BCB 444/544 Fall 07 Oct 29 ... BCB 444/544
advertisement

BCB 444/544 Fall 07 Oct 29
BCB 444/544
Homework 5
Due Mon Nov 5
HW 5
Name _______________________________________
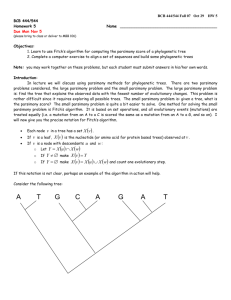
1. Use Fitch’s Algorithm to determine the parsimony score of the tree shown below.
must:
a. Label each node in the tree with the set of possible states
For full credit, you
b. Give the parsimony score (number of evolutionary steps) for the tree Parsimony score is 8
G
T
C A
{CT}*
G
{AG}*
A
T
A
T
A
A
G
{AT}*
{AT}*
{A}
C
{GC}*
{AGC}*
{GCT}*
{A}
{A}
{A}
{AGCT}*
Questions
1. Are the trees produced by the various methods identical? Describe the major differences (if any) between the
trees you produced.
The trees from the different programs have similar groupings with a distinct HV2 group with the 2 SIV
sequences clustered just outside (except for Fitch where the fell within) and a HV1 group that is very
distinct in some trees (UPGMA and Kitch) and less distinct in others. The different trees have different
topologies in the sense that the Fitch and NJ trees have branch lengths that represent evolutionary
distances, whereas Kitch, UPGMA and Parsimony show equal branch lengths. Since these trees are rooted
the roots vary but it doesn’t change the relationships between the species among the trees.
2. From your consensus tree made by the protpars bootstrapping, What grouping of species had the highest
bootstrap support value? What grouping of species had the lowest bootstrap support value?
There were several branch points (internal nodes) with a bootstrap support of 100%, see the parsimony tree
below. The lowest bootstrap support was between HV2NZ and HV2RO (36.2%).
BCB 444/544 Fall 07 Oct 29
Fitch
NJ-UPGMA
Kitch
HW 5
NJ-NJ
+------HV1JR
+-77.6-|
+-100.0-|
+------HV1B1
|
|
+-57.8-|
+-------------HV1MN
|
|
+-100.0-|
+--------------------HV1EL
|
|
+-100.0-|
+---------------------------HV1Z8
|
|
|
+----------------------------------SIVCZ
|
|
+------HV2NZ
+-100.0-|
+-36.2-|
|
|
+-56.1-|
+------HV2RO
|
|
|
|
|
|
+-72.5-|
+-------------HV2G1
+------|
|
|
|
|
|
+--------100.0-|
+--------------------HV2BE
|
|
|
|
|
+---------------------------HV2D1
|
|
|
+------------------------------------------------SIVM1
|
+-------------------------------------------------------SIVML
parsimony
Parsimony