Document 10524989
advertisement

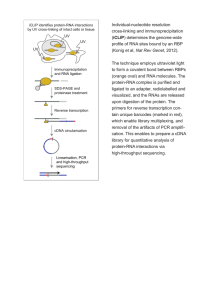
TITLE: A Method for Single Cell Sequencing of miRNAs and Other Difficult to Obtain Cellular RNAs INVENTORS: David Corey and Yongjun Chu TECHNOLOGY: Biological UTSD: 2763 SUMMARY: This invention is a method that can sequence short RNA, preferably less than 200 base pairs, from 10 pg of sample (a single cell). There are methods for long RNA sequencing starting from a single cell. This new technology is different from DNA harvesting and amplifying. First, total RNA is extracted from a single cell and circularized without introduction of any nucleic acid to the sequence. Random 5’-primer is annealed to the circular template. Linear cDNA is created by reverse transcriptase. The transcription stops randomly after 5-10 (circular) repeats. Then, random 3’- adaptor is added to create double stranded cDNA through PCR amplification with indexed primers. The resulting library is sequenced. Software to handle and analyze the sequencing data has been developed. It will identify the origin of a RNA molecule sequence. The technology is currently at final stage of development mostly for research purpose with potential for clinical diagnostic purpose. Please contact the Office for Technology Development for more details: Phone: 214-648-1816 Email: TechnologyDevelopment@utsouthwestern.edu Please reference UT Southwestern Case Number: 2763