1 23 Isolation, characterization and predicted genome locations of ruff (
advertisement

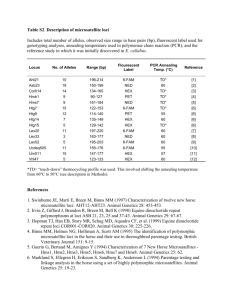
Isolation, characterization and predicted genome locations of ruff (Philomachus pugnax, AVES) microsatellite loci Lindsay L. Farrell, Deborah A. Dawson, Gavin J. Horsburgh, Terry Burke & David B. Lank Conservation Genetics Resources ISSN 1877-7252 Conservation Genet Resour DOI 10.1007/s12686-012-9639-0 1 23 Your article is protected by copyright and all rights are held exclusively by Springer Science+Business Media B.V.. This e-offprint is for personal use only and shall not be selfarchived in electronic repositories. If you wish to self-archive your work, please use the accepted author’s version for posting to your own website or your institution’s repository. You may further deposit the accepted author’s version on a funder’s repository at a funder’s request, provided it is not made publicly available until 12 months after publication. 1 23 Author's personal copy Conservation Genet Resour DOI 10.1007/s12686-012-9639-0 TECHNICAL NOTE Isolation, characterization and predicted genome locations of ruff (Philomachus pugnax, AVES) microsatellite loci Lindsay L. Farrell • Deborah A. Dawson Gavin J. Horsburgh • Terry Burke • David B. Lank • Received: 26 March 2012 / Accepted: 28 March 2012 Ó Springer Science+Business Media B.V. 2012 Abstract We identified 247 unique ruff (Philomachus pugnax) microsatellite sequences. Primer sets were designed from 102 selected loci and tested in 12–24 individuals from a captive population. Sequence homology was used to assign locations in the chicken (Gallus gallus) and/ or zebra finch (Taeniopygia guttata) genome for the majority of these loci. Fifty-two loci were found to be polymorphic and 47 of these were typed in known families. Forty-six loci displayed Mendelian inheritance including Ppu058, which was confirmed to be Z-linked by the complete absence of any heterozygous females. Keywords Lek Male morphs Microsatellite Scolopacidae Shorebird Simple tandem repeat (STR) We have isolated and characterized microsatellite markers for the ruff (Philomachus pugnax). The ruff is a lekking shorebird with an autosomal genetic polymorphism for male mating behaviour (Lank et al. 1995) and belongs to the Scolopacidae family. A proportion of the ruff microsatellites isolated will be of utility in closely related species (Primmer et al. 1996), including other species of Scolopacidae (n = 88, Sibley and Monroe 1990). Within this family, several species are of conservation concern, including the critically endangered spoon-billed sandpiper L. L. Farrell D. A. Dawson G. J. Horsburgh T. Burke Department of Animal and Plant Sciences, University of Sheffield, Western Bank, Alfred Denny Building, Sheffield S10 2TN, UK L. L. Farrell (&) D. B. Lank Department of Biological Sciences, Simon Fraser University, Burnaby, BC V5A 1S6, Canada e-mail: lfarrell@sfu.ca (Eurynorhynchus pygmeus; IUCN Red List, Birdlife International). Blood was collected from captive ruff individuals at Simon Fraser University, Canada (population maintained by DBL since 1985), and stored in absolute ethanol. Genomic DNA was extracted using ammonium acetate (Nicholls et al. 2000). Two ruff microsatellite-enriched libraries were created following Armour et al. (1994). For both libraries, genomic DNA from a single female ruff (Bird ID 6233) was digested with MboI, size selected (178–856 bp) and enriched for (CA)n, (CAG)n, (GCC)n, (TTTC)n and their complements. The first library was additionally enriched for (AT)n. Clones were sequenced in both directions using BigDye terminators and a consensus sequence created. We used BlastN 2.2.4 software (Altschul et al. 1997) to check for duplication of new and published ruff microsatellites (n = 9 published, Thuman et al. 2002). In total, 247 unique microsatellite sequences were identified and primers designed for 102 loci using PRIMER3 (Rozen and Skaletsky 2000; Tables 1, 2). Primers were designed for just three of the 104 sequences that contained only mononucleotide (A or T) repeats (Ppu017, Ppu033 and Ppu071). One hundred and two markers were tested in 12–24 known-sex individuals using multiplex PCR. Each 2-ll PCR contained approximately 10 ng of genomic DNA, 0.2 lM of each primer and 1 ll Qiagen Multiplex PCR Mix (Qiagen Inc.; Kenta et al. 2008). PCR amplification was performed using a DNA Engine Tetrad 2 Thermal Cycler (MJ Research, BioRad UK) with the profile: 15 min at 95 °C, followed by 35 cycles of 94 °C for 30 s, annealing temperature (Tables 1, 2) for 90 s, 72 °C for 1 min, and a final step of 60 °C for 10 min. Products were loaded on an ABI3730 Genetic Analyzer (Applied Biosystems) using ROX500 size standard and genotypes scored with GENEMAPPER v4.0 123 123 Ppu022 Ppu021 Ppu020 Ppu019 Ppu018 Ppu017 Ppu016 Ppu015 Ppu014 Ppu013 Ppu012 Ppu011 Ppu010 Ppu009 Ppu008 Ppu007 Ppu006 Ppu005 Ppu004 Ppu003 HE616911 Ppu001 24C08 HE616932 24C05 HE616931 24B05 HE616930 24A01 HE616929 6G09 HE616928 6F12 HE616927 5E02 HE616926 5B12b HE616925 5A10 HE616924 4H10 HE616923 4G03 HE616922 4F08 HE616921 4D09 HE616920 4D05 HE616919 4D01 HE616918 4C09 HE616917 4A08 HE616916 3H04 HE616915 3E05 HE616914 3E04 HE616913 3C02 EMBL acc. no. & clone name Locus 2 2 2 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 Lib 2 2 1 1 11 11 – 3 2 2 – 4 1 1 2 2 24 – 3 3 3 3 3 3 4A 4 4 4 – 3 3 5 5 8 8 2 2 1 1 1A 1 CH chr ZF chr 79772128 75106465 61096046 156510069 71134 19600964 – 84720681 71904153 61061152 – 52883524 65490386 166535076 22065196 19888521 3112345 – 18163845 18786830 56310381 51033809 49883043 57218799 2561871 6796049 29737180 23020195 – 76352543 76776326 16041469 17130526 19350034 22771586 92082626 89244831 13670975 122413141 50776302 52975585 Chicken chr loc. T – Zebra finch chr loc. T – (ATAGAT)9 6-FAM R: GGGAAACATCATGCAACAAC F: TGAATGCATGAATTAGGTAGTGG 1.20e-37 8.90e-20 F: AAAGCTTGTAAGCTCTAAGCAATACC 6-FAM R: AGGCTATTGACACTTCACAAAGG (CTAT)12 3.90e-57 4.00e-81 R: GCGGTATTTCTGGCCTAGC F: TCCTGTCCTGTCCTTGGAAC HEX 3.10e-20 6.70e-25 R: GCTACTGGGTGCTGTTACTTCC – (GT)13 F: TAACCCACGAGTGGCTCTG HEX 7.60e-10 (AC)11 R: AGAGATACAGTAAGCTTCGTATGACAGACAC F: TGCCTTCTTACTTTCTCAATATTTGTGG HEX 1.90e-28 4.90e-38 R: GTGCTACTGAAATCGTCTGATGTTGG – (AGAT)13 F: GTTGGCCTGGACTCCGTCTG HEX 6.00e-06 (T)14 R: TCAAAGACTTCTGCAAAGTTATTCTTCTAAGC HEX F: TCAGGCAGTGGGACTAGATGATTG (TC)12 1.10e-22 5.70e-13 R: TGACTTTGGAGGTTGTTACTTATTGTTGTC F: GGTCCAGTTCTGTGTGCCAGTTT HEX 1.70e-54 (AC)5 F: CAACCCCATCTCCTGGCTTTT 9.80e-27 6-FAM R: TGCTCCATGGAATCAAACATGG R: CAGCTCGGTACATTGGTGCTTG (GT)10 HEX 1.20e-30 – (AG)6 F: ACATGCTCCTCTTCCATTTGCAG 3.80e-63 1.40e-61 F: TGCAAGGCCAGGTAGAAACAAG 6-FAM R: ACCAAGGCATTACTGTGTTGGAAG (AC)6 4.90e-03 1.20e-32 R: TGGACTGAAGGTGACTATTCTGCTG HEX F: CGCACATCTGCTGTTGAGAAATC (AC)5 3.40e-63 6.40e-43 R: GATGTGAACTACCTGCAAATCCACAG F: GGGAGCTCAGGGATGCAGTG HEX F: TCTTTATGATGCTATTTGAGGGTTTGG 2.20e-37 (GT)12 F: GAAGTTCCTCTTACCAATTTGCTTGC R: TGACCTGCTGGTACTCCACCAC 6.90e-17 HEX 6-FAM R: CCTATTCATGTCTCCAAGTTCAATCC R: AATGCCACTGCACCAGAAGTAGTC (AC)12 (AC)12 6-FAM 3.70e-46 5.90e-64 – (GT)5 F: GCCAGAGTAGCAACAGTCAGTGC 4.50e-52 4.00e-27 F: TGGAAGTGGAAGGAGGTCTGTG 6-FAM R: TCCACTCAGGTGCAGGCTTC (GT)9 8.10e-70 2.70e-53 F: GGAGCAATGTGATACCACTAAGGACTG 6-FAM R: CTCCTGACCTCCACCGCAAC (TC)5 4.40e-73 8.00e-108 F: TTGGCACCAATAGTTGCCTCA 6-FAM R: CCTCTTCAGAGGAACAAAGCAAGA (TCTA)10 9.50e-79 8.80e-84 R: AGCATGTAGTGCTTCAGTTATTTAGATGC F: CAGGATTGCTTTGGCTGGAG HEX 1.20e-27 (CTAT)11 F: ACCAGGCTTCTTCCCTCTGGA 5.60e-61 HEX Primer sequence 50 –30 R: TGAAACTTCACATTTTGGGGATGA (TAGA)12 1.00e-07 Fluoro label (F) 3.30e-27 Repeat motif E-value in Gga E-value in ZF 722 791 367 407 536 414 584 693 431 542 495 371 530 436 427 577 534 480 646 337 494 Seq. length (bp) Table 1 Characterization and predicted chromosome locations of 52 ruff (Philomachus pugnax) microsatellite loci Y Y Y Y Y Y Y Y Y Y Y Y N Y Y Y Y Y N Y Y MI 58.83 59.02 59.70 59.92 59.82 60.09 60.19 59.84 63.51 63.74 65.42 65.16 65.15 64.74 64.80 64.51 65.07 64.61 64.86 65.17 63.55 63.69 63.99 64.29 65.06 65.55 63.91 64.36 64.68 64.41 63.66 63.20 63.80 63.90 65.42 65.13 62.9 60.8 63.18 63.13 64.67 64.65 Tm (°C) 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 56 PCR Ta (°C) 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 24 n1 23 15 23 20 24 10 24 20 24 17 23 21 23 24 24 13 22 24 10 23 24 n2 7 5 7 5 8 2 6 3 6 2 2 3 5 12 3 2 2 2 3 4 4 A 297 319 246 150 250 223 299 248 201 230 219 222 371 226 300 293 250 227 230 274 197 Exp allele size (bp) ° 267–302 (296, 296) 312–329 (321, 321) 245–302 (243, 245) 145–156 (151, 154) 242–270 (250, 258) 207–218 (227, 227) 212–227 NT 244–278 (247, 247) 194–204 (214, 220) 223–228 (229, 229) 214–222 NT 216–223 (223, 223) 221–233 NT 317–474 (436-436) 298–302 (299, 299) 281–290 (294, 294) 241–246 (245, 251) 216–220 (221, 221) 192–233 NT 367–373 (366, 371) 275–287 (275, 279) Obs. allele size (ruff 6233) & range (bp) ¥ 0.91 0.40 0.52 0.45 0.79 0.10 0.62 0.40 0.62 0.35 0.69 0.57 0.17 0.95 0.20 0.38 0.31 0.12 0.10 0.65 0.62 HO 0.80 0.70 0.44 0.69 0.78 0.52 0.62 0.51 0.72 0.47 0.48 0.51 0.71 0.89 0.19 0.40 0.48 0.11 0.59 0.70 0.57 HE 0.61 0.04§ 1.00 0.02§ 0.04 0.02§ 0.05 0.65 0.19 0.50 0.19? 1.00 0.00§ 1.00 1.00 1.00 0.06 1.00 0.11§ 0.30 0.12 pHWE* Author's personal copy Conservation Genet Resour Ppu046 Ppu043 Ppu042 Ppu041 Ppu040 Ppu039 Ppu038 Ppu037 Ppu036 Ppu035 Ppu034 Ppu033 Ppu032 Ppu031 Ppu030 Ppu029 Ppu028 Ppu027 Ppu025 Ppu024 HE616933 Ppu023 3C03 HE616956 5A12 HE616953 3A02 HE616952 6D06 HE616951 4G04 HE616950 3E11 HE616949 3D04 HE616948 6A11 HE616947 4A01 HE616946 4H11 HE616945 67A09 HE616944 65A09 HE616943 65A08 HE616942 64C01 HE616941 64A02 HE616940 63G02 HE616939 63D04 HE616938 63D02 HE616937 63C04 HE616935 63A05 HE616934 33C07 EMBL acc. no. & clone name Locus 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 Lib Table 1 continued 4 4 2 2 11 11 11 11 5 5 1A 1 2 2 26 26 10 10 3 3 10 10 8 8 2 2 13 13 2 2 10 10 1 1 7 7 – 1 – 13 7 7 CH chr ZF chr 37989476 41459422 33590280 50681577 15017303 9474078 15593478 10001495 321716 2524315 49840927 52147383 101346526 98252724 1368934 2193405 895537 4306840 104595407 104574877 10268721 12101849 27910568 30604045 49636829 29908799 16024608 1071128 122215464 120354672 7745116 9185554 108220670 130142524 19299021 15230709 – 24109016 – 9775634 3954280 29587083 Chicken chr loc. T – Zebra finch chr loc. T – 6-FAM R: TGACACACAGGTTTGGAA F: TCGTCTGATTTGTATTGTTCTT (GT)10 1.90e-138 2.20e-42 R: GACTTTGGTAGGTCTTCTGGTTCCTG HEX F: CTCTCCAGGTTGCCTAAAGTCTATGG (GT)7 8.30e-54 2.10e-05 R: TTGTTCACATTGAGAAGTTGATGAC HEX F: TGGTCCCTTCCAACTCTAGAAA (TTTC)48 1.0e-139 3.80e-07 R: AGCAGACCGGAGAAGCAACA HEX F: TGATTTTCCGAAACAAGTTTTAATCG (AC)9 6.50e-56 1.90e-71 R: GGAACGATGTGGGTTACTTCCAG HEX F: CTCCTGGCTGCGTTGTTCTG (GT)9 1.20e-59 1.90e-58 R: CTTGCCATTCAGGTTAAGTACACTTCC 6-FAM F: GCAACTGCTGCACTCCCAAC (TGAT)6 1.50e-73 1.30e-52 R: TTAATATGGCAGCCTTACCTAACGAAAC HEX F: CATGACTACCTATCGAATCCTCTTTGG (GAAA)5 1.70e-42 4.10e-22 R: TCCATATTTATTACAGCCCAGAAGACC HEX F: CTCTTGTGGTACCTGGAAGAGGTG (GT)6 1.10e-19 3.30e-50 R: TTTCCCAGCATGACATACATTGC 6-FAM F: AGACCCGGGTGTTCAAGGTG (GT)7 2.10e-33 1.20e-46 R: TTGAACGTTTGGGACAGGTGAC 6-FAM F: AAGCTTGTCTTCTGGAATGAAGCAG (GT)9 2.00e-14 7.90e-37 R: CCACCCTTCATATTGACTCG HEX F: CTCCATGGACCAGAAATGAG (AAT)6 6.40e-37 3.20e-56 R: GATCATCTAGTCTAACCATCAACTCTG HEX F: AGTTGTTGAGGCACCATCC (G)12 1.80e-07 9.00e-27 R: TAAGAGGTTGCCAGGTTGTG 6-FAM F: CATTTCTTGTTGTGATTAATAGTCTCC (AC)18 6.50e-10 1.70e-05 R: TGAGGACTGTGGTTTAAGAGC 6-FAM F: TGATTCTTATTAGGATTATTTGATGC (GT)10 2.30e-25 8.90e-41 R: CTCGTTGGTCATAATTTGAGG HEX F: CAGGCTTAACACTCTTTCTTCC (GT)11 2.30e-77 1.30e-110 R: CTAACCTGGATGGCTGTTTG HEX F: AGGGTATTGTTGGAGAAATGG (GT)10 1.50e-50 1.10e-62 R: GCACCAGAACTGCCACATAG HEX F: CCTGAACCATTAGTTTACTTGCTG (TGAT)6 2.50e-12 2.30e-14 R: TCCTGTGAGCTGTTAATTCTGAG F: TGTTAGCAGGCTGATGTGTG 6-FAM 1.20e-28 6.60e-43 R: GCATCACAAATGCAACTTCAG – (GAAA)8 6-FAM F: GATCCAGACTGCCTAAACAGC (AC)17 2.20e-18 F: GGAAACCTTCCCATCAACAG R: GCCAGGGCAGAGACATAAAG F: CAGCTATGAGGTTACCAGAGAGG Primer sequence 50 –30 R: GAAGGGATGCATGGTTGG HEX 6-FAM Fluoro label (F) – 1.80e-12 (TCTA)7 (AGAGAGGAG AAGAGAGA)7 4.70e-13 5.80e-52 Repeat motif E-value in Gga E-value in ZF 306 255 413 614 600 563 422 454 673 576 548 491 461 437 713 567 266 480 421 294 592 Seq. length (bp) Y N Y Y Y Y Y Y Y N Y Y Y Y Y Y Y Y Y Y Y MI 55.34 55.22 64.54 64.51 60.44 60.10 63.40 63.39 63.96 64.38 64.23 64.30 64.38 64.21 63.55 63.43 64.15 64.53 64.33 64.40 58.02 58.07 58.85 58.50 58.77 58.69 57.07 57.14 57.23 57.32 58.23 57.92 59.32 59.28 58.66 58.44 59.34 58.95 59.85 59.38 59.84 59.44 Tm (°C) 56 64 60 63 64 64 64 63 64 56 56 56 56 56 56 56 56 56 56 56 56 PCR Ta (°C) 12 12 12 12 12 12 12 12 12 24 24 24 24 24 24 24 24 24 24 24 24 n1 12 12 11 12 12 12 12 12 12 22 15 24 19 17 21 18 24 24 14 17 21 n2 3 2 2 2 3 2 3 2 3 2 2 3 8 3 3 2 3 10 7 9 11 A 170 186 377 173 207 193 279 234 203 235 136 156 262 321 136 166 182 301 348 138 294 Exp allele size (bp) ° 173–180 (177, 177) 185–187 NT 360–363 (127, 127) 172–174 (173, 173) 209–213 (207, 209) 190–194 (194, 194) 276–283 NT 235–237 (236, 236) 200–208 (200, 204) 178–214 NT 132–149 (132, 132) 135–162 NT 250–271 (258, 260) 320–325 (321, 321) 131–140 (136, 136) 168–170 (168, 168) 185–193 (185, 185) 277–320 NT 331–346 (332, 346) 122–161 (135, 157) 287–406 (287, 287) Obs. allele size (ruff 6233) & range (bp) ¥ 0.58 0.16 0.18 0.50 0.25 0.58 0.33 0.33 0.50 1.0 0.00 0.95 0.57 0.11 0.90 0.11 0.58 0.58 0.85 0.82 0.23 HO 0.62 0.15 0.17 0.46 0.23 0.50 0.30 0.29 0.42 0.51 0.12 0.55 0.77 0.11 0.62 0.35 0.58 0.72 0.87 0.86 0.78 HE 0.29 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.00§? 0.04§ 0.00? 0.04§ 1.00 0.21? 0.34§ 0.44 0.54§ 0.72 0.38 0.00§ pHWE* Author's personal copy Conservation Genet Resour 123 123 5C04 HE616970 4F01 HE616969 4C06 HE616968 3F07 HE616967 4H06 HE616962 3F11 HE616961 6F01 HE616960 6B02 HE616959 5C08 HE616958 1 1 1 1 1 1 1 1 1 1 Lib 1 1 1 1 Z Z 6 6 – 4 3 3 4 4 1 1 1A 1 6 6 CH chr ZF chr 67624179 136476610 12955004 121754800 37214802 6052811 18076829 20079681 – 38001317 26505233 2063655 41075858 64774867 65490386 163634502 3982690 4758111 27151910 28674879 Chicken chr loc. T – Zebra finch chr loc. T – 1.80e-14 R: TTGTGATGCACAAGTCTTTCAAGG F: TGAATGCAAATACAGCATCAGTGAG 6-FAM R: CTGACTCATGATGCCTCATCTCG (GT)7 2.60e-34 6.10e-27 HEX F: TCTACTGAGCTCACAGAAACAAAGGAAC (GT)8 2.20e-51 F: AGTAGCTGCCAATCCACAGG 6-FAM R: TCTCCTGCTTGGCCTCTTT (GT)14 1.30e-27 1.30e-45 R: CCTGCTGTGAAATCTACCCATCC F: TGCAGTGCAATGTGTGTGACC 6-FAM 3.70e-21 8.50e-28 R: AATGCTTGTGGGTGGCAATG – (AG)8 F: TTGCACGTGTCCTTAGCTTGC 6-FAM 2.30e-02 (GT)6 R: GTTGGGTGTTCCTGCTGACG HEX F: CCCGTCAGCGAATATAAGAGCAG (AC)10 1.60e-53 3.10e-41 F: TCAGCACTGAAACTGAGGAAATTATTG 6-FAM R: GAGCATTCCTCCCGCTGTG (AC)7 2.00e-58 4.20e-09 F: AACTTCAAAGACTTCTGCAAAGTTATTCTTC 6-FAM R: TGAACTTACACTGGTGAACTAACTTTCTCTC (AG)12 3.00e-08 2.10e-14 R: AACACACTGAGCGTCGTTTTATCA F: TGCAGCATTCTTCGCAGCTA HEX 2.70e-75 (TC)10 F: TGCAGCTTTAATTGCAACAGCTAATC 2.20e-70 HEX Primer sequence 50 –30 R: AGCGCTCAGGTCTGAATGAGTTC (TC)10 1.60e-103 Fluoro label (F) 1.60e-61 Repeat motif E-value in Gga E-value in ZF 493 331 477 564 641 593 640 522 659 496 Seq. length (bp) N Y Y Y N N N Y Y Y MI 63.60 63.51 64.42 64.36 60.08 60.28 64.07 64.28 64.11 63.67 64.02 64.26 64.26 63.80 63.48 63.61 63.09 63.22 64.35 64.66 Tm (°C) 64 63 60 64 63 64 63 63 63 64 PCR Ta (°C) 12 12 12 12 12 12 12 12 12 12 n1 9 9 6 (F) 6 (M) 8 12 10 12 12 12 12 n2 2 2 6 6 8 3 3 4 10 4 3 A 214 263 224 360 238 198 276 389 225 291 Exp allele size (bp) ° 213–214 NT 263–265 (262, 262) 221–233 (227, 227) 328–381 (329, 358) 238–242 NT 192–204 NT 225–275 NT 280–366 (368, 376) 223–231 (226, 230) 288–292 (290, 292) Obs. allele size (ruff 6233) & range (bp) ¥ 0.44 0.33 0.00 0.85 1.00 0.08 0.20 0.16 0.75 0.50 0.58 HO 0.36 0.50 0.00 0.87 0.90 0.23 0.24 0.23 0.87 0.53 0.62 HE 1.00? 0.49§ – 0.06 0.82 0.05§ 0.04 0.13 0.05 0.45 0.21 pHWE* Deficiency of heterozygotes observed; ? excess of heterozygotes observed, determined by Chi squared test (two tailed p \ 0.05). Deficiencies and excesses of heterozygotes are likely attributable to non-random population structure caused by captive breeding * HWE for all loci was assessed in 12 individuals, which may contain second order relatives (half-sibs and closer relatives were removed) ¥ the allele sizes presented in parentheses are those of the female ruff individual (Bird ID 6233) from which the genomic library was created, NT not tested ° Expected allele size based on the sequenced clone allele of the female ruff individual (Bird ID 6233) from which the genome library was created MI Mendelian inheritance displayed when genotyped in a pedigree of 64 families, consisting of both parents and 5–33 offspring per family; Y yes, N no pHWE calculated from a maximum of 12 individuals, using GENEPOP v.4.0 (Rousset 2008) § n1 Number of individuals tested in a captive population. n2 Number of individuals amplified and genotyped. A Number of alleles observed, M male, F female. HO observed heterozygosity (calculated from n, using CERVUS v3.0). HE expected heterozygosity (calculated from n, using CERVUS v3.0), HWE Hardy–Weinberg equilibrium T – The location of each microsatellite sequence was assigned in the chicken (v 2.1, May 2006 ENSEMBL release) and zebra finch (December 2011 ENSEMBL Release 65) based on sequence homology (see Dawson et al. 2006, 2007). Of these 52 polymorphic loci characterized, 50 could be assigned a location in the chicken genome and 46 in the zebra finch genome. Ppu060 Ppu059 Ppu058 (Zlinked) Ppu057 Ppu052 Ppu051 Ppu050 Ppu049 Ppu048 HE616957 Ppu047 3B01 EMBL acc. no. & clone name Locus Table 1 continued Author's personal copy Conservation Genet Resour Ppu104 Ppu101 Ppu095 Ppu092 Ppu086 Ppu083 Ppu079 Ppu071 Ppu070 Ppu064 Ppu056 Ppu055 Ppu054 Ppu053 Ppu045 Ppu044 Ppu026 HE616912 Ppu002 6H07 HE617014 5F03 HE617011 5C07 HE617005 6C11 HE617002 6B07 HE616996 4G10 HE616993 5G10B HE616989 4G02 HE616981 6E05 HE616980 5H06 HE616974 66F02 HE616966 65H06 HE616965 65F04 HE616964 3H05 HE616963 4D02 HE616955 3E10 HE616954 63C05 HE616936 3D09 EMBL acc. no. & clone name Locus 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 Lib 5 5 Z Z 9 9 6 6 Z Z 9 9 – 4 2 2 5 5 5 5 22 22 1 1 8 8 Z Z 2 2 – – 1A 1 – CH chr ZF chr 53923720 54678545 45558110 14416326 13825337 13341088 12562519 14776618 22314343 72933854 12079437 11757764 – 39272313 19900285 17900039 33970154 36514685 48965965 50276059 1590011 690245 85953917 196624265 11832348 15987120 59515109 32113785 141544149 140526702 – – 36868616 39405325 – Chicken chr loc. T – Zebra finch chr loc. T – (GT)5 HEX R: TGTGAAAGCCCTTGAAGGCATC F: CCCAATGTGCAGAAACCCTCAC 6.40e-46 1.10e-70 F: TTCACTTACACTTGAGGATGGTTG 6-FAM R: AAACTAAACTACTGCTGGGCATTG (GT)5 6.10e-33 7.60e-82 F: GGTGTGTGTTTCTGTCTCCCTCT 6-FAM R: ATGGGAGTTATGCACACATCAAA (TC)5 1.70e-35 2.00e-71 F: AAAGGAGTTCCCTGGCACAGTTT HEX R: CGGCACAATTAATACTACCGGCTTA (AT)5 1.30e-67 1.90e-55 F: TCTGCAGTTGGAAGGCTGTAA 6-FAM R: TCTTGATGTTGCCAAAATATACATACA (GT)5 2.90e-76 1.00e-81 R: TCCATCAAGAACTCATTAGCTTAGTAGCC F: CTGGGTGTTGTGCCTTCAGC 6-FAM 7.10e-96 5.70e-92 R: GTTGGGCGGAGAATGGACTG – (GT)5 HEX F: AGCTGAATGAACATCTGCATATTAGAACAC (GA)7 2.10e-19 F: AAATCTGAACCAGAAATAATATAGTCAATCC 6-FAM R: TGAAGTGTTCTCCACTTCTTTGATG (T)10 1.20e-43 3.50e-61 F: TTCTCCTCACCCTCCTCTAGCATTTAG 6-FAM R: GCGCATGGCTTTATACCATATTTCC (AT)5 1.80e-44 5.70e-62 F: TGTTGCTGTTGTTGTCGTGGTC 6-FAM R: GAGTGCTCTGCACCCTTCACC (AC)5 5.20e-43 4.20e-135 F: CCTCTGGCAAATACTCAATGC HEX R: CACTGGAAAGGTCAGGAAGC (AC)8 1.50e-06 1.00e-22 F: TGGAGCTTAACATCTACAAATGC 6-FAM R: TTGGCTTTCTCTTATCCATCAC (GAAA)14 7.90e-12 8.80e-13 F: GCACCGCAGAAGTTGATAAG 6-FAM R: CTGAGGTGCTCATGGTTACAG (GT)5 7.50e-48 2.00e-25 F: TTGGCACCAATAGTTGCCTCAT 6-FAM R: CCTCTTCAGAGGAACAAAGCAAGA (AC)10 9.00e-49 4.30e-84 F: TCATTTCTCCTCCTAACGCTGAAG R: ACTGTCCTTGCATCTTCCTCTCC HEX 7.10e-61 (AC)10 F: TCAGGCACATTACATTATTCACTATCTATG R: GCTGCCACTTACTGAAATCTG F: CTTTGTCATTATGTAGGGTCCTG R: AACATACAACACCTCTCTCTCTTTCTTT F: GAAGCTGAGATTCCCATATAATCACT Primer sequence 50 –30 6.60e-70 HEX 6-FAM HEX Fluoro label (F) R: CCACCCTCCTAGCAAACACC (TGTT)6 (AC)10 (GAAA)44 Repeat motif – – 5.40e-43 3.90e-19 – E-value in Gga E-value in ZF 471 367 504 606 524 562 324 585 401 357 305 375 505 407 561 543 414 538 Seq. length (bp) 65.74 66.02 60.88 60.44 61.51 61.87 64.15 64.44 60.81 60.95 64.16 64.15 65.12 64.72 61.87 61.42 65.41 65.17 64.58 64.41 59.84 59.72 58.33 58.47 58.38 58.53 63.32 63.81 63.22 63.12 61.83 61.77 57.65 57.70 60.45 60.23 Tm (°C) 56 60 56 56 60 60 56 56 56 56 56 56 56 56 63 61 56 56 PCR Ta (°C) Table 2 Additional ruff microsatellite loci with assigned predicted genome locations but that were abandoned from further testing 24 6 (F) 6 (M) 24 24 6 (F) 6 (M) 12 24 24 24 12 24 24 24 12 (F) 12 (M) 12 12 24 24 n1 23 6 6 19 24 6 6 11 17 1 21 11 4 2 7 8 11 11 23 21 n2 1 1 1 1 1 1 1 1 1 1 1 1 2 2 3 1 1 1 1 1 11 A 246 205 247 280 196 212 245 241 198 248 143 336 284 233 326 193 294 384 Exp allele size (bp) ¥ 243 205 205 244 279 196 196 213 245 241 198 245 249–283 276–283 322–338 230 230–238 324 195 291 313–378 Obs allele size (bp) Monomorphic Monomorphic Monomorphic Monomorphic Monomorphic Monomorphic Monomorphic Monomorphic Monomorphic Monomorphic Very poor amplification Very poor amplification Very poor amplification Very poor amplification Monomorphic Monomorphic Monomorphic Unreliable poor reverse primer Marker status Author's personal copy Conservation Genet Resour 123 123 Ppu121 Ppu107 Ppu093 Ppu085 Ppu080 Ppu068 Ppu067 Ppu062 Ppu242 Ppu221 Ppu199 Ppu181 Ppu172 Ppu164 Ppu148 Ppu140 Ppu138 HE617015 Ppu105 65E06 HE617031 6F04B HE617017 5C11 HE617003 6A05 HE616995 6H10 HE616990 3E09 HE616978 4G09 HE616977 5E09 HE616972 67C06 HE617152 63C10 HE617131 67D10 HE617109 63D03 HE617091 66G11 HE617082 64B02 HE617074 64A03 HE617058 21A01 HE617050 36B04 HE617048 6H11 EMBL acc. no. & clone name Locus Table 2 continued 1 1 1 1 1 1 1 1 2 1 2 1 2 2 2 2 2 1 Lib 6 6 1 1 3 3 2 – 1A 1 Z Z Z – 9 9 1 1 26 26 10 10 Z Z 3 3 1 1 12 12 18 18 5 5 – 22 CH chr ZF chr 32662952 33702631 77944747 188022341 106323359 106292912 31937001 – 58008408 60285776 37374037 6212608 43145669 – 16851809 15940752 81416718 191524107 273083 1257327 18002891 19602417 7124071 42994287 110357795 110633741 51091377 178594445 13993026 13482713 9810117 5211094 2549845 4932892 – 1024052 Chicken chr loc. T – Zebra finch chr loc. T – (GT)6 7.60e-12 HEX R: ACGAGAGGCCAAATATGGTG F: TTCCCTAACCAGCAATTAGCTATC (AC)5 7.60e-25 1.80e-33 R: CACAGACACTTCTTTAACAGTTATTCCAACC 6-FAM F: CTTTGGACATGCCAGCAGGA (TTGTT)9 4.20e-09 3.40e-68 F: AGCCATTTGCTTAATCCTACCTTCTG R: ATGGGAGGCCATGCAACAC HEX 7.50e-40 (TAGA)6 F: TGCCCAGGGAAGTGGTTG 4.20e-60 HEX R: AGTCCAACCATCAACCTAACTCTGAC (GT)6 R: CCACAGCTCTGCCTCTGTTAATATG 6.90e-09 – HEX F: TTTCAGATGTGATTGGTGTTCAGC (GA)7 6.90e-13 3.00e-19 R: GGCAGAAGCAAGAAACAACAGAAC F: CTTGCCAAAGGGCAAGGTG 6-FAM 1.20e-32 (AGAT)11 F: CTGACTGCTGCCTGCCTCTC 4.20e-80 6-FAM R: GGTGTCACCTCCTATACCTACATGCTC (AGAAG)55 R: TGTGGAATTAAAGTTCAACCCATTAGC 3.00e-25 – HEX F: TCAGTGTTTCTACTTGGATGGAATGG (AC)7 3.40e-98 1.40e-37 R: TTCTTCTGGAGCCCTCTCTG 6-FAM F: GATCAGGTTGTGAGCAGCAG (GT)7 7.30e-12 5.20e-18 R: TGCTCTGCACTTGTGTAACG 6-FAM F: GCGAGACATCTGAATCTCCTC (GT)8 2.10e-60 9.90e-22 F: TGCTGAAATCCATTCTACTAACG HEX R: CCTATTAATTGCACACAAATTGC (GT)5 1.90e-35 7.10e-39 R: AGCTGCAATTGATGCCAGTG 6-FAM F: TTGGAGGGTTCTTTGGGTTG (GT)10 2.80e-41 7.20e-05 F: CAGGAAGAGGTGAGCTGGAG HEX R: CTAAGGCACACAGCTGAACG (GA)6 2.10e-48 4.50e-52 R: CTGGAAAGGCTGAAATCCTG 6-FAM F: CTGCTCATCTCTCCTTTCTTCC (TC)5 5.20e-62 1.50e-69 R: TCAGGATGACTCCAGACGTG 6-FAM F: TGGCTTGTGCAAAGTAGATTTC (AC)7 1.20e-132 1.70e-106 F: TAGTAGCCAATCGGGTGAGC HEX R: AGCCAGCTGGAGTAGTGTGTG (AC)6 7.00e-85 4.30e-105 F: TTGGATATTCTGCCGAGATG R: GGGAAGCTTGGGATTCTATG 7.60e-42 1.00e-11 F: AAATTAGCATAAAGACGGAGAAGTTGC Primer sequence 50 –30 R: GCCTTCTTGCTGTATTCAGGTGAG HEX 6-FAM Fluoro label (F) – (ATAG)4 Repeat motif E-value in Gga E-value in ZF 593 316 813 450 570 669 606 400 323 468 557 677 305 609 756 713 531 304 Seq. length (bp) 59.96 60.02 64.61 65.04 64.27 63.93 63.02 63.12 63.49 63.36 63.96 63.57 63.79 64.27 64.28 64.50 59.67 59.58 58.65 58.98 58.63 58.47 62.35 62.15 59.66 60.13 59.81 59.60 59.82 59.41 60.52 60.24 58.60 58.67 63.56 63.14 Tm (°C) 56 56 60 60 56 56 56 60 56 56 56 56 56 56 56 56 56 60 PCR Ta (°C) 24 24 12 12 24 12 (F) 12 (M) 12 (F) 12 (M) 12 24 24 24 12 (F) 12 (M) 24 24 24 24 24 12 n1 0 0 0 0 0 0 0 0 5 18 4 4 23 21 23 23 15 10 n2 – – – – – – – – 1 1 1 0 1 1 1 1 1 1 1 A 256 215 230 203 447 263 509 243 315 356 148 – 262 155 329 340 256 257 249 Exp allele size (bp) ¥ – – – – – – – – 126 357 148 – 265 155 331 341 255 284 251 Obs allele size (bp) Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Monomorphic Monomorphic Monomorphic Monomorphic Monomorphic Monomorphic Monomorphic Monomorphic Monomorphic Monomorphic Marker status Author's personal copy Conservation Genet Resour 67A12 HE617150 66F08 HE617146 64E01 HE617094 67G04 HE617083 66E10 HE617080 65H12 HE617078 67F11 HE617073 67B07 HE617071 66A03 HE617065 66A01 HE617064 65C11 HE617060 66C09 HE617044 66E12 HE617039 1 1 1 1 1 1 1 1 1 1 1 1 1 1 Lib 1A – – 3 1A 1 1A 1 2 2 2 2 1 1 4 4 4 4 2 2 15 15 6 6 3 3 1A 1 CH chr ZF chr 48527357 – – 97005426 6252519 7368828 56094038 58206176 39737798 70828859 92057916 89221409 112179239 104968881 12229126 32471914 68748861 91803651 147316308 146180882 994445 3838997 6728242 2107306 31383766 31663341 61734639 73188968 Chicken chr loc. T – Zebra finch chr loc. T – 6.20e-23 – (GT)10 6-FAM 6-FAM R: TCTCATGTACTGTGATGTCTACTGTG F: CCCAGGCTAGGAGCTTTG R: CCAAGACATGCACTGTGTCC F: AACTCACCATATGTGCCAAGG (GT)6 4.00e-29 – R: AAACTGTAAGTTATTCCAAAGGAACC HEX F: GGGTTACAGCTGTTTGTATTTCC (GT)6 1.30e-09 7.00e-13 R: AGAGTATGCTGGGCTTGTGG 6-FAM F: ACTTGGGCAGAACCAGAATG (GA)8 1.50e-39 2.30e-80 R: AACTTTCCAACTGTTTCAGTTCC HEX F: AAATATGTGAAATTGGTCCAACAG (CTTTCT)8 7.30e-27 2.10e-80 R: TCCAGGGTTACAAACAAGAGC 6-FAM F: TCAGCATTGCCTTTACCTTTC (CTTC)10 1.40e-56 1.10e-68 R: AATAGCAAGCTGCATCACAAAG 6-FAM F: GCCAGTGAGTAGTGGGTTCC (CA)9 7.50e-27 9.90e-18 R: TTTCTCTTCCTGCTAAATCCAAC 6-FAM F: ACTGATGGCAGAGCAGATTG (CA)8 7.90e-44 4.00e-86 R: GCTCCAAGCTCTTAGTCATGG HEX F: GATCTAAGTTTCCTGGGATTGC (CA)10 7.00e-18 8.00e-30 R: GAGGTTTATGGTAATGAAGAAGTTGC HEX F: ATCACACGTGCACCAAACAC (CA)5 5.00e-28 3.30e-42 R: CTGGCACTAATGTGTAGTCGTG HEX F: GATCGGAGCCTTCTTACCTG (CA)6 3.70e-39 9.40e-23 R: GCTGGAGGGAAGCTTGTTC HEX F: TTGTGTCATTTGGATGCCTTAG (AG)6 6.80e-54 8.60e-34 R: GCTCTCCTTGTGCTTGTTTG F: ACACCCACTATTTGTGAAGAGC HEX 2.10e-47 (AC)7 F: TCTATGCCTTAAATCCTATTTACAAAC 1.40e-36 HEX Primer sequence 50 –30 R: CAGCACTGTCATTGTTCTGC (AC)5 5.70e-30 Fluoro label (F) 4.60e-28 Repeat motif E-value in Gga E-value in ZF 338 398 206 507 856 721 441 621 326 561 307 808 460 401 Seq. length (bp) 58.41 58.54 60.16 59.87 59.35 58.99 60.28 60.11 58.73 59.21 59.23 59.35 59.56 59.58 58.94 58.99 59.09 59.09 60.59 60.50 58.39 58.89 59.94 60.00 58.65 58.25 57.98 57.68 Tm (°C) 56 56 56 56 56 56 56 56 56 56 56 56 56 56 PCR Ta (°C) 24 24 24 24 24 24 24 24 24 24 24 24 24 24 n1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 n2 – – – – – – – – – – – – – – A 259 263 132 319 175 358 284 287 154 131 232 166 159 199 Exp allele size (bp) ¥ – – – – – – – – – – – – – – Obs allele size (bp) Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Failed to amplify Marker status ¥ Expected allele size based on the sequenced clone allele of the female ruff individual (Bird ID 6233) from which the genomic library was created n1 Number of individuals tested in a captive population. n2 Number of individuals amplified and genotyped. A Number of alleles observed, M male, F female T – The location of each microsatellite sequence was assigned in the chicken (v 2.1, May 2006 ENSEMBL release) and zebra finch (December 2011 ENSEMBL Release 65) based on sequence homology (see Dawson et al. 2006, 2007). Of these 50 additional loci tested, 45 could be assigned a location in the chicken genome and 46 in the zebra finch genome. Ppu240 Ppu236 Ppu184 Ppu173 Ppu170 Ppu168 Ppu163 Ppu161 Ppu155 Ppu154 Ppu150 Ppu134 Ppu129 HE617035 Ppu125 65F08 EMBL acc. no. & clone name Locus Table 2 continued Author's personal copy Conservation Genet Resour 123 Author's personal copy Conservation Genet Resour Gga1 Tgu1A Gga2 Tgu2 Gga3 Ppu048* Ppu184 Ppu048* Ppu184 Ppu025 Ppu026 Ppu240 Ppu039* Ppu001 Ppu173 Ppu080 Ppu125 Ppu026 Ppu039* Ppu001 Ppu173 Ppu080 Ppu125 Ppu059* Ppu003 Ppu163 Ppu059* Ppu003 Ppu028 Ppu060* Tgu3 Gga4 Ppu010 Ppu010 Ppu071 Ppu015 Ppu032 Ppu071 Ppu015 Ppu085* Ppu043* Ppu170 Ppu032 Ppu043* Ppu018 Ppu170 Ppu022 Ppu129 Ppu013 Ppu051* Ppu129 Ppu012 Ppu011 Ppu011 Ppu012 Ppu013 Ppu009 Ppu161 Ppu079 Ppu046* Ppu161 Ppu017 Ppu050* Ppu168 Ppu004 Ppu038* Ppu168 Ppu004 Ppu038* Ppu030 Ppu030 Ppu045 Ppu154 Ppu045 Ppu154 Ppu007 Ppu007 Ppu019 Ppu236 Ppu035 Ppu093* Ppu172 Tgu5 Ppu040* Ppu138 Ppu040* Ppu138 Ppu006 Ppu006 Ppu070 Ppu070 Ppu064* Ppu104 Ppu064* Ppu104 Ppu009 Ppu046* Ppu050* Ppu018 Ppu022 Gga5 Tgu4A Ppu051* Ppu155 Ppu052* Ppu035 Ppu093* Ppu172 Ppu155 Ppu044 Ppu164 Ppu021 Ppu016 Ppu049* Ppu060* Ppu107 Ppu242 Ppu055 Ppu021 Ppu049* Ppu016 Ppu164 Ppu107 Ppu242 Ppu055 Ppu028 Ppu163 Gga6 Ppu134 Ppu092 Ppu057* Ppu047* Ppu121 Gga11 Tgu6 Gga7 Ppu134 Ppu092 Ppu057* Ppu047* Ppu121 Gga22 Ppu056 Ppu105* Ppu042* Ppu041* Tgu22 Ppu056 Gga9 Ppu027 Ppu023 Ppu148 Tgu24 Ppu014 Tgu13 Gga13 Gga15 Ppu031 Ppu024 Ppu148 Ppu083* Ppu095 Ppu062* Gga10 Tgu10 Ppu036* Ppu029 Ppu034 Ppu199 Ppu036* Ppu029 Ppu034 Ppu199 Ppu033 Ppu033 Tgu12 Tgu9 Ppu083* Ppu095 Ppu062* Ppu054 Ppu005 Ppu054 Ppu005 Ppu020 Ppu042* Ppu041* Ppu020 Tgu8 Gga8 Ppu023 Ppu027 Gga12 Tgu11 Tgu7 Tgu15 Ppu150 Ppu150 Gga18 Ppu140 Tgu18 Ppu140 Ppu031 Gga26 Ppu221 Ppu037* Tgu26 Ppu221 GgaZ Ppu058* Ppu068 Ppu101* TguZ Ppu181 Ppu086* Ppu053 Ppu181 Ppu058* Ppu068 Ppu067 Ppu101* Ppu053 Ppu086* Fig. 1 Chromosomal locations in the chicken and zebra finch genomes of 102 ruff microsatellite loci. Locations were assigned as in Table 1. Gga, chicken (Gallus gallus) chromosome; Tgu, zebra finch (Taeniopygia guttata) chromosome; Underlined loci are polymorphic, loci listed in bold were monomorphic (*polymorphism assessed in 12 individuals; all other loci were tested in 24 individuals); Loci listed in italics failed to amplify a product or amplified a stutter/unreliable product (Table 2) and the loci in plain text have not been tested for amplification or polymorphism software (Applied Biosystems). Observed and expected heterozygosities were calculated using CERVUS v3.0 (Kalinowski et al. 2007; Table 1). Deviations from Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium were assessed using GENEPOP v.4.0 (Rousset 2008). All loci were assessed for Hardy–Weinberg equilibrium and linkage disequilibrium in captive individuals, selected to avoid parent–offspring relationships and full and half sibs. Polymorphic loci were suspected of Z-linkage if no females were heterozygous. Mendelian inheritance was assessed in 64 families, consisting of both parents and 5–33 offspring per family, comprising 381 known-sex individuals. Any Z-linked loci were tested for HWE in males only. Of the 102 markers tested, 23 were monomorphic, 27 failed to amplify or amplified a stuttery/unreliable product, and 52 were polymorphic (Tables 1, 2). Five polymorphic autosomal loci (Ppu001, Ppu003, Ppu009, Ppu010 and Ppu016) amplified a product [70 bp larger than the cloned sequence but were all inherited in a Mendelian fashion (Table 1). In total, 47 of the 52 polymorphic loci were checked for Mendelian inheritance, which was confirmed for all except Ppu002 (Tables 1, 2). A high proportion of sequences (85 %) could be assigned locations in both the zebra finch and chicken genomes based on sequence homology (following Dawson et al. 2006, 2007). Of the 102 newly-isolated ruff microsatellites, 95 could be assigned to a chromosome location 123 Author's personal copy Conservation Genet Resour in the chicken, 92 in the zebra finch and 87 in both chicken and zebra finch (Fig. 1). Only two loci could not be assigned a chromosomal location in either chicken or zebra finch (Ppu002 and Ppu008, Tables 1, 2). There was no statistical difference in the number of sequences that could be assigned a location to each genome (Fisher’s two-tailed Exact test p = 0.66). One locus (Ppu058) assigned to both the zebra finch and chicken Z chromosomes (Fig. 1), was homozygous in all of the 127 females genotyped, but heterozygous in 27 of the 140 males typed, confirming its sex-linked status (Fisher’s two-tailed Exact test p \ 0.001). Twelve autosomal loci, assessed in 12 individuals, showed significant deviation from Hardy–Weinberg equilibrium (p \ 0.05) or a deficiency of heterozygotes (Table 1). Three groups of loci showed evidence of linkage disequilibrium (p \ 0.05; Ppu010–Ppu016, Ppu036–Ppu038–Ppu041– Ppu042–Ppu051–Ppu052, Ppu037–Ppu040; Table 1), however following FDR correction (Benjamini and Hochberg 1995) no p-values were significant. All loci in each group were assigned to different chromosomes, except Ppu041–Ppu042, which are closely neighbouring on chromosome 11 in chickens and zebra finches, and therefore may be physically linked (Table 1, Fig. 1). The deviation from Hardy–Weinberg equilibrium and linkage disequilibrium displayed by some groups of loci is probably due to the non-random structure of our captive population. However, 46 of the 47 loci checked displayed a pattern consistent with Mendelian inheritance (Tables 1, 2). The microsatellite loci developed during this study are suitable for the analysis of parentage and population structure and will be used to construct a linkage map for the ruff. The utility of these loci in other shorebird species can be predicted based on the BLAST E-value recorded from a comparison of their sequences similarity with the chicken genome (Küpper et al. 2008, but see also Dawson et al. 2010). The sequences of three loci (Ppu004, Ppu005 and Ppu030) displayed particularly high sequence homology to both the genetically distant chicken and zebra finch (BLAST E-values \ E-70; Table 1). These loci are therefore expected to amplify in a wide range of species, including those of conservation interest. Additionally, if required, these specific loci would be the most suitable to develop into conserved markers, which would further enhance their cross-species utility (Dawson et al. 2010). Any primer sets designed as a consensus among all three species (ruff, zebra finch and chicken) are expected to display the highest cross-species utility. Acknowledgments This work was performed at the NERC Biomolecular Analysis Facility–Sheffield supported by the UK Natural Environmental Research Council. The captive ruff colony was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC; to DBL) and LLF was supported by an NSERC studentship. References Altschul SF, Madden TL, Schäffer AA et al (1997) Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 Armour JAL, Neumann R, Gobert S, Jeffreys AJ (1994) Isolation of human simple repeat loci by hybridization selection. Hum Mol Genet 3:599–605 Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300 Dawson DA, Burke T, Hansson B et al (2006) A predicted microsatellite map of the passerine genome based on chicken– passerine sequence similarity. Mol Ecol 15:1299–1320 Dawson DA, Åkesson M, Burke T, Pemberton JM, Slate J, Hansson B (2007) Gene order and recombination in homologous regions of the chicken and a passerine bird. Mol Biol Evol 24:1537–1552 Dawson DA, Horsburgh GJ, Küpper C et al (2010) New methods to identify conserved microsatellite loci and develop primer sets of high utility—as demonstrated for birds. Mol Ecol Res 10: 475–494 Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106 Kenta T, Gratten J, Hinten G, Slate J, Butlin RK, Burke T (2008) Multiplex SNP-SCALE: a cost-effective medium-throughput SNP genotyping method. Mol Ecol Res 8:1230–1238 Küpper C, Burke T, Székely T, Dawson DA (2008) Enhanced crossspecies utility of conserved microsatellite markers in shorebirds. BMC Genomics 9:502–522 Lank DB, Smith CM, Hanotte O, Burke T, Cooke F (1995) Genetic polymorphism for alternative mating behaviour in lekking male ruff Philomachus pugnax. Nature 378:59–62 Nicholls JA, Double MC, Rowell DM, Magrath D (2000) The evolution of cooperative and pair breeding in thornbills Acanthiza (Pardalotidae). J Avian Biol 31:165–176 Primmer CR, Møller AP, Ellegren H (1996) A wide-range survey of cross-species microsatellite amplification in birds. Mol Ecol 5:365–378 Rousset F (2008) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Res 8:103–106 Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Bioinformatics methods and protocols: methods in molecular biology, pp 365–386. Humana Press, Totowa, NJ Sibley CG, Monroe BL (1990) Distribution and taxonomy of birds of the world. Yale University Press, New Haven, CT Thuman K, Widemo F, Piertney SB (2002) Characterization of polymorphic microsatellite DNA markers in the ruff (Philomachus pugnax). Mol Ecol Notes 2:276–277 123


![1. • void sum(double A[][M], double B[][M], double C[][M], double D[][M]) {](http://s2.studylib.net/store/data/010408584_1-eca3970d67c1b7f10382ba55a6f82240-300x300.png)