p ˆ
2
− d
) /n
1
+ ˆ
2
(1 − p
2
) /n
2
λ
XY ij
= β u i v j
λ
XY ij
= β i v j
λ
XY ij
= β j u i
λ
XY ij
= β i
β j
�
¯
− t ·
σ
√ n
,
¯
+ t ·
σ
√ n
�
H o
: π ijk
= π i ++
π
+ j +
π
++ k
H o
: π ijk
= π i ++
π
+ jk
H o
: π ijk
= π ij +
π
+ jk
/ π
++ k
ˆ
1
− p
2
± z ·
�
ˆ
1
(1 − ˆ
1
) /n
1
+ ˆ
2
(1 − p
2
) /n
2
1
Example: Running in
OZ
• http://www.statsci.org/data/oz/ms212.html
•
Students in an introductory statistics class participated in a simple experiment:
The students took their own pulse rate. They were then asked to flip a coin. If the coin came up heads, they were to run in place for one minute. Otherwise they sat for one minute. Then everyone took their pulse again. The pulse rates and other physiological and lifestyle data are given in the data.
Linear Model
• y = a + b
1 x
1
+ b
2 x
2
+ b
3 x
3
+ ... + b p x p
•
Matrix definition
Y = Xß where Y is column vector of length n and X is model matrix of dimension n by (p+1)
• some considerations w.r.t. X
40
20
0
80
60
40
20
0
60
80
First step: look at difference in Pulse2 - Pulse1
80
60
40
20
0
1 2 1.1
2.1
1.2
interaction(factor(Gender), factor(Ran))
2.2
factor(Gender)
40
20
0
80
60
20 25 30
Age
35 40 45 40 60
Weight
80 100
Running in OZ
• graphically not much support for any variable but “Ran”
lm( formula = dPulse ~ . Pulse1 Pulse2 , data = fitness )
Residuals :
Min 1 Q Median 3 Q Max
-41.687
-3.319
0.022
5.505
42.394
Coefficients :
Estimate Std. Error t value Pr(>| t |)
( Intercept ) 47.8000
82.1489
0.582
0.562
Height 0.1325
0.1100
1.205
0.231
Weight -0.0349
0.1327
-0.263
0.793
Age -0.1142
0.3752
-0.304
0.761
Gender2 -0.6911
3.5736
-0.193
0.847
Smokes2 1.8033
4.8125
0.375
0.709
Alcohol2 2.2119
3.1862
0.694
0.489
Exercise2 0.8202
4.5202
0.181
0.856
Exercise3 0.7009
4.9284
0.142
0.887
Ran2 -52.6999
2.8767
-18.319
< 2e-16 ***
Year -0.1791
0.8203
-0.218
0.828
---
Signif. codes : 0 ‘ *** ’ 0.001
‘ ** ’ 0.01
‘ * ’ 0.05
‘.’ 0.1
‘ ’ 1
Residual standard error : 14.45
on 98 degrees of freedom
( 1 observation deleted due to missingness )
Multiple R squared : 0.7823
, Adjusted R squared : 0.7601
F statistic : 35.21
on 10 and 98 DF , p value : < 2.2e-16
Goodness of Fit
• for simple linear model:
R 2 is square of correlation between X and Y
• if Model 1 is simpler form of Model 2:
R 2 of model 1 is smaller than R 2 of model 2
•
Instead: use adjusted R 2 for model with p variables X
1
, X
2
, ..., X p
R 2 adj
= 1 - (SSE/(n-p-1)) / (SST/(n-1))
Parameter Estimates
•
ß = (X’X) -1 X’Y
•
(X’X) -1 exists, if X has full column rank, i.e. no dependencies between columns of
X
•
we can use a generalized inverse if X does not have full column rank
Individual Parameters
• in the model result we see that no other variable is significant
•
How do we get significances?
Distributional Assumption
•
Y = X ß + error
•
Normal Model: errors are independent, identically distributed with N(0, sigma 2 )
•
Then (Y-Xß) ~ N(0, sigma 2 ) and Y ~ N(Xß, sigma 2 ) and (X’X) -1 X’Y ~ N( ß, (X’X) -1 sigma 2
)
Confidence Intervals
• for ß i
i=1, ..., p+1:
b i
/Var(b i
) ~ t n-1
•
R commands: coef, vcov, confint
Compare Models
•
Often we want to test hypothesis of the form:
H
0
: ß
1
= ß
2
= ... = ß k
=0
•
Equivalent to comparison of two models M
1
and
M
2
, where M
1
is sub-model of M
2
•
M
1
is sub-model of M
2
, when M
2
has all parameters that M
1
has and additional parameters
•
H
0
: parameters in M
2
but not in M
1
are 0
H
1
: at least one parameter is not 0
F-test
•
Assume model M
1
has p parameters and model M
2
has p+k parameters
• let SSE
1
be the sum of squared errors in model M
1
and SSE
2 the errors in M
2
, then
•
[(SSE
1
- SSE
2
)/k]/[SSE
2
/(n-p-k)] ~ F k,n-p-k
• i.e. for a small p-value we reject the null hypothesis in favor of model M
2
; a large p-value makes us keep the simpler model
F distribution
•
F has two parameters: n1 and n2, called degrees of freedom
• domain is R +
• for n1=1 F = t 2
• use tables/applets for significant values
Example: Running
•
M1: Ran
•
M2: Ran and all other variables
> base <lm( dPulse ~ Ran , data = fitness )
> anova( base , model )
Analysis of Variance Table
Model 1 : dPulse ~ Ran
Model 2 : dPulse ~ ( Height + Weight + Age + Gender + Smokes + Alcohol +
Exercise + Ran + Pulse1 + Pulse2 + Year ) Pulse1 Pulse2
Res.Df RSS Df Sum of Sq F Pr(> F )
1 107 20969
2 98 20454 9 514.59
0.2739
0.9803
Conclusion: no variable besides Ran contributes significantly
Residual Plots: fitted vs residuals
• under model assumptions fitted values are independent of residuals - we should not see trends or patterns
• residuals should have same error variance - we should see a “band” around zero of same height across Y
• only 5% of residual values should be above
2 or below -2
Residual Plots: explanatory vs residuals
• under model assumptions X values are independent of residuals - we should not see trends or patterns
• residuals should have same error variance - we should see a “band” around zero of same height across X
• only 5% of residual values should be above
2 or below -2
0.1
0.0
-0.1
-0.2
-0.3
0.3
0.2
Problematic residual plots
0.2
0.1
0.0
-0.1
-0.2
0.0
0.5
fitted(m)
1.0
1.5
0.2
0.4
fitted(m)
0.6
0.8
From Fitness Data
40
•
Two Groups with non-homogeneous variance
•
Large Residuals
-20
20
0
60 80 100 120 fitted(model)
140 160 180
Variable Selection
•
Forward Selection: include most significant terms until upper limit is reached or no significant improvement can be found
•
Backward Selection:
Start at complex model, prune least significant terms
Next time:
•
Interaction Effects