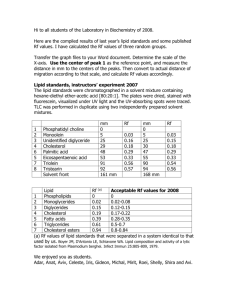
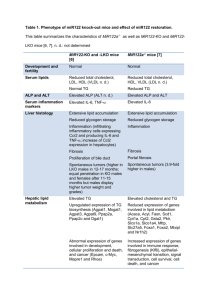
424 R&R and PHS-398 Specific Table Of Contents
advertisement

Principal Investigator/Program Director (Last, first, middle): 424 R&R and PHS-398 Specific Table Of Contents Page Numbers SF 424 R&R Face Page------------------------------------------------------------------------------------------ 1 Table of Contents--------------------------------------------------------------------------------------------- 3 Performance Sites--------------------------------------------------------------------------------------------- 5 Research & Related Other Project Information------------------------------------------------------------------ 6 Project Summary/Abstract (Description)---------------------------------------- 7 Public Health Relevance Statement (Narrative attachment)---------------------------------------- 8 Bibliography & References Cited---------------------------------------- 9 Facilities & Other Resources---------------------------------------- 11 Other Attachments---------------------------------------- 12 List of referees---------------------------------------- 12 Research & Related Senior/Key Person-------------------------------------------------------------------------- 13 Biographical Sketches for each listed Senior/Key Person---------------------------------------- 17 Research & Related Budget - Year 1---------------------------------------------------------------------------- 49 Research & Related Budget - Year 2---------------------------------------------------------------------------- 52 Research & Related Budget - Year 3---------------------------------------------------------------------------- 55 Research & Related Budget - Year 4---------------------------------------------------------------------------- 58 Budget Justification---------------------------------------- 61 Research & Related Budget - Cumulative Budget----------------------------------------------------------------- 62 PHS 398 Specific Cover Page Supplement------------------------------------------------------------------------ 63 PHS 398 Checklist--------------------------------------------------------------------------------------------- 65 PHS 398 Career Development Award Supplemental Form------------------------------------------------------------ 67 Candidates Background---------------------------------------- 69 Career Goals and Objectives---------------------------------------- 70 Development Activities During Award Period---------------------------------------- 71 Training in the Responsible Conduct of Research---------------------------------------- 72 Statements by Mentor, Co-Mentors, Consultants, Contributors---------------------------------------- 73 Institutional Environment---------------------------------------- 78 Institutional Commitment to Career Development---------------------------------------- 79 Specific Aims---------------------------------------- 80 Research Strategy---------------------------------------- 81 Protection of Human Subjects---------------------------------------- 90 Women &Minorities---------------------------------------- 91 Planned Enrollment Table---------------------------------------- 92 Children---------------------------------------- 93 Vertebrate Animals---------------------------------------- 94 Select Agent Research---------------------------------------- 95 Table of Contents Page 3 Principal Investigator/Program Director (Last, first, middle): Consortium/Contractual---------------------------------------- 96 Resource Sharing Plan---------------------------------------- 97 Table of Contents Page 4 Principal Investigator/Program Director (Last, first, middle): Project Summary: Non-alcoholic fatty liver disease (NAFLD) is an increasingly prevalent medical problem that affects approximately a third of the US population. Patatin-like phospholipase domain containing 3 (PNPLA3) was first implicated in the metabolism of hepatic triacylglycerides (TAGs) when our lab found that a missense mutation that substitutes isoleucine at position 148 with methionine (I148M) was associated with non-alcoholic fatty liver disease in humans. Homozygotes with the variant allele (148M) have a 2-fold higher risk of hepatic TAG accumulation than homozygotes with the wild type allele (148I). The function of PNPLA3 and the cause of I148M induced hepatic TAG accumulation remains unclear. Since fatty liver represents an accumulation of fatty acids in the form of triglycerides, I will adapt and/or develop targeted liquid chromatography-tandem mass spectrometry (LC-MS-MS) methods to measure the concentrations of different classes of fatty acid-containing lipids in liver, adipose tissue, and plasma from mouse models of PNPLA3-associated steatosis. Mice will be studied on both a normal chow diet and on diets designed to exacerbate or alleviate PNPLA3-I148M associated accumulation of hepatic TAG. I will then extend these LC-MS/MS-based methods to determine the flux of each lipid class by measuring the rate of incorporation of deuterium from deuterated water (D2O) in cultured cells that overexpress PNPLA3, and in genetically manipulated mouse models. Alternative stable isotope labeling strategies, such as 13C-glycerol or 13C-fatty acid incorporation, will also be employed to compliment the D2O incorporation data. Execution of this research proposal will provide training in the application of metabolomics technology to study human disease and shed insight into the function of PNPLA3 and its role in the progression of fatty liver disease. Project Description Page 7 Principal Investigator/Program Director (Last, first, middle): Project Narrative: Fatty liver disease is a growing epidemic that afflicts a third of the US population and leads to steatosis, cirrhosis, and liver failure. We have identified a gene named PNPLA3 that is strongly associated with fatty liver disease in human, but the function of this gene remains unclear. I am proposing experiments designed to determine the function of PNPLA3 and ultimately aid in the development of effective medical therapies to prevent fatty liver disease. Public Health Relevance Statement Page 8 Principal Investigator/Program Director (Last, first, middle): RESOURCES FACILITIES: Specify the facilities to be used for the conduct of the proposed research. Indicate the performance sites and d escribe capacities, pertinent capabilities, relative proximity, and extent of availability to the project. Under “Other,” identify support services such as machine shop, electronics shop, and specify the extent to which they will be available to the project. Use continuation pages if neces sary. - The mentor and co-mentor Drs. Hobbs and Dr. Jonathan Cohen, have 1,050 sq. ft. of laboratory space in the McDermott Center for Human Growth and Development. Dr. Xie has 900 sq. ft. of laboratory space adjacent to the laboratory of Drs. Hobbs and Cohen. The laboratories are equipped with an Amino DW2C Dual wavelength spectrophotometer; a Beckman DU-70 scanning spectrophotometer; a Fluostar Optima Multi-functional Fluorimeter; a Bio-Rad Biologic FPLC system; a Beckman HPLC system; Pharmacia column chromatography and electrophoresis apparatus; Bio-Rad electrophoresis and blotting systems; two Forma Scientific CO2 incubators, a sterile hood, an inverted microscope, liquid N2 storage chambers, a Beckman scintillation counter, PCR machines, orbital shaking-incubators, bacterial incubators, -80 C freezers, -20 C freezers, a chromatography refrigerator, refrigerators, low speed, high speed and ultraspeed centrifuges, rotors, including 70.1Ti, 70Ti, 50.2Ti, 45Ti, type65, SW60, SW41Ti, SW40Ti, NVT65 rotors, and JA 20,17,14, and 10 rotors. The investigators and their personnel have access to core facilities, including high speed and ultra-centrifuges, cold/warm rooms, tissue culture rooms and a cleaning facility. Laboratory: Dr. Hobbs also has a laboratory (~600 sq. ft.) in the Department of Molecular Genetics. The Department of Molecular Genetics has a real-time PCR machine and as Agilent gas chromatography/mass spectrometer. Dr. Cohen also has ~600 sq. ft. of laboratory space in the Center for Human Nutrition. Clinical: Not applicable The mouse colony will be housed in a secure vivarium requiring a combination card-key access to approved users. A staff of veterinarians specializing in laboratory animal care provides expert assistance with care and maintenance of the mouse colony. The facility has a total of 16,500 sq ft floor space, is self contained with respect to physical plant, and occupies the entire second floor of the investigators’ building, providing convenient access to animals. Excellent surgical and barrier facilities are available as needed. Animal: The investigators’ laboratories are equipped with personal computers connected by local area networks. The McDermott Center and the Medical School have computer support personnel and equipment to support our additional computer needs. Computer: Office: The PI and co-PIs have personal offices of 110 sq. ft. each adjoining their laboratories. The McDermott Center provides administrative support and access to office equipment (fax, photocopier, etc.). The McDermott Center has core facilities for DNA sequencing available on a cost-recovery basis to Center faculty. Other: MAJOR EQUIPMENT: List the most important equipment items already available for this project, noting the location and pertinent capabilities of each. Core Facilities available at UT Southwestern (which are subsidized) 1. Protein Chemistry Technology Core peptide mapping - A fee-for-service facility for protein sequencing, and protein modification analysis 2. Transgenic/Knockout- Technology Center Core Facility - A fee for service facility run by Dr. Robert Hammer for the development of genetically modified mice. 3. Structural Biology Core Facility – A fee for service facility directed by Dr. Diana Tomchick, provides a unique platform for biochemists and molecular biologists to engage in structural biology projects by making macromolecular crystallography available as a general tool. Facilities Page 11 Principal Investigator/Program Director (Last, first, middle): List of Referees: Jeffrey McDonald, Ph.D. Associate Professor of Molecular Genetics UT Southwestern Medical Center 5323 Harry Hines Blvd. Dallas, TX 75390-9046 E-mail: Jeffrey.mcdonald@utsouthwestern.edu Telephone: 214-638-8663 Elizabeth Parks, Ph.D. Associate Professor of Nutrition UT Southwestern Medical Center 5323 Harry Hines Blvd, Dallas, TX, 75390-9046 E-mail: Elizabeth.parks@utsouthwestern.edu Telephone: 214 648-2054 Ralph Deberardinis, M.D., Ph.D. Assistant Professor of Pediatrics and Genetics UT Southwestern Medical Center 5323 Harry Hines Blvd., Dallas, TX, 75390-8502 E-mail: ralph.deberardinish@utsouthwestern.edu Telephone: 214-633-1803 Donald Small, M.D. Professor of Physiology and Biophysics Boston University Medical Center 700 Albany Street, Boston, MA, 02118-2526 E-mail: dmsmall@bu.edu Telephone: 617-638-4002 List of referees Page 12 Principal Investigator/Program Director (Last, first, middle): BIOGRAPHICAL SKETCH Provide the following information for the Senior/key personnel and other significant contributors in the order listed on Form Page 2. Follow this format for each person. DO NOT EXCEED FOUR PAGES. NAME POSITION TITLE Postdoctoral Researcher II eRA COMMONS USER NAME (credential, e.g., agency login) EDUCATION/TRAINING (Begin with baccalaureate or other initial professional education, such as nursing, include postdoctoral training and residency training if applicable.) DEGREE INSTITUTION AND LOCATION MM/YY FIELD OF STUDY (if applicable) University of Delaware, Newark DE B.S. 06/06 Chemical Engineering Boston University, Boston MA Ph.D 01/12 Biophysics University of Texas Southwestern Medical Center, Dallas TX Postdoctoral Lipid Physiology A. Personal Statement I will serve as the principle investigator on this research training grant. My primary research interested is using metabolomics and stable isotope labeling to study lipid metabolism and metabolic disease. I obtained a bachelor’s degree in Chemical Engineering, where I specialized in Biochemical Engineering, and a doctoral degree in Physiology and Biophysics under the mentorship of Dr. Donald Small. This education provided multidisciplinary training in quantitative biochemistry and physiology. In 2011, I began a post-doctoral fellowship at UT Southwestern under the mentorship of Dr. Helen Hobbs and Dr. Jonathan Cohen in 2011. During my postdoctoral training I have specializing in using LC-MS/MS based metabolomics technology to study lipid physiology, sterol metabolism and the progression of fatty liver disease. I hope to continue this training under the Mentored Research Scientist Development Award in Metabolomics. B. Positions and Honors Positions and Employment: 2004-2006 Undergraduate Researcher, Department of Chemical Engineering, University of Delaware, ………………..Newark DE 2006-2011 Graduate Research Fellow, Department of Physiology and Biophysics, Boston University, Boston ………………..MA 2011Post-Doctoral Fellow, Department of Molecular Genetics, UT Southwestern Medical Center, ………………..Dallas, TX Honors: 2006 2009,2012 2010 2010 2010 2011 Graduate Research Assistant Scholarship Kern Aspen Lipid Conference Travel Award Ruth Kirschstein Scholar Henry J. Russek Student Achievement Award in Physiology and Biophysics NIH-HGSRF Award Deuel Lipid Conference Travel Award Biosketches Page 17 Principal Investigator/Program Director (Last, first, middle): C. Publications Interfacial Properties of a Complex Multi-Domain 490 Amino Acid Peptide Derived from Apolipoprotein B (Residues 292-782); Mitsche, M.A., Wang, L, Jiang, G.Z., McKnight, C.J., Small, D.M; Langmuir, 25(4) pp 2322-2330, 2009 The adsorption of biological peptides and proteins at the oil/water interface. A potentially important but largely unexplored field; Small, D.M., Wang, L., Mitsche, M.A.; Journal of Lipid Research, Vol. 50, S329-S334, 2009 Deposition of egg-PC to an Air/Water and Triolein/Water Interface; Mitsche, M.A, Small, D.M.; Biophysical Journal, 96(3) pp.461a, 2009 Adsorption of Egg-PC to an Air/Water and Triolein/Water Interface: Use of the 2-Dimensional Phase Rule to Estimate the Surface Composition of a Phospholipid/Triolein/Water Surface as a Function of Surface Pressure; Mitsche, M.A., Wang, L., Small, D.M.; Journal of Physical Chemistry B, 114(9) pp 3276-3284 C-terminus of Apolipoprotein A-I Removes Phospholipids (PL) from a Triolein/PL/Water Interface, but the N-Terminus does not: A possible mechanism for nascent HDL Assembly; Mitsche, M.A., Small, D.M.; Biophysical Journal, 101(2) pp. 353-361, 2011 Feature Article Interfacial Properties of the N-terminal Lipid –Binding Domains of ApolipoproteinB and Their Role in Triacylglyceride-Rich Lipoprotein Assembly; Mitsche, M.A.; Ph.D. Thesis, Boston University 2012 Surface Pressure Dependent Remodeling of Amphipathic α-Helices at a Triolein/Water Interface; Mitsche, M.A., Small, D.M.; Accepted to the Journal of Lipid Research on 1/23/13 D. Research Support Ongoing Research Support None Completed Research Support None Biosketches Page 18 Principal Investigator/Program Director (Last, first, middle): Background Statement: I began my technical education by studying chemical engineering at the University of Delaware where I specialized in biochemical engineering. Studying engineering gave me a strong fundamental education in physical and bio-chemistry, physics, mathematics, computer science, and analytical reasoning, which have served me well since transitioning into biomedical research during graduate school. During my undergraduate years, I came to realize that a traditional scientific graduate education would be required if I wished to work in research and development. Therefore, I decided to pursue a doctoral degree in biophysics at Boston University. My graduate work, under the direction of Dr. Donald Small, focused on elucidating the mechanism of lipoprotein remodeling by studying the surface chemistry of apolipoprotein fragments at a triacylglyceride/water interface. Working with Dr. Small provided me with extensive exposure to the field of lipid physiology, where I became particularly interested in biology of intracellular lipid droplets and lipid metabolism in general. I came to the conclusion that studying lipid droplet biology would require additional training in molecular genetics and lipidomics. Lipidomics, and metabolomics in general, is of particular interest to me because it will reveal fundamental aspects of lipid physiology that have been unapproachable to date and my engineering training makes me uniquely suited to assist in the development of this technology. Therefore, the opportunity to learning mass spectroscopy based lipidomics was my primary objective in choosing a lab for my post-doctoral training. In 2011 I joined the Hobbs-Cohen lab at the University of Texas Southwestern Medical Center as a post-doctoral fellow. My post-doc began with training on a liquid chromatographytandem mass spectrometer (LC-MS/MS) and a method for measuring sterols developed by Dr. Jeff McDonald. To date, my post-doctoral research has focused on using LC-MS/MS to study sterol metabolism. My first project is aimed at defining the post-squalene biosynthetic pathway of cholesterol. In the 1960’s two alternative post-squalene cholesterol biosynthetic pathways were proposed, however the relative utilization of these two pathways remains unknown. With the LC-MS/MS based sterol methods, most of the post-squalene sterol biosynthetic intermediates can be measured, and the turnover of these intermediates can be measured by labeling newly synthesized sterols with deuterated water (D2O). I have developed a method to monitor sterol biosynthetic intermediates turnover in vivo using D2O and have applied it to culture cells and mice to measure the kinetics of this pathway. Stable isotope labeling with D2O will be directly translatable to other lipid classes, as proposed in this grant. My second project is focused on measuring sterol metabolism and clearance in ABCG5/G8 mice. Our lab has recently developed liver and intestine specific ABCG5/G8 knock-out mice. To measure the tissue specific contribution of ABCG5/G8 to sterol clearance, I have been developing and implementing stable isotope labeling experiments to measure the absorption and clearance of cholesterol and plant sterols in ABCG5/G8 genetically manipulated mice. In addition to continuing these projects, I began to develop methods for measuring lipid class relevant to other research projects in the lab. Specifically I was interested in using LC-MS/MS to measure triacylglycerides (TAG) to measure lipid species in mice over expressing PNPLA3 in the liver. After successfully developing a method to measure individual TAG species, I observed a large reduction in the percentage of poly-unsaturated fatty acid TAGs when PNPLA3 was over expressed. I also used this method to measure turnover of TAG species using D2O. In this proposal, I have developed a research plan to pursue these findings and develop methods to measure other lipid classes to understand the function of PNPLA3 and, in the future, to use these methods to study other proteins involved in intracellular lipid droplet biology. Candidates Background Page 69 Principal Investigator/Program Director (Last, first, middle): Career Goals and Objectives: The long-term goal of my career is to run a laboratory that focuses on using mass spectroscopy for biomedical research and medical diagnosis. The primary objective of the lab will be two-fold. Firstly, the lab will focus its independent research on using metabolomics and stable isotope labeling to elucidate the molecular mechanism of the progression of metabolic disease. In particular, we will aim to advance our understanding of the genetic and dietary contributions to lipid metabolism. Secondly, the lab will use metabolomics and stable isotope labeling to both assist in the diagnosis of patients presenting complex metabolic phenotypes and provide a service to other biomedical researchers. Reaching this goal will require significant training in the next four years. I will need to become an expert in development of LC-MS/MS based metabolomics methods and understand how to apply these techniques to biomedical research. In addition, I will need training in molecular biology, genetics, and animal husbandry to establish an independent lab studying lipid metabolism. This research proposal outlines a series of experiments to elucidate the function of PNPLA3 that will provide excellent training in the application of metabolomics and stable isotope labeling to biomedical research. The HobbsCohen lab and UT Southwestern is an ideal environment to develop these skills because many of the required techniques are of molecular biology, genetics, and animal handling are already used in the lab In addition, there is a growing group of metabolomics researchers in the department and throughout the university. Career Goals and Objectives Page 70 Principal Investigator/Program Director (Last, first, middle): Career Development/Training Activities During Award Period: The majority of the training associated with Sub-Aims Year 1 Year 2 Year 3 Year 4 this grant will be accomplished through the Develop Lipidomics Methods 1A successful execution of the research Lipidomics of PNPLA3 Mice 1B proposal. A timeline for the execution of this Lipidomics on Custom Diets 1C proposal is shown in Figure 1. This research Human Plasma 1D will give me training in LC-MS/MS based D2O Labeling in Cells 2A metabolomics, cell culture, and mouse Alternative Labeling in Cells 2B handling in addition to other skills required D2O Labeling in PNPLA3 Mice 2C to manage an independent laboratory. To D2O Labeling on Custom Diets assist in the execution of this proposal, an 2D advisory committee of six members has been Figure 1: Timeline of research proposal assembled. This team includes Drs. David Russell and Jeff McDonald who are experts in LC-MS/MS based lipidomics, Drs. Ralph DeBerardinis and Elizabeth Park who are experts in measuring metabolite flux using stable isotopes, and Drs. Helen Hobbs and Jonathan Cohen who are experts in lipid metabolism and PNPLA3. Regular attendance of seminars at UTSW in metabolism, nutrition, and obesity and University Lecture will be essential for my training as well. In addition, I will attend two conferences per year to be exposed to progress in lipid metabolism and metabolomics. Development Activities During Award Period Page 71 Principal Investigator/Program Director (Last, first, middle): Responsible Conduct of Research Training for Postdoctoral Scholars at UT Southwestern I am currently enrolled in post-doctoral responsible conduct in research training at UT Southwestern. This training will be completed in May of 2013. The format of the training is outlined below: 1. Format a. Introductory Presentation on the Postdoctoral Certificate Training Program - attended by all postdoctoral scholars within two months of postdoctoral appointment b. Eight-week course of faculty led discussions - The eight-week course meets a requirement for the Postdoctoral Certificate in Research, which must be completed within the first two years of training at UT Southwestern. The course is offered in the Spring term. Participation is monitored and pass/fail grade is assigned by faculty of the graduate school’s Postdoctoral Affairs Office in conjunction with registration and academic certificate completion. Format: 90 minutes every week for eight weeks - 45-minute lecture followed by 45-minute faculty-led discussion. 2. Subject Matter a. Introductory Presentation i. UT Southwestern Graduate School of Biomedical Science Policies on Academic Integrity ii. Mentor/mentee responsibilities and relationships iii. Importance of learning and following lab notebook policy b. Eight-week course i. Human Research Ethics and Genetics ii. Use of Animals in Research iii. Research misconduct, plagiarism, and peer review iv. Record Keeping and Research Notebooks; v. Authorship vi. Data and Image Manipulation vii. Conflict of Interest viii. Collaboration, Technology Transfer, and Intellectual Property 3. Faculty Participation a. Introductory Presentation – Deirdre Brekken, Ph.D. b. Eight-week course i. Humans: Fred Grinnell, Ph.D. ii. Animals: Bart Carter, D.V.M. iii. Misconduct: Jerry Neiderkorn, Ph.D. iv. Record keeping: David Russell, Ph.D. v. Authorship: Melanie Cobb, Ph.D. vi. Images: Sandra Schmid, Ph.D. vii. Conflicts: Stuart Ravnik, Ph.D. viii. Technology: Philip Thomas, Ph.D. 4. Duration of Instruction a. Introductory Presentation – 60 minutes b. Eight-week course – 90 minutes: 45-minute lecture followed by 45-minute faculty-led discussion 5. Frequency of Instruction a. Introductory Presentation – one 60-minute session b. Eight-week course – once per week for eight weeks 12/04/2012 Training in the Responsible Conduct of Research Page 72 Principal Investigator/Program Director (Last, first, middle): Statements by Mentor, Co-Mentors, Consultants, Contributors Page 73 Principal Investigator/Program Director (Last, first, middle): Statements by Mentor, Co-Mentors, Consultants, Contributors Page 74 Principal Investigator/Program Director (Last, first, middle): Statements by Mentor, Co-Mentors, Consultants, Contributors Page 75 Principal Investigator/Program Director (Last, first, middle): Statements by Mentor, Co-Mentors, Consultants, Contributors Page 76 Principal Investigator/Program Director (Last, first, middle): Merck Research Laboratories Merck & Co., Inc. 2015 Galloping Hill Road Kenilworth, NJ 07033-1310 T 908-298-4000 merck.com February 27, 2013 To Whom It May Concern, This letter affirms my willingness to serve as a consultant to mass spectrometry methods he is developing as part of a KO1 application. on the I am currently the Director of Analytical Biochemistry, which is a group of 18 mass spectrometry scientists focused on cardiometabolic research. I have considerable experience with the analysis of lipid concentration and turnover by mass spectrometry and also have several Ph.D. experts in the field in my group as regular consultants in this area of research. As part of an ongoing collaboration with the Hobbs-Cohen laboratory, my group has already provided data and technical advice on lipids measurement to , and we look forward to continuing this relationship. Sincerely, Thomas P. Roddy, Ph.D. Director of Analytical Biochemistry in Molecular Biomarkers TPR/bf Statements by Mentor, Co-Mentors, Consultants, Contributors Page 77 Principal Investigator/Program Director (Last, first, middle): Description of Institutional Environment UT Southwestern Medical Center has a vibrant academic environment sustained by over 1,200 full time faculty, including five Nobel laureates, nineteen members of the National Academy of Sciences, nineteen members of the Institute of Medicine, thirteen members of the American Academy of Arts and Sciences and twelve investigators of Howard Hughes Medical Institute. UTSW supports more than 3,500 research projects with annual funding over $400 million. These laboratories occupy approximately 582,000 square feet of space with more construction under way. As part of the total amount of funding in 2001, the Texas legislature appropriated $9 million annually to the Institute for Innovations in Medical Technologies. The Institute will include centers for genetics, molecular and cellular biotechnology, new drug and vaccine development, computational biology, and advanced medical devices and imaging technology. UTSW also has several core facilities including those for microarray, transgenic, DNA sequencing, live cell imaging, flow cytometry, structural biology, and rapid biochemical kinetics technology, in addition to a comprehensive library. Nobel Laureates Drs. Joseph Goldstein and Michael Brown oversee the Department of Molecular Genetics, which primarily focuses on the human genetics and molecular mechanisms underlying human body energy homeostasis, including cholesterol metabolism, triglyceride metabolism and glucose metabolism. This Department has been funded with a P01 grant >4 million/year for more than 30 years. The department, where the applicant will perform research, has well-established programs related to the applicant’s area of research, including faculty members and resources. Dr. Helen Hobbs, a HHMI investigator, and Dr. Jonathan Cohen (co-mentors on this project) are active in NAFLD research. In addition, the Department of Molecular Genetics is establishing a small molecule mass spectroscopy based metabolomics facility. This includes three faculty members, Dr. Jeff McDonald, Dr. David Russell, and Dr. Ruth Gordillo, who are currently working on expanding metabolomics research and four modern tandem mass spectrometers. The applicant will have full-time access to a triple quadrupole mass spectrometer. Furthermore, there are other investigators whose NAFLD research is funded by NIH, for example Dr. Jay Horton, Dr. Jeff Browning, and Dr. Elizabeth Parks. These investigators all work closely with Dr. Helen Hobbs and Dr. Jonathan Cohen, the applicant’s mentors. These resources provide a unique environment for Dr. training in NAFLD and metabolomics research. The facilities and resources outlined in the resources section will be freely available to the applicant and will allow the applicant to perform all of the experiments outlined in the grant proposal. The applicant will also have many opportunities for his intellectual development. He will have access not only to faculty members in the Department of Molecular Genetics, but also to accomplished investigators in many other departments. He will attend multiple basic science seminars including the weekly University Lecture Series, in which Nobel-caliber scientists (from within and outside the institution) present their research. He will also attend weekly seminars on Nutrition, Metabolism and Obesity hosted by Center for Human Nutrition, which attract world-renowned scientists. The applicant will present his work periodically to other post-doctoral fellows and graduate students in weekly laboratory meetings. Institutional Environment Page 78 Principal Investigator/Program Director (Last, first, middle): Institutional Commitment to Career Development Page 79 Principal Investigator/Program Director (Last, first, middle): Specific Aims: Genetic studies provide compelling evidence that the patatin-like phospholipase domain containing protein 3 (PNPLA3) confers susceptibility to fatty liver disease, a burgeoning health problem throughout the world (1-3). A sequence variation in PNPLA3 (I148M) is associated with the full spectrum of nonalcoholic fatty liver disease, including steatosis, nonalcoholic steatohepatitis, cirrhosis and hepatocellular carcinoma, but the function of the protein remains enigmatic (4). The overall objective of this proposal is to define the physiological role of PNPLA3 using liquid chromatography-tandem mass spectroscopy (LC-MS/MS)-based lipidomics and stable isotope labeling. Since fatty liver represents an accumulation of fatty acids in the form of triglycerides (TAG), targeted LC-MS-MS methods will be adapted, optimized and/or developed to measure concentrations of fatty acid-containing metabolites in liver, adipose tissue, and plasma from mouse models of PNPLA3-associated steatosis. LC-MS/MSbased methods will also be used to determine the flux of each lipid class by measuring the rate of incorporation of deuterium from deuterated water (D2O) into lipids in cultured cells and genetically-manipulated mouse models that have been developed in our laboratory. Execution of this research proposal will provide training in the application of metabolomics technology to study a prevalent human disease and shed insight into the function of PNPLA3 and its role in the progression of fatty liver disease. Specific Aim 1: Determine the effect of PNPLA3-wt and the I148M on the lipidome of mice Aim 1A: Develop LC-MS/MS methods to quantitate acyl-lipids Aim 1B: Compare lipidomes of plasma, liver and adipose tissue in PNPLA3 mouse models Aim 1C: Determine effects of changes in dietary composition on hepatic lipid profile Aim 1D: Compare lipidomes of plasma from humans homozygous for the non-risk (148I) and the risk (148M) PNPLA3 variant. Specific Aim 2: Define the metabolic basis of PNPLA3-associated fatty liver Aim 2A: Measure lipid turnover using D2O in cultured hepatocytes expressing wt or mutant PNPLA3 Aim 2B: Compare D2O with isotope labeled fatty acids and glycerol in cultured cells Aim 2C: Determine lipid flux in PNPLA3 models using D2O Aim 2D: Examine effects of changes in diet composition on lipid flux in mice IMPACT: Our previous work provided compelling evidence that genetic variation in PNPLA3 is a major cause of fatty liver disease in the US population. The studies outlined in this proposal will elucidate the metabolic mechanism by which the risk allele causes liver fat accumulation and may provide the basis for new therapeutic options to prevent this potentially life-threatening chronic disease. Specific Aims Page 80 Principal Investigator/Program Director (Last, first, middle): RESEARCH STRATEGY (a) Significance: Non-alcoholic fatty liver disease (NAFLD) is an increasingly prevalent medical problem that affects approximately a third of the US population (1). Hepatic steatosis alone has no known medical consequences. However, the disease progresses to steatohepatitis and cirrhosis in a subset of individuals with steatosis. Soon cirrhosis due to NAFLD will overtake hepatitis C as the major indication for liver biopsy (2). Homozygosity for the PNPLA3-148M is associated with a 1.5-2-fold higher hepatic TAG content (3). The 148M allele is associated with the full progression of NALFD, as well as alcoholic liver disease. In the Hispanic population, where the prevalence of cirrhosis due to fatty liver disease is strikingly high, the frequency of the PNPLA3 risk allele is 49% (4). Combatting this disease will require elucidation of the physiological role of PNPLA3 and the mechanism by which the 148M allele results in liver disease. A series of studies using LCMS/MS based metabolomics and stable isotope labeling to elucidate the in vivo function of PNPLA3 and the mechanism of 148M induced hepatic TAG accumulation is proposed. Understanding the functional role of PNPLA3 in the progression of fatty liver disease is essential to designing effective treatment strategies of NAFLD and to understand the progression of this disease. (b) Innovation: This proposal deploys innovative methods in novel animal models to determine the metabolic basis of fatty liver disease, a major clinical disorder. A central aspect of the proposal is the development of LC-MS/MS based methods to simultaneously determine both the concentrations and the flux of many lipid classes, including triacylglycerides, phospholipids, cholesteryl esters, free fatty acids, diacylglycerides, monoacylglycerides, and lysophospholipids. Currently, the overwhelming majority of biomedical studies assay the concentrations of only a small number of individual metabolites. Such measurements provide limited mechanistic insight into the underlying biological processes. Measurement of multiple metabolites in metabolic pathways, referred to as metabolomics, is largely confined to a few specialized laboratories. Similarly, conclusions regarding the flux of metabolites are frequently inferred from their concentrations, or from indirect indices such as enzyme activities or mRNA levels assessed by real-time PCR. Here we will focus on lipid metabolism, although the conceptual approach and methodology can be readily applied to other analytes. LC-MS/MS metabolomics methods for the major lipid classes have been developed (for example by the LIPID MAPS consortium (5)). These methods allow the rapid quantitation of hundreds of individual lipid species, but extension of these techniques to determine rates of lipid flux has been very limited. We will combine LC-MS/MS based lipidomics with stable isotope labeling, using deuterium or 13C to monitor the incorporation of hydrogen or carbon into newly synthesized lipids. Hydrogen from water is incorporated into most metabolites during biosynthesis. Therefore labeling with deuterated water (D2O) allows in vivo flux to be determined on multiple metabolites simultaneously (6). This method can be used to determine the rate of synthesis and turnover of essentially any molecule that can be measure by LC-MS/MS. The second major innovation in this project is the and utilization of an animal model that recapitulates a common form of human fatty liver disease. In 2008, the Hobbs-Cohen laboratory identified the first DNA sequence variation associated with fatty liver disease (3). The variant substitutes methionine for isoleucine at codon 148 in the gene encoding patatin-like domain-containing protein 3 (PNPLA3), and is associated with the full spectrum of disease from steatosis to cirrhosis. Despite several mechanistic studies in genetically manipulated mice that either overexpress or lack PNPLA3, the mechanism by which sequence variation in PNPLA3 impacts liver fat content remains equivocal. Here we will pursue the metabolic underpinnings of PNPLA3-associated fatty liver-disease using a novel mouse model in which the isoleucine codon at position 148 of the endogenous gene has been replaced with methionine. mRNA measurements using real-time PCR indicate that expression of the modified allele is indistinguishable from that of the wild-type transcript. On high carbohydrate diets, mice expressing the I148M allele have a two-fold increase in liver fat content, quantitatively recapitulating the human phenotype. The application of powerful metabolic flux measurements to a mouse model engineered to precisely mimic the human disease-causing genetic variant should provide significant mechanistic insights into this important disorder. Research Strategy Page 81 Principal Investigator/Program Director (Last, first, middle): (c) Approach: Background and Rationale: Patatin-like phospholipase domain containing protein 3 (PNPLA3) was first implicated in the metabolism of hepatic triacylglycerides (TAGs) when our laboratory discovered that a missense variant in PNPLA3 is strongly associated with non-alcoholic fatty liver disease (NAFLD). The culprit variant substitutes methionine for isoleucine at position 148 (I148M) (3). PNPLA3 is a member of the patatin-like phospholipase domain containing protein family and is primarily expressed in liver and adipose tissue in humans and mice (7). The protein is tightly associated with membranes and partitions between the endoplasmic reticulum and lipid droplets (8). PNPLA3 consists of a highly conserved N-terminal domain that shares sequence similarity with patatin, a plentiful protein expressed in plants that has nonspecific acyl hydrolase activity. This domain contains the catalytic dyad with a consensus serine lipase motif (Gly-X-Ser-X-Gly)(9). Substituting alanine for the catalytic serine in the consensus motif (S47A) is predicted to yield a catalytically dead enzyme. PNPLA3 is dramatically regulated by nutritional intake both at the transcriptional and the post-transcriptional level. During fasting, the transcription of PNPLA3 is suppressed and the protein is actively degraded. Upon re-feeding, PNPLA3 transcription is increased and degradation of the protein is retarded (10). The phenotypic effect of the PNPLA3-I148M substitution is consistent with PNPLA3 playing a role in fatty acid metabolism. Two hypotheses have been proposed for the molecular function of PNPLA3, which are based primarily on in vitro assays. The first, suggested by Huang et al, proposes that PNPLA3 functions as an acylglycerol hydrolase(11). Biochemical characterization of the purified protein reveals that the I148M mutation causes a loss of hydrolytic activity. Arguing against a loss of function in PNPLA3 causing hepatic steatosis is the finding that PNPLA3 knockout mice do not have increased hepatic TAG levels, even on a high sucrose diet (12). The second hypothesis, suggested by Kumari et al, proposes that PNPLA3 has lysophosphatidic acid acyl-transferase (LPAAT) activity (13). The I148M substitution increases LPAAT activity when the protein is expressed in bacteria as a fusion protein with trigger factor. LPAAT catalyzes the conversion of lysophosphatidic acid to phosphatidic acid, which is a key step in TAG biosynthesis. Evidence that this activity is not responsible for the increase in hepatic TAG associated with the PNPLA3-I148M variant is the finding that overexpression of the mutant (or wt) form of PNPLA3 in the livers of mice fails to promote LPAAT activity (14). Thus, additional studies will be required to test the function of PNPLA3. Our lab has established several model systems to investigate the function of PNPLA3. We have adenoviral constructs to express wild type (wt), I148M, or S47A isoforms of PNPLA3 in cultured human hepatocytes (HuH-7 cells) (11). We also have developed transgenic mice expressing similar amounts of wt (PNPLA3wtTg) or mutant (PNPLA3148MTg) human PNPLA3 in either the liver or adipose tissue (14). The liverspecific PNPLA3148MTg mice have increased hepatic TAG content on a chow diet. The increase in hepatic TAG is further exacerbated when the mice consume a high sucrose diet, which drives lipogenesis. In contrast, overexpression of similar amounts of wt PNPLA3 in the liver does not alter hepatic TAG levels (14). Since overexpression of a heterologous protein may introduce artifacts, we have recently developed mice in which either the I148M or the catalytically inactive S47A variant are knocked in to the mouse Pnpla3 gene. Neither of these new mouse models develop hepatic steatosis on a chow diet, but ingestion of a diet high in sucrose is associated with a ~2-fold increase in hepatic TAG content in both strains of genetically-modified mice (data not shown). These mouse models, including the knockout (12), transgenic, and knock-in models, will be referred to collectively as PNPLA3 genetically manipulated mice. In this research proposal, high pressure liquid chromatography (HPLC) coupled to tandem mass spectroscopy (LC-MS/MS) will be used to identify and measure metabolites that are influenced by expression of PNPLA3 or the 148M variant. Then, a comprehensive analysis of the in vivo flux of fatty acid-containing metabolites by stable isotope labeling will be performed. These experiments will reveal the molecular mechanism of action of PNPLA3 and how the I148M mutation leads to hepatic lipid accumulation. Preliminary Results: Established methods to analyze TAG composition of tissues in PNPLA3 genetically manipulated mice: The most robust phenotype associated with the I148M variant is accumulation of hepatic TAGs(3). Therefore, we developed a LC MS/MS method to measure individual TAG species in cultured cells, plasma and tissues. First, TAGs were separated from other lipids using a C-18 column with a linear solvent gradient (98% methanol (MeOH)/2% dichloromethane to 86% MeOH/14% dichloromethane over 20 min). Under these conditions, TAGs differing by a single double bond elute ~two minutes (min) apart with a peak width of ~20 seconds (sec), shown in Figure 1. Using electrospray ionization, TAGs ionize without fragmentation so the number of Research Strategy Page 82 Principal Investigator/Program Director (Last, first, middle): carbons and double bonds in each TAG can be determined based on the mass to charge ratio in the first quadrapole (Q1) of the MS/MS. Upon collision induced fragmentation in Q2, a single fatty acid is lost from the TAG ion. The fatty acid composition is determined by scanning the mass of all fatty acids that could potentially be lost from the ion in Q3. A library of all measureable TAGs was created by pairing the intact ion mass in Q1 with the mass of the TAG missing one fatty acid in Q3. The library generated from mouse liver included ~270 different TAG species. The fatty acid Figure 1: Example chromatograph of selected 54 composition of each TAG was determined. A carbon TAGs separated by HPLC. The TAGs are similar approach was used to measure 24 cholesteryl esters species. identified based on the number of carbons (1st number), number of double bonds (2nd number), and Altered TAG profile associated with hepatic expression of PNPLA3: the identity of the fatty acids (shown in parentheses). The concentrations of 271 TAG species were measured in the livers of wt, PNPLA3wtTg and PNPLA3148MTg mice (six 8-10 week old male mice/group). The peak intensity of each species was normalized to an internal standard of 45:0 (15:0,15:0,15:0) and the ratio of the mean peak intensity for the TAG in the PNPLA3wtTg to the wt mice was determined. No net change in TAG level was observed between the two strains, but there were significant differences in the relative amounts of 47 of the TAG species that were measured (defined as a t-test p-value <0.05 with 6 mice per group) relative to the wt mice. In general, the TAG that had an odd number of carbons or contained exclusively shorter chained fatty acids (≤16 carbons) were increased while the levels of the TAG containing more than 4 double bonds were decreased in the livers of the PNPLA3wtTg mice. The livers of PNPLA3148MTg mice had an increase in the total TAG content and more than 60% of the TAG species were increased relative to both the wt and wt-transgenic mice. The only species that were not increased in PNPLA3wtTg mouse livers relative to wt were TAGs that contained more than 4 double bonds. This trend of decreased polyunsaturated hepatic TAGs observed in the livers of the PNPLA3wtTg mice was accentuated in the PNPLA3148MTg mice (Figure 2). In vivo turnover of TAGs in cultured hepatocytes (HuH7 cells) expressing recombinant PNPLA3: The rate of TAG synthesis in HuH-7 cells infected with recombinant adenoviruses expressing wt or mutant PNPLA3 was measured using D2O labeling. Cells were grown in DMEM plus 10% FCS and then 5% of the medium was substituted with D2O and grown for additional 24 hrs. Cells were collected at the indicated time points (Figure 2) and the TAG was Figure 2: A) Example of a PUFA containing TAG that is isolated, and 32 TAG species were decreased in PNPLA3-ITg liver. B) Example of TAG increased in assayed using LC-MS/MS as PNPLA3-ITg liver. C) Fold change of hepatic TAGs in PNPLA3described above. The rate of IMTg with the specified number of double bonds. Each point increase in labeled triolein was represents a different TAG species. D) Average fold change of greater in the cells expressing hepatic TAGs with a 0-10 double bonds in livers of PNPLA3PNPLA3-WT and slower in cells WTTg (blue) and PNPLA3-IMTg (red) relative to wt mice. expressing PNPLA3- I148M than in Research Strategy Page 83 Principal Investigator/Program Director (Last, first, middle): those infected with an empty virus. A higher rate of deuterium incorporation implies a faster rate of TAG synthesis if the cells have equivalent concentrations of TAGs. The cells over expressing I148M-PNPLA3 had a A) TAG content that was 80% higher than the cells expressing PNPLA3-wt or the empty vector. The relative TAG synthesis associated with expression of either PNPLA3-WT or PNPLA3-I148M was equivalent and about 25-30% higher than cells infected with an empty vector (Figure 2B). Therefore, increased TAGs caused by PNPLA3-148M cannot be solely attributed to increased synthesis. Additional studies, such as measuring turnover in the tissues of genetically-modified mice and using alternative stable isotope labels (for example fatty acids or glycerol), will be required to B) elucidate the function of PNPLA3. Specific Aim 1: Determine the effect of PNPLA3-wt and PNPLA3-I148M on the lipidome of mice Rational: PNPLA3 plays a role in the metabolism of a diverse set of metabolites comprising thousands of chemically distinct molecules. The most abundant acyllipids in human tissues are phospholipids (PL), TAGs, sphingolipids, and cholesteryl esters (CE). The complexity of these lipid classes is due primarily to the diversity of fatty acids. At least 50 different fatty acids can be detected in human plasma (15). These fatty acids Figure 3: Turnover of TAGs in cells vary in length from 12 to 24 carbons and can contain up overexpressing PNPLAS measured with to 6 double bonds at any location in the fatty acid. D2O. The turnover was measured in Huh-7 Molecules containing multiple fatty acid chains can cells over expressing PNPLA3-wt or I148M. incorporate a multitude of combination of fatty acids, The fraction of TAGs newly synthesized (dresulting in thousands of fatty acid combinations for a TAG/Total TAG) was determined using mass single class of lipids. Traditional methods for isolating isotopomer distribution analysis (MIDA). and quantifying lipid species using thin layer chromatography (TLC), gas chromatography (GC), and/or biochemical assays have traditionally been insensitive, non-specific, and time consuming. LC-MS/MS provides an excellent alternative to these approaches that will be more accurate, more sensitive and less laborious. Modern triple quadrapole mass spectrometers permit single mass unit selectivity and sub-nanogram sensitivity. Lipids that differ by only a single double bond or carbon can be distinguished based on their mass to charge ratio (m/z), and each component fatty acid can be identified based on its neutral loss by collision induced fragmentation. However, MS/MS alone is not sufficient to accurately quantify individual lipid species. Due to natural isotopic natural abundance there is overlap between a lipid of interest and the M+2 isotopic peak of a similar lipid with one additional double bond. This results in convolution of multiple lipid species at a single mass. These lipids can be deconvoluted by incorporating a chromatographic separation (HPLC) prior to the MS/MS. The HPLC eluent can be introduced directly into the MS via an atmospheric-pressure ionization or electrospray source. Therefore, by combining HPLC and MS, individual lipid species can be quantified accurately. This approach has been used by multiple investigators to develop LC-MS/MS techniques that can quantify the concentration of many lipids in a biological sample. Aim 1A: Develop LC-MS/MS Methods to Quantitate Acyl-Lipids Here I will implement methods that have been developed by the LIPID MAPS consortium and others to measure the concentration of fatty acid-containing lipids in plasma, liver and adipose tissue, including TAGs, CEs, PLs, diacylglycerides (DAG), monoacylglycerides (MAG), lysophospholipids, acyl-CoAs, and free fatty acids. An LC-MS/MS system (ABSciex Qtrap 4000 triple quadrapole with a Shimadzu LC20A HPLC) will be used for these experiments. Since the LC-MS/MS methods for each class of lipids have been developed and reported, it is anticipate that establishing these assays in our laboratory will take between 4 and 8 weeks per lipid class. If problems arise during the implementation of methods, Dr. Jeff McDonald, a member of the LIPID Research Strategy Page 84 Principal Investigator/Program Director (Last, first, middle): MAPs consortium and a faculty member in my department, will be consulted (see letter). If we hit roadblocks, I will consult Drs. Jose Castro and Tom Roddy, who are in the small molecule mass spectroscopy division at Merck; these investigators are collaborating with our laboratory on another project and have agreed to help troubleshoot any problems I encounter in setting up these assays or interpreting the data generated. Each method will be designed to fully analyze a sample in less than 30 min, which will assist in rapid implementation of methods to measure each lipid class in many samples. Potential Problems and Alternative Strategies: The methods that will be implemented have several potential drawbacks. First, it is impractical to obtain authentic internal standards for each lipid species, thus it is not possible to calculate the absolute concentration of each lipid. Where practical, standards will be used to quantify the absolute concentration of a lipid species. Lipids for which no internal standard is available will be normalized to a single internal standard (specific to the lipid class). This analysis will provide relative concentrations of each lipid species. Second, the fatty acid double bond location (i.e. ω3, 6, or 9 fatty acids) cannot be differentiated using the proposed LC-MS/MS method. If it is necessary to identify the location of the double bond, an orthogonal method, such as GC-flame ionization detection (GC-FID), can be used. Our laboratory has an established method using an HP6890 instrument to measure saponified TAG composition using TLC and GC-FID (14). Lastly, using different LC MS/MS methods for each lipid class can potentially be cumbersome if different columns and mobile phase combination are used. To minimize this challenge, methods that utilize identical HPLC solvents and columns will be developed. Aim 1B: Compare lipidomes of plasma, liver and adipose tissue in the PNPLA3 mouse models As described previously, our laboratory has established several lines of transgenic and knock-in mice that express various forms of PNPLA3. This collection includes transgenic mice overexpressing PNPLA3-wt or -148M in liver (under the control of the ApoE promoter) or in adipose tissue (under the control of the AP2 promoter) (14), and PNPLA3 148M and 47A knock-in mice (unpublished data). We also have Pnpla3-/- mice that were obtained from Erin Kershaw and have been characterized extensively in our laboratory (12). To glean insights into the effects of PNPLA3 on lipid composition, we will characterize and compare the lipidomes of these different mouse strains. To determine the effect of PNPLA3 expression on the lipidome of tissue, we will extract the lipids from the liver, adipose tissue and plasma using the LC-MS/MS methods developed in Aim 1A. For each experiment, we will use six 12-week-old male, chow-fed genetically-modified mice and an equal number of wild-type littermate controls. Mice will be sacrificed and liver, WAT, and plasma will be collected and store at 80C. Tissues will be homogenized and lipids extracted using a Bligh-Dyer extraction. The concentration of each lipid species will be measured by LC-MS/MS. The lipid level in each mouse model will be compared to the wt littermate controls and significant differences between means will be determined by a t-test. In addition, trends in the lipids most affected by PNPLA3 will be identified, such as a reduction in PUFA containing TAGs shown in preliminary data. Aim 1C: Determine effects of changes in dietary composition on hepatic lipid profile The accumulation of hepatic TAGs in the PNPLA3148MtTg mice is increased when the mice are fed a highsucrose diet (14). Although these mice also develop steatosis on some chow diets, they do not develop steatosis on all chow diets (unpublished data). When the mice are fed a chow diet that contains animal fat source, the PNPLA3148MtTg have 50% higher hepatic TAGs than PNPLA3wtTg mice. However, when vegetable fat replaces the animal fat, the PNPLA3148MtTg and PNPLA3wtTg mice have similar levels of hepatic TAG. These differences are not caused by any differences in PNPLA3 mRNA or protein expression (unpublished data). This suggests that PNPLA3 I148M induced NAFLD can potentially be alleviated by dietary modification. To investigate the role of diet in PNPLA3 function, the lipidome of the PNPLA3 mouse models on the two different chow diets will be characterized. For these experiments, I will use 8-10 week-old male mice (6/group) that have been fed either of the two chow diets for four weeks. The tissues will be processed and the results analyzed exactly as described above. I will also examine the effect of a high-sucrose,+/- polyunsaturated fatty acids (PUFA) on tissue lipid composition. Since over-expression of PNPLA3 results in a reduction in TAG: PUFAs, supplementing PUFAs into a high-sucrose diet may result in less accumulation of hepatic TAGs in the PNPLA3148MtTg mice. Other dietary modifications may also be tested, depending on results obtained from the studies described in Aim 1B. The choice of mouse models will also depend on the results of Aim 1B. Likely, I will use the I148M knock-in mice for these studies. Research Strategy Page 85 Principal Investigator/Program Director (Last, first, middle): Aim 1D: Lipidome of human plasma with PNPLA3 II or MM We have collected plasma samples from age-, sex- and race-matched subjects who are homozygotes for the PNPLA3-148I or the PNPLA3-148M variant (100/group). We will analyze the lipidome of these samples using the methods developed in Aim 1A. The lipids from 100 μL of plasma will be extracted with a Bligh-Dyer extraction and concentrated. After extraction, the samples will be analyzed by LC-MS/MS and the level of each lipid species will be quantified. Metabolites that differ significantly between the two genotypes will be identified using a t-test. A separate set of samples taken from the Dallas Heart Study will be analyzed to confirm any observed associations. These results will then be compared to the plasma lipidomes of the PNPLA3 mouse models, which will allow us to address two questions: 1) Are there any changes in the profile of acyl-containing lipids that relate to the susceptibility allele in PNPLA3? 2) How well do the lipid distributions in the plasma of the mouse models expressing the PNPLA3-I148M variant reflect those of humans with the same variant? Specific Aim 2: Role of PNPLA3 in Lipid Flux Rationale: Regulated synthesis and catabolism of metabolites, known as metabolic flux, is the central function of metabolism. Measuring metabolic flux has traditionally been a technically challenging aspect of metabolic research. A standard method to measure metabolic flux is with radioactive isotope labeling. For example, the flux of TAGs in vivo can be measured by injecting tritium-labeled water into the circulation of an animal. The tritium is incorporated into TAGs. After an hour of labeling the animal is sacrificed, the organs of interest are collected, the lipids are extracted, TAGs are isolated from a TLC plate, and the radioactivity of the TAGs is measured. Using this methodology, the rate of TAG synthesis can be measured and compared between groups of animals. Usually, flux is not measured directly, but rather is inferred from the measurement of the concentration of a metabolite. It is assumed that an increased in the amount of a metabolite is the result of either an increase in synthesis or a decrease in catabolism. This assumption is derived from the law of first order kinetics, which states that for a molecule to accumulate in a steady state system there must either be an increased rate of appearance (i.e. synthesis) or a decreased rate of disappearance (i.e. catabolism or clearance). In some biological systems this is a valid assumption. However, this assumption is not valid for PNPLA3148MTg mice. When PNPLA3 is overexpressed in mouse liver (the PNPLA2wtwtTg mice) there is no excess accumulation of hepatic TAGs relative to wt mice (14). The PNPLA3148MTg mice have ~60% more hepatic TAG than PNPLA3wtTg mice. Assuming first order kinetics, one would predict that the PNPLA148MTg mice have a higher rate of TAG synthesis than the PNPLA3wtTg or wt mice, and the PNPLA3wtTg and wt mice have similar TAG synthesis rates. When tritiated water was used to measure TAG synthesis, the two strains of transgenic mice had similar rates of hepatic TAG synthesis, and both transgenic models had ~30% higher synthesis rate than wt mice (14). Therefore, a more refined model is required to describe the mechanism of PNPLA3-I148M induced NAFLD. Using radioisotope labeling has numerous technical limitations that hinder the ability to develop and validate models of the mechanism of PNPLA3. To limit exposure to radioactivity, labeling experiments are generally performed on a very short time scale, for example, after only one hour for TAG synthesis. The halflife of mouse hepatic TAGs is ~3 days. Therefore, the labeling protocol only samples the most rapidly turningover pool of TAGs. Tritiated water labeling also cannot be directly translated into human due to the toxicity of radioactivity. Finally, the radio-labeling methods for measuring flux are inherently non-specific and cannot utilize modern metabolomics technologies. Stable isotope labeling is an alternative approach to measuring metabolic flux in vivo and does not have the same technical drawbacks associated with radio-labeling. The principle of stable isotope labeling is similar to radio-labeling. A stable isotope labeled substrate, such as deuterated water (D2O), is administered to an animal. The isotope label, which has a higher molecular weight than a non-labeled substrate, will incorporate into metabolites via biosynthesis causing an increase in the molecular mass of the newly synthesized metabolites (5). The mass shift is measured using a mass spectrometer and the fraction of newly synthesized metabolites is calculated using mass isotopomer distribution analysis (MIDA) (16). Combining LC-MS/MS based metabolomic methods with stable isotope labeling will allow the measurement of metabolic flux of thousands of molecules simultaneously and will be a powerful tool for understanding the mechanism of PNPLA3. In this aim, in vivo stable isotope labeling will be used to measure the role of PNPLA3 on lipid flux in cultured hepatoma cells overexpressing PNPLA3 and in PNPLA3 genetically manipulated mouse models. Research Strategy Page 86 Principal Investigator/Program Director (Last, first, middle): Aim 2A: Measure Lipid Flux using D2O in Cultured Cells Over-Expressing PNPLA3 In this set of experiments, cultured hepatocytes overexpressing recombinant PNPLA3-wt, PNPLA3-I148M and PNPLA3-S47A will be used to assess the effect of the I148M variants on lipid flux. D2O will be used to monitor the flux of fatty acid-containing metabolites in these cells. HuH-7 cells will be infected with recombinant Figure 4: Experimental plan for in vivo lipid labeling using D2O in HuH-7 adenoviruses expressing the cells infected with PNPLA3-wt, PNPLA3-148M, PNPLA3-47A, or an various forms of human empty virus, as described in Aim 2A. PNPLA3. Cells will be grown for 3 days in DMEM plus 10% FCS medium. On day 4 the medium will be supplemented with 5% D2O. The cells will be harvested at several time points over the next 48 hours. Twelve time points in duplicate will be harvested for each condition. The time points will be exponentially spaced with the majority of the cells harvested in the first 8 hours. After harvesting the cells, the lipids will be isolated using a Bligh-Dyer extraction and then the lipids will be analyzed by LCMS/MS using the methods developed in Specific Aim 1. This experimental plan is illustrated in Figure 4. Five isotopomers (M=0 to M=4) will be monitored for each lipid species to determine the change in the isotopic distribution. If any problems arise during the development of these experiments, I will consult with Drs. Elizabeth Parks and Ralph Deberadinis (see letters), who are experts in using stable isotopes to measure metabolic flux. The relationship between the fraction of molecules newly synthesized and time is based on the change in the isotopic distribution. Based on this relationship, flux parameters (e.g., metabolite turnover, halflife and rate of synthesis) will be quantified. The effect of PNPLA3 on the flux of lipid will be determined by comparing the quantitated rate of synthesis and half-life in each group of cells infected with different viruses containing PNPLA3. To assist in data analysis and quantification, Matlab algorithms have been programed for MIDA and kinetic parameter regression. This approach will be applied to most metabolites measured by the LC-MS/MS methods developed in Aim 1A. With these tools, it is feasible to monitor the turnover of hundreds, if not thousands, of individual metabolite species in vivo. Potential Limitations and Alternative Strategies: Quantifying the rate of synthesis of each lipid species requires using MIDA or other similar techniques. To apply MIDA, the number of label incorporation sites in a molecule (N; i.e. the number of deuterium atoms that can incorporate into a single lipid) must be assigned (17). Deuterium can be metabolically incorporated at numerous steps during lipid assembly. For example, deuterium can be incorporated in TAGs during fatty acid synthesis, fatty acid elongation, or during esterification. Therefore, there is uncertainty about the value of N for most lipids. To overcome this limitation the fraction of the water pool that is labeled with D2O (p) must be assigned. The value of p can be calculated based on a molecule with a known N value, for example cholesterol, or determined by GC-MS. After assigning a value of p, N of each lipid species can be calculated using a non-linear regression. The uncertainty associated with this regression complicates direct comparison between different lipids species. To overcome this uncertainty, alternative stable isotope labels with a known N value, such as 13C labeled fatty acids or glycerol, will be used (see Aim 2B). These alternative labeling strategies will also provide an orthogonal method to confirm that any results are not due to artifacts associates with using D2O labeling. Aim 2B: Alternative stable isotope labeling strategies in cultured cells over-expressing PNPLA3 While D2O is an excellent isotope label for monitoring the flux of most metabolites, there are some limitations to this approach (see above). Glycerolipids can also be labeled by providing stable isotope labeled metabolites, such as 13C-fatty acids or 13C-glycerol, which will be incorporated into a lipid species in a known stoichiometric ratio (18). These labels also monitor a subtly different process than water labeling. Lipid biogenesis involves two broad steps: 1) de novo fatty acid synthesis and 2) assembly and reassembly of lipids. Research Strategy Page 87 Principal Investigator/Program Director (Last, first, middle): The rate-limiting step in de novo lipid biogenesis is fatty acid synthesis. Since deuterium from D2O incorporates primarily during fatty acid synthesis, D2O only monitors this process. Lipid assembly and reassembly, commonly known as “futile cycling”, occurs at a faster rate. By labeling with the preformed lipid components, such as fatty acids and glycerol, the rate of lipid assembly and fatty acid exchange can be measured in vivo. To address the functional mechanism of PNPLA3, 13C-3-glycerol, 13C-U-palmitate, 13C-U- oleate, and 13C-Uarachidonate will be used to monitor lipid flux in cultured hepatoma cells overexpressing wt and mutant PNPLA3. HuH-7 cells will be infected with adenoviruses expressing PNPLA3-wt, PNPLA3-148M, PNPLA347A, or an empty virus and cultured for 3 days in DMEM with 10% FCS. On day 4, stably isotope label substrate will be added to each dish of cultured cells. After adding the labeled substrate, cells will be harvested over a time course. Appropriate timing of sampling and concentration of stable isotope labeled substrate will be determined empirically. After harvesting, lipids will be extracted and analyzed by LC-MS/MS. Rate of incorporation will be determined by analyzing the fraction of each lipid species with incorporated label. Each group of infected cells will be compared to each other and to the incorporation when D2O was used for labeling. If necessary, these experiments will be extended to primary hepatocytes or to the PNPLA3 genetically-manipulated mouse models. Aim 2C: Lipid flux in PNPLA3 genetically manipulated mouse using D2O Although many facets of PNPLA3 biology can be addressed in cultured cells, an animal model must be utilized to define the bona fide physiological function of the protein. D2O will be used to measure the metabolic flux in genetically manipulated mouse models. The experiments will be conducted in a similar manner to the cell culture experiments proposed in Aim 2A. The body water pool of the mice will be enriched by injecting 500 μL of D2O (in physiological saline) into the circulation of 6 mice of each genotype. After injection, the water source of the mice will be supplemented with 6% D2O. This causes a rapid enrichment of D2O in body water to around 5%(6). After injection, tissue samples from the animals will be collected in a time course with exponentially timed sampling. For example collections may be made at 0, 1, 2,4, 7, 10, 14, 24, 32 and 48 hours after injection of D2O. In the first series of experiments, 10 μL of plasma per mouse will be collected at each time point. Lipids will be extracted from plasma and analyzed by LC-MS/MS. This experimental plan is illustrated in Figure 5. The rate of synthesis and half-life of each measureable lipid will be determined based on the rate of incorporation of deuterium. Each mouse model will be compared based on the quantified rate of synthesis and on the qualitative characteristics of the incorporation. Plasma will be analyzed before individual tissues because fewer mice are Figure 5: Experimental plan for in vivo lipid labeling using D2O in needed for each experiment, PNPLA3 genetically manipulated mice, as described in Aims 2C and 2D allowing more efficient optimization of the dosing and collection procedures. Once the optimal time course of sampling is determined, the experiment will be repeated using 10 to 20 mice per group. Each mouse will be injected with 500 μL D2O and tissues collected at the pre-specified time points. Lipids from the liver and WAT will be analyzed by LC-MS/MS and the rates of synthesis of selected lipids will be determined based on rate of deuterium incorporation. The exact number of mice per group and timing of the sampling will be dependent on the rate of incorporation in plasma and the availability of genetically manipulated mice. The results obtained from the different groups of mice will be compared based on the quantitative rate of synthesis and the qualitative characteristics of deuterium incorporation. This experiment will reveal the influence PNPLA3 and the I148M variant on lipid flux. The metabolic role of the protein should be apparent from these results. Aim 2D: Dietary Influence on Lipid Flux and the Contribution of PNPLA3 PNPLA3 function is influenced by diet; therefore dietary modification may provide a potential therapeutic strategy to alleviate NAFLD in homozygote MM patients. In Aim 1C, we will examine the effect of differences in diets on the lipidome in the PNPLA3 genetically-modified mouse models. Ideally, a dietary Research Strategy Page 88 Principal Investigator/Program Director (Last, first, middle): modification will be identified that lowers hepatic TAGs in either the PNPLA3148MTg or the PNPLA3-I148M knock-in mice. When such a diet is identified, in vivo lipid flux in mice consuming that diet will be measured. This experiment will be conducted as described in Aim 2C. Six animals per group per diet will be injected with 500 μL D2O and the water will be supplemented with D2O. At 10 time points, 10 μL of plasma per mouse will be collected. The plasma lipids will be extracted and analyzed by LC-MS/MS to determine the rate of deuterium incorporation. The rate of synthesis will be determined based on the rate of deuterium incorporation. Mouse models on different diet will be quantitatively compared based on the rate of lipid synthesis. If necessary, a time course of tissue collection after D2O administration. This will depend on the correlation between liver and plasma lipid turnover, established in Aim1C. By measuring the lipid flux in these animals the metabolic mechanism by which dietary change modifies PNPLA3 148M pathology will be determined. Research Strategy Page 89 Principal Investigator/Program Director (Last, first, middle): Protection of Human Subjects: In sub-aim 1D of this research proposal I will analyze 200 human plasma samples. These samples include 100 homozygote patients with PNPLA3 II and MM. I will use approximately 100 μL of plasma from each subject to determine the effect of PNPLA3 I148M on the human lipidome. The samples were collected last year, so analysis of the plasma samples will cause no additional risk to the subjects. The cohort of subjects includes approximately 50% female and 33% African Americans, 33% Hispanics, and 33% whites. No children were used in this study because they are not afflicted with atherosclerosis. Protection of Human Subjects Page 90 Principal Investigator/Program Director (Last, first, middle): Inclusion of Women and Minorities: The subjects for this proposal comprise an equal portion of women and men. In addition, the study is racially diverse including equal portions of African America, Hispanic, and White subjects. No gender or racial background was excluded from this study. Women &Minorities Page 91 Principal Investigator/Program Director (Last, first, middle): Targeted/Planned Enrollment: This proposal uses a databank of plasma samples that have already been collected from a gender and ethnically diverse population of subjects. No additional enrollment of patients is expected for this research plan. Planned Enrollment Table Page 92 Principal Investigator/Program Director (Last, first, middle): Inclusion of Children: Children were excluded from this research study. Children were excluded because they do not generally exhibit risk of atherosclerosis, so the profile of their lipidome will be irrelevant to this study. Children Page 93 Principal Investigator/Program Director (Last, first, middle): Vertebrate Animals Description of Animals: In my research plan, I propose using wild type C57BL/6 mice or genetically modified mice that have been bred into a C57BL/6 background. The genetically modified mice will include PNPLA3 knock-out mice, PNPLA3 liver and adipose tissue specific transgenic mice, and PNPLA3 I148M and S47A knock-in mice. The mice will be at least 8 weeks old for all experiments. I will use both female and male mice. I will use approximately 500 mice for this research proposal. Why Animals: Lipid metabolism is a complex process that is intrinsically linked to systematic biological factors such as intracellular signaling, diet, immune system, etc. Although cell culture models can provide some insight into the function of PNPLA3, animal models will be required to determine the true function. Lower organisms, such as flies or c. elegans, do not recapulate human lipid physiology therefore a rodent model will be required. Veterinary care of animals: The animals will be housed in UT Southwestern animal housing facility located in the Y building 2nd floor and exposed to 12 hour light and 12 hour dark cycle. Normal chow or specialized diets and water will be freely available to the animals. Cages will be routinely changed to ensure a clean environment for the animals. Procedures to minimize discomfort, stress or pain of anmials: Mice will be sacrificed anaesthetized by inhaling isoflurane in a big chamber. All animal procedures will be approved by UT Southwestern’s institutional animal care and use committee (IACUC) before executing the experiment. Animal Euthasnasia: Surplus mice that do not qualify for proposed experiments will be euthanized via either carbon dioxide or isoflurane inhalation. These two methods are consistent with the AVMA Guidelines of Euthanasia, 2012. Vertebrate Animals Page 94 Principal Investigator/Program Director (Last, first, middle): Select Special Agents: No select agents will be used in this research proposal Select Agent Research Page 95 Principal Investigator/Program Director (Last, first, middle): Consortium/Contractual Arrangement: This research proposal does not involve a consortium of researchers. All research will be performed at UT Southwestern. Consortium/Contractual Page 96 Principal Investigator/Program Director (Last, first, middle): Resource Sharing Plan: A resource sharing plan is not required for this proposal because it does not involve developing model organisms, genome wide association studies, and it does not request more than $500,000 per year. Resource Sharing Plan Page 97 SUMMARY STATEMENT ( Privileged Communication ) PROGRAM CONTACT: Richard Okita 301-594-3827 okitar@nigms.nih.gov Release Date: Application Number: Principal Investigator 05/29/2013 1 K01 GM109317-01 PHD Applicant Organization: UT SOUTHWESTERN MEDICAL CENTER Review Group: ZRG1 IMST-K (50) Center for Scientific Review Special Emphasis Panel Mentored Research Scientist Development Award in Metabolomics Meeting Date: 05/22/2013 Council: AUG 2013 Requested Start: 09/01/2013 Project Title: SRG Action: Next Steps: Human Subjects: Animal Subjects: Gender: Minority: Children: Project Year 1 2 3 4 ___________ TOTAL RFA/PA: RM12-025 PCC: P140RO Dual PCC: 1ME201 Dual IC(s): RM Determining the Function of PNPLA3 Utilizing Metabolomics and Stable Isotope Labe Impact Score: 38 Visit http://grants.nih.gov/grants/next_steps.htm 30-Human subjects involved - Certified, no SRG concerns 44-Vertebrate animals involved - SRG concerns 1A-Both genders, scientifically acceptable 1A-Minorities and non-minorities, scientifically acceptable 3A-No children included, scientifically acceptable Clinical Research - not NIH-defined Phase III Trial Direct Costs Requested 95,559 96,926 98,334 99,783 _______________ 390,602 Estimated Total Cost 103,204 104,680 106,201 107,766 _______________ 421,850 ADMINISTRATIVE BUDGET NOTE: The budget shown is the requested budget and has not been adjusted to reflect any recommendations made by reviewers. If an award is planned, the costs will be calculated by Institute grants management staff based on the recommendations outlined below in the COMMITTEE BUDGET RECOMMENDATIONS section. 1 K01 GM109317-01 2 ZRG1 IMST-K (50) 1K01GM109317-01 SCIENTIFIC REVIEW OFFICER'S NOTES VERTEBRATE ANIMAL UNACCEPTABLE RESUME AND SUMMARY OF DISCUSSION: This application for a K01 mentored research scientist development award in metabolomics proposes to extend the principal investigator’s training in biochemical engineering, biophysics, molecular genetics and lipidomics by utilizing targeted liquid chromatography-tandem mass spectrometry (LC-MS-MS) methods to determine the flux of different classes of fatty acid-containing lipids in liver, adipose tissue, and plasma in mouse models of PNPLA3associated steatosis. This project will be carried out under the direction of Professors Helen Hobbs and Jonathan Cohen at the University of Texas Southwestern Medical Center. Reviewers noted that the applicant is highly regarded by his advisors and mentors and is a productive and enthusiastic research scientist. While early in his career, he was considered to have demonstrated innovation by developing methods for the analysis of multiple triglyceride species and also has demonstrated an appreciation for the need to combine metabolomics concentration measurements with flux measurements. The mentoring team was viewed as having significant experience in training PhD and postdoctoral students as well as excellent publication and funding records. While the career development plan addresses several areas of importance to establishing an independent research career, it was felt that it did not address the PI’s lack of background in biological sciences and did not define a plan to establish an independent career. The research plan was considered to be outstanding and was noted to do an excellent job of combining metabolomics with flux analysis. The complementarity of the in vitro and in vivo uses of labeling and metabolomics to evaluate the pathways of lipid metabolism with changes in PNPLA3 function was specifically cited as a strength of the application. While the environment at UTSW was considered to be outstanding, the institutional commitment to was noted to be weak. At the conclusion of the discussion, enthusiasm for the strengths of the sponsors, the applicant, the research plan and the institutional environment was dampened somewhat by weakness described for the training plan and institutional support and as a result, the panel noted the potential overall impact/merit of the proposal to be excellent. DESCRIPTION (provided by applicant): Non-alcoholic fatty liver disease (NAFLD) is an increasingly prevalent medical problem that affects approximately a third of the US population. Patatin-like phospholipase domain containing 3 (PNPLA3) was first implicated in the metabolism of hepatic triacylglycerides (TAGs) when our lab found that a missense mutation that substitutes isoleucine at position 148 with methionine (I148M) was associated with non-alcoholic fatty liver disease in humans. Homozygotes with the variant allele (148M) have a 2-fold higher risk of hepatic TAG accumulation than homozygotes with the wild type allele (148I). The function of PNPLA3 and the cause of I148M induced hepatic TAG accumulation remains unclear. Since fatty liver represents an accumulation of fatty acids in the form of triglycerides, I will adapt and/or develop targeted liquid chromatography-tandem mass spectrometry (LC-MS-MS) methods to measure the concentrations of different classes of fatty acidcontaining lipids in liver, adipose tissue, and plasma from mouse models of PNPLA3-associated steatosis. Mice will be studied on both a normal chow diet and on diets designed to exacerbate or alleviate PNPLA3-I148M associated accumulation of hepatic TAG. I will then extend these LC-MS/MSbased methods to determine the flux of each lipid class by measuring the rate of incorporation of deuterium from deuterated water (D2O) in cultured cells that overexpress PNPLA3, and in genetically manipulated mouse models. Alternative stable isotope labeling strategies, such as 13C-glycerol or 13Cfatty acid incorporation, will also be employed to compliment the D2O incorporation data. Execution of this research proposal will provide training in the application of metabolomics technology to study human disease and shed insight into the function of PNPLA3 and its role in the progression of fatty liver disease. 1 K01 GM109317-01 3 ZRG1 IMST-K (50) PUBLIC HEALTH RELEVANCE: Fatty liver disease is a growing epidemic that afflicts a third of the US population and leads to steatosis, cirrhosis, and liver failure. We have identified a gene named PNPLA3 that is strongly associated with fatty liver disease in human, but the function of this gene remains unclear. I am proposing experiments designed to determine the function of PNPLA3 and ultimately aid in the development of effective medical therapies to prevent fatty liver disease. CRITIQUE 1: Candidate: 2 Career Development Plan/Career Goals & Objectives/Plan to Provide Mentoring: 2 Research Plan: 1 Mentor(s), Co-Mentor(s), Consultant(s), Collaborator(s): 1 Environment and Institutional Commitment to the Candidate: 2 Overall Impact: This is an application from a postdoctoral scholar who wants to receive training to be able to run a service metabolomics lab with expertise in stable isotope technology. This will allow metabolomic data to be complemented by flux measurements. This variation of metabolomics has great potential for the discovery of new regulatory processes. This training will take place in a Genetics lab which generated very interesting data on the mechanism of liver steatosis. The involvement of experts in lipidomics, metabolism and stable isotope technology, as well as the rich environment of UTSW, bodes well for the success of this project. 1. Candidate: Strengths The applicant, , has received excellent training in Biophysics of lipids with Dr Donald Small. He should have no problems mastering the metabolomics techniques of his project. Weaknesses It is not clear how much has learned about metabolism and isotopic techniques before starting his postdoctoral scholarship. He will not get such training from the experts in Genetics who will co-mentor him (Dr Hobbs and Cohen who initiated this excellent project on the consequences of mutation in PNPLA3). It will thus be important that get training in metabolism from Drs Parks and DeBerardinis, and from other metabolic experts at UTSW. On page 70, acknowledges that he needs training in a number of disciplines (molecular biology, genetics and animal husbandry) but does not mention metabolism. 2. Career Development Plan/Career Goals & Objectives/Plan to Provide Mentoring: Strengths career plan (briefly outlined page 70) is to run a service lab which will assist other investigators. “Independent research” is just mentioned passing by. There is nothing wrong in running a service lab. Actually, many metabolomics units are set up to provide services to biological investigators. In fairness to the applicant, it is a bit early in his career to have a clear idea of a possible future independent research. Weaknesses None noted 3. Research Plan: 1 K01 GM109317-01 4 ZRG1 IMST-K (50) Strengths The applicant correctly points out that classical metabolomic studies yield only vague conclusions on the impact of a perturbation on metabolomics fluxes. It is erroneous to interpret variations in relative concentrations as reflecting corresponding variations in fluxes. Assessment of variations in metabolic fluxes requires isotopic tracer studies conducted under rigorous conditions to avoid artifacts and errors in data interpretation. The association of metabolomics and mass isotopomer analysis provides a powerful tool to identify new regulatory mechanisms and new pathways. The applicant understands this very well, and benefits from the mentorship of an expert in lipidomics, Dr Jeffrey McDonald, and experts in mass isotopomer analysis, Drs Elizabeth Parks and Ralph DeBerardinis. The planning of the components and phases of the project is very logical with the right mixture of in vivo and in vitro experiments, using rodent and human materials. The project centers on lipidomics. Although there a many types of lipids, the number of their building blocks is reasonable, and is made up of fairly few types of compounds. Thus the analytical techniques to be used can crank out data rapidly without having to identify totally unknown compounds. Weaknesses One would have liked to see more detail on the measurements of isotopic enrichments of lipids containing many carbons, where the naturally-enriched M1, M2 mass isotopomers have similar abundances as the unlabeled M peak. The section on the use of other tracers than D2O is only sketchily outlined. 4. Mentor(s), Co-Mentor(s), Consultant(s), Collaborator(s): Strengths Dr McDonald has much expertise in lipidomics Drs Parks and DeBerardinis have great expertise in mass isotopomer analysis applied to lipogenesis and to pathway regulation Should the need arise, there is much expertise in metabolic NMR at UTSW (Drs Craig Malloy and Shawn Burgess) Weaknesses Because the synthesis of complex lipids involves low concentrations of the CoA esters of the fatty acids, metabolomics and labeling of acyl-CoAs might provide useful information on the synthesis of complex lipids. 5. Environment and Institutional Commitment to the Candidate: Strengths The environment at UTSW is outstanding Weaknesses None noted Protections for Human Subjects: Acceptable Risks and Adequate Protections Deidentified human plasma samples will be used. 1 K01 GM109317-01 5 ZRG1 IMST-K (50) Data and Safety Monitoring Plan (Applicable for Clinical Trials Only): Not Applicable (No Clinical Trials) Inclusion of Women, Minorities and Children: G1A - Both Genders, Acceptable M1A - Minority and Non-minority, Acceptable C3A - No Children Included, Acceptable Vertebrate Animals: Acceptable Biohazards: Not Applicable (No Biohazards) Training in the Responsible Conduct of Research: Acceptable Comments on Format (Required): Appropriate Comments on Subject Matter (Required): Appropriate and extensive Comments on Faculty Participation (Required; not applicable for mid- and senior-career awards): Excellent distribution of faculty expertise Comments on Duration (Required): adequate Comments on Frequency (Required): adequate Resource Sharing Plans: Not Applicable (No Relevant Resources) Budget and Period of Support: Recommend as Requested CRITIQUE 2: Candidate: 2 Career Development Plan/Career Goals & Objectives/Plan to Provide Mentoring: 4 Research Plan: 2 Mentor(s), Co-Mentor(s), Consultant(s), Collaborator(s): 2 1 K01 GM109317-01 6 ZRG1 IMST-K (50) Environment and Institutional Commitment to the Candidate: 4 Overall Impact: Although relatively new to the use of LC/MS/MS for metabolomics research, e is off to a good start, has demonstrated his skill and innovation in developing methods for the analysis of multiple triglyceride species. He has demonstrated an appreciation for the need to combine metabolomics concentration measurements with flux measurements. The project describes the use of deuterium labeling as a means for making these flux measurements. This methodology is generalizable to other metabolites and is translatable to human investigation. The project encompasses in vitro cell studies, in vivo transgenic mouse studies, and evaluation of human blood samples of patients known to have genetic variation relevant to this study. Thus, this project strongly supports the career development in the use of metabolomics. The mentors are outstanding with experience in mentorship to successful transition of previous mentees to independent careers. However, the closely knit goals of this project with those of mentors may make it difficult down the road for to assert his independence of thought in the execution of this study. The proposal suffers from a lack of any formal training in LC/MS/MS, metabolomics, statistical analyses of metabolomics data, or career development courses. As well, institutional support is ambiguous in its commitment to dedicated time or independence after the K01 is completed. 1. Candidate: Strengths is well regarded by his advisors and mentors as a highly motivated and productive researcher with a strong potential to become an independent investigator. Track record in the hands-on use of LC/MS/MS with the ability to develop new analytical approaches is shown in the preliminary data on the TG assays. Good grasp of the use of the metabolite labeling from deuterium oxide as a means to measure metabolic fluxes. Weaknesses None noted 2. Career Development Plan/Career Goals & Objectives/Plan to Provide Mentoring: Strengths Advisory team covering all aspects of the research plan will provide guidance in meeting the goals of the research and in transitioning to an independent career. Drs Hobbs and Cohen experience with mentorship is well established and provides training in lab etiquette, integrity in data collection and recording, data presentation, manuscript and grant preparation, and scheduled evaluation of progress towards short and long-term goals. Weaknesses The research is intertwined closely with the work of Hobbs and Cohen, and does not establish a unique niche for that is distinct from his mentors. There are no plans for formal training in the field of metabolomics and the statistical tools currently used. 3. Research Plan: Strengths 1 K01 GM109317-01 7 ZRG1 IMST-K (50) The use of metabolomics analysis to evaluate changes in lipid metabolism in transgenic mice targeting the relevant pathways is a powerful approach to understand the nuances of metabolic regulation. Metabolomics, when combined with the use of deuterium labeling to assess metabolic fluxes significantly strengthens the project. A much better understanding of metabolic regulation is obtained when flux measurements are combined with concentration measurements. Includes provisions for translation to humans, with aim 1D to compare lipid profiles in humans with the PNPLA3 mutant mice. In vitro and in vivo complement each other in the use of the metabolomics and labeling approaches to evaluate the pathways of lipid metabolism with changes in PNPLA3 function. The experimental plan is relevant to goal of developing his expertise in the use of metabolomics and isotopic tracers for the study of human metabolic diseases. Weaknesses None noted 4. Mentor(s), Co-Mentor(s), Consultant(s), Collaborator(s): Strengths The mentorship by Drs. Hobbs and Cohen is a strength in that the lab has a good track record in mentoring of post-docs on their way to establishing independent research. Mentorship in mass spectroscopy and metabolomics will be provided by Jeff McDonald, Ph.D. This will be a continuation of the training already provided by Dr. McDonald for the use of LC/MS/MS to measure triglycerides, and deuterium incorporation into sterols. This has been a productive training period and it can be expected to continue in that vein. Weaknesses None noted 5. Environment and Institutional Commitment to the Candidate: Strengths UTSW is an outstanding research institution with renowned experts in the field of research that will pursue during this K01 award period. Weaknesses The amount of protected time for research is unspecified, and is stated only as “will provide protected time for to pursue his research. No indication that the institution has a commitment to independence of research following this K01. Protections for Human Subjects: Acceptable Risks and Adequate Protections Data and Safety Monitoring Plan (Applicable for Clinical Trials Only): Not Applicable (No Clinical Trials) 1 K01 GM109317-01 8 ZRG1 IMST-K (50) Inclusion of Women, Minorities and Children: G1A - Both Genders, Acceptable M1A - Minority and Non-minority, Acceptable C3A - No Children Included, Acceptable Samples are from previously approved study of PNPLA3 patients. Vertebrate Animals: Unacceptable Justification for the numbers of mice is not given. Biohazards: Not Applicable (No Biohazards) Training in the Responsible Conduct of Research: Acceptable Comments on Format (Required): Format is 45-min lecture followed by 45-min faculty led discussion. Comments on Subject Matter (Required): Covers ethical and regulatory considerations for use of animals and humans in research. Authorship, data, conflict of interests, and collaboration issues are discussed. Comments on Faculty Participation (Required; not applicable for mid- and senior-career awards): 8 faculty members lead the discussions Comments on Duration (Required): 8-week course Comments on Frequency (Required): Once per week. Resource Sharing Plans: Not Applicable (No Relevant Resources) Budget and Period of Support: Recommend as Requested CRITIQUE 3: Candidate: 3 Career Development Plan/Career Goals & Objectives/Plan to Provide Mentoring: 3 Research Plan: 3 Mentor(s), Co-Mentor(s), Consultant(s), Collaborator(s): 3 1 K01 GM109317-01 9 ZRG1 IMST-K (50) Environment and Institutional Commitment to the Candidate: 3 Overall Impact: It is impressive to see a chemical engineer geared up to pursue a career in clinical biology. The proposal is organized well with two specific aims to reach the goal. Both strategies are laid out well to secure the productive outcome of the proposed plan. Metabolomics flux analysis further strengthens this proposal. 1. Candidate: Strengths Candidate possesses needed expertise to reach the proposed goal. Already actively involved in the same project and showed the credibility with one recent paper in J. Lipid Research. Weaknesses Given that the stable isotope labeling experiments require critical assessment of reaction conditions; it would be helpful to know the experience candidate possesses on this aspect. 2. Career Development Plan/Career Goals & Objectives/Plan to Provide Mentoring: Strengths Applicant understands the areas of expertise that he/she need to gain. The time line is proposed well in the figure 1. Weaknesses In order to conduct meaningful metabolism studies on any particular platform, it is necessary to have either the knowledge or a plan for pathway analysis. This proposal will be benefited much more if the lipid biosynthetic pathway analysis was included. Candidate is planning on conducting flux analysis may be that will provide the hints needed. 3. Research Plan: Strengths Aims are defined well in the proposal Weaknesses If possible include mass spec imaging in parallel with flux analysis. This would strengthen the outcome to seek pathway analysis. 4. Mentor(s), Co-Mentor(s), Consultant(s), Collaborator(s): Strengths Productive team with all necessary expertise. Weaknesses None noted 5. Environment and Institutional Commitment to the Candidate: Strengths 1 K01 GM109317-01 10 ZRG1 IMST-K (50) Contain all the support needed. Weaknesses None noted Protections for Human Subjects: Acceptable Risks and Adequate Protections Data and Safety Monitoring Plan (Applicable for Clinical Trials Only): Not Applicable (No Clinical Trials) Inclusion of Women, Minorities and Children: G1A - Both Genders, Acceptable M1A - Minority and Non-minority, Acceptable C3A - No Children Included, Acceptable Vertebrate Animals: Unacceptable Number of animals are not justified by a power analysis Biohazards: Acceptable Training in the Responsible Conduct of Research: Acceptable Comments on Format (Required): Appropriate Comments on Subject Matter (Required): Appropriate Comments on Faculty Participation (Required; not applicable for mid- and senior-career awards): Appropriate Comments on Duration (Required): Appropriate Comments on Frequency (Required): Appropriate Resource Sharing Plans: Not Applicable (No Relevant Resources) 1 K01 GM109317-01 11 ZRG1 IMST-K (50) THE FOLLOWING RESUME SECTIONS WERE PREPARED BY THE SCIENTIFIC REVIEW OFFICER TO SUMMARIZE THE OUTCOME OF DISCUSSIONS OF THE REVIEW COMMITTEE ON THE FOLLOWING ISSUES: PROTECTION OF HUMAN SUBJECTS (Resume): ACCEPTABLE The application proposes the use of existing samples which previously were collected for use in other unrelated studies. All samples and data cannot be linked to identifiable living beings and no code can allow re-identification by investigators involved in the studies described in this application. The E4 exemption applies. INCLUSION OF WOMEN PLAN (Resume): ACCEPTABLE Both genders are adequately recruited and this was noted to be acceptable. INCLUSION OF MINORITIES PLAN (Resume): ACCEPTABLE Both minorities and non-minorities are adequately recruited and this was noted to be acceptable. INCLUSION OF CHILDREN PLAN (Resume): ACCEPTABLE Children were not recruited for this study and this was noted to be acceptable VERTEBRATE ANIMAL (Resume): UNACCEPTABLE The five issues concerning protections of vertebrate animals were not adequately addressed. While the number of animals to be used in each Aim was stated, the application did not justify this number and this was noted to be Unacceptable. SCIENTIFIC REVIEW OFFICER'S NOTES: The Training in the Responsible Conduct of Research Section was complete and was noted to be Acceptable. The application stated that a Resource Sharing Plan was not required and this was noted to be Acceptable. COMMITTEE BUDGET RECOMMENDATIONS: The budget was recommended as requested. NIH has modified its policy regarding the receipt of resubmissions (amended applications). See Guide Notice NOT-OD-10-080 at http://grants.nih.gov/grants/guide/notice-files/NOT-OD10-080.html. The impact/priority score is calculated after discussion of an application by averaging the overall scores (1-9) given by all voting reviewers on the committee and multiplying by 10. The criterion scores are submitted prior to the meeting by the individual reviewers assigned to an application, and are not discussed specifically at the review meeting or calculated into the overall impact score. Some applications also receive a percentile ranking. For details on the review process, see http://grants.nih.gov/grants/peer_review_process.htm#scoring.