Receptors of the innate immune system

Receptors of the innate immune system
06.04.2009
Nadine Sündermann
Receptors of innate immunity
- are germline encoded
consist of a ‘‘recognition’’ domain and a protein-proteininteracting region for downstream signaling
- act as a molecular switch to trigger innate immune activation
- tightly regulate the subsequent adaptive immune responses to microbial infections
- recognize specific components of microbes
- are widely distributed in or on a variety of cell types
- can detect a wide range of ligands derived from either pathogenic or nonpathogenic microbial infections
Receptors of innate immunity
Mainly 4 types of PRRs are involved:
TLRs: Toll – like Receptors
NLRs: nucleotide-binding and oligomerization domain
(NOD) – like receptors
RLRs: retinoic acid-inducible gene I (RIG-I) – like receptors
CLRs: C-type lectin receptors
The 4 types of PRRs
Change in gene expression in response to invasive microbes
Toll – like receptors
type 1 integral membrane glycoproteins
key components of the host’s first line of defence
recognizing a variety of microbial products and initiating a complex immune response
designed to eliminate the invading pathogen
extracellular domain is made up of multiple LRRs (leucinerich repeats) providing a platform for pathogen binding, giving a specific pattern-recognition site
NOD – like receptors
are exclusivly intracellularly and are part of a family of 23 members:
Nod1 (CARD4) and Nod2 (CARD15) detect distinct substructures from bacterial peptidoglycans synergizes with extracellular recognition with TLRs, helping to clear the pathogen by up-regulating synthesis of proinflammatory factors
NALPs (NACHT/LRR/PYD-containing proteins), which include Nalp2 ,
Nalp3 and Ipaf , are important components of caspase-1-containing inflammasomes
NOD – like receptors
RIG-I – like receptors
are cytosolic
family of cytoplasmic RNA helicases
are exclusively viral sensors
complement the TLR viral sensor system of DCs made up by TLR3, TLR7,
TLR8 and TLR9
RIG-I = retinoic-acidinducible protein 1
MDA-5 = melanomadifferentiation-associated gene 5
C-type lectin receptors
bind to carbohydrates in a calcium-dependent manner
lectin activity is mediated by conserved carbohydraterecognition domains (CRDs)
involved in fungal recognition and the modulation of the innate immune response
expressed by most cell types including macrophages and
DCs
Dectin-1 is a specific receptor for β-glucans
• infectious diseases and their secondary effects have always been one of the biggest threats to humans and are still the second major cause of death
• economic and environmental changes in human lifestyles
more than 30 emerging pathogens during the past 30 years
• large number of studies in immunology and microbiology have revealed pivotal roles of host innate immune system in sensing microbial infections via specific innate immune receptors
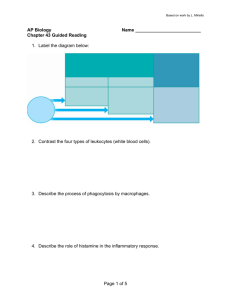
Bacterial recognition of host innate immune receptors
+MD-2
TLRs and NLRs:
have distinct intracellular localizations and independent signaling pathways
but share their ligands, such as bacterial cell wall components and flagellin but maintaining concurrent signaling pathways
can interact at the level of their intracellular signaling cascades (e.g. by CARD) but only cell type specific,
NOD1 and NOD2 ligands and TLR ligands can induce self-tolerance, but not crosstolerance, to each other in vivo (Kim et al ., 2008)
Virus recognition of host innate immune receptors
host immune system has evolved to detect and interfere with viral infections at the levels of cell entry, replication, packaging and exit
their contributions to either protective or pathological immune responses largely depend on the type of virus, route of infection, and other host factors (Finberg et al ., 2007)
viral nucleic acids recognized by these innate immune receptors can be divided into 4 groups: ssRNA, double-stranded (ds) RNA, ssDNA, and dsDNA (Ishii and Akira, 2005)
Virus recognition of host innate immune receptors
Viral RNA: sensed by TLR3 (TLR3 recognizes dsRNA generated during viral replication),
TLR7 or TLR8 (recognize genomic ssRNA, all being in the endosome of specialized immune cells (DCs) and are thought to detect viral RNA delivered from outside of cells
3 homologous DExD/H box RNA helicases designated RLRs sense cytosolic viral RNA within both immune and nonimmune cells
TLR9 recognize viral DNA as well as a TLR9-independent, but uncharacterized recognition machineries in the cytoplasm
DAI (ZBP1 or DLM1) containing 2 Z-DNA binding domains might be a potential cytoplasmic DNA sensor (Takaoka et al ., 2007)
Parasite recognition of host innate immune receptors
Symbiotic relationships between hosts and eukaryotic microbes are often silent and chronic, in part by their sophisticated machineries that can manipulate the host immune system
Glycosylphosphatidylinositols (GPIs) are found in all eukaryotes, but some parasites such as Leishmania and Plasmodium species express 10
– 100x more GPIs, structurally distinct from those of the host cells (Gowda, 2007)
– recognized by
TLR2 and/or TLR4
Fungi recognition of host innate immune receptors
TLR2, TLR4, and TLR9 are involved in sensing fungal components, such as zymosan, phospholipomannan,mannan, and fungal DNA
But: although MyD88 -/- mice are highly susceptible in vivo to infections with various fungi, the physiological roles of the in individual TLRs in fungal infection are still controversial
dectin-1 is the major receptor for soluble and particulate β-1,3- and/or β-1,6-linked glucans – induces phagocytosis, production of reactive oxygen species in macrophages, and cooperation with TLR2, resulting in optimal immune activations such as the production of proinflammatory cytokines
Molecular basis of TLR – Ligand interaction
Structure:
Ectodomain containing horseshoe-like solenoid shapes (LRR motifs spaced by hydrophobic amino acid residues)
transmembrane region
cytoplasmic signaling domain homologous to IL-1R termed the Toll/IL-1R homology (TIR) domain
Functionality:
dimerisation through the C-terminal region of the extracellular domain
LRR motifs responsible for TLR ligand binding (only agonist, not antagonist induces conformational changes of extracellular domain) subsequent activation through intracellular TIR domain
Factors that influence Microbial Signature of
TLR ligands
TLRs can distinguish microbes as nonself and host cells as safe self
infections change microenvironments of infected sites which can modify the characteristic of TLR ligands:
E. g. TLR9 activation by host-derived DNA can be enhanced by the presence of cofactors such as HMGB1, a nuclear DNA-binding protein released from necrotic cells during tissue damage, or the antimicrobial peptide cathelicidin
(LL37), or in the presence of anti-DNA or antinuclear (chromatin) antibodies (Ab).
HMGB1’s receptor RAGE and the FcgR receptor (CD32) for IgG
Intracellular Trafficking of TLRs and their ligands
VSV
Recognition of viral ssRNA
by TLR7 from pDCs
by RIG-I from most other cell types
Viral RNA was delivered to a TLR7-expressing vesicle via endosomal uptake, but pDCs have constitutive autophagy induction (capture viral RNA replicated in the cytosol via an ATG5-dependent manner and deliver the RNA into a TLR7- expressing late endolysosome)
ATG5 acts as a negative regulator of RIG-I-mediated viral RNA recognition, and type I IFN is subsequently produced through IPS-1
UNC93B is a necessary molecule for recruiting TLR7
(TLR3 and TLR9) from ER to the endosome
Once ssRNA is recognized by TLR7, NF-kB and
IRF7
Conclusion
Immune system is complex mechanism – still many interactions are not known
it is impossible to cover all the new findings that demonstrate novel mechanisms or the critical importance of innate immune receptors for host defense against microbial infections
distinct mechanisms of sensing ‘‘microbial signatures’’ create unique molecular switches to trigger the immune system
continued mapping of the complex networks of host-microbe interactions may improve the understanding of self/non-self discrimination in immunity and its intervention
Thank you for your attention!
NOD – like receptors
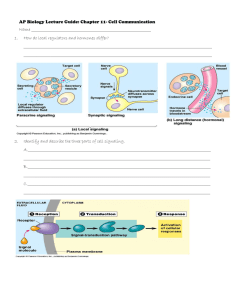
Toll-like receptors (TLRs) and Nod-like receptors (NLRs) are key initiators of inflammation during host defence. Acting as dimers, the ten different TLRs display differential recognition of microbial products.
TLR4 detects lipopolysaccharide (LPS), which is specific to Gram-negative species. TLR1/2 and TLR2/6 recognize triacylated and diacylated lipoproteins, respectively, from mycobacteria. TLR5 responds to flagellin from the flagella of multiple bacteria, TLR7 and TLR8 detect single-stranded RNA (ssRNA) from viruses or synthetic compounds such as imiquimod, and TLR9 detects CpG motifs that are common in bacterial and viral pathogens. Each of these receptors signal via MyD88, leading to transforming growth factor--activated kinase 1 (TAK1) activation, which, in turn, induces the nuclear factor-B (NF-B) and p38/c-
Jun N-terminal kinase (JNK) pathways. TLR3 recognizes double-stranded RNA (dsRNA) and can also activate these pathways, but does so via the adaptor protein TRIF (Toll/interleukin-1 receptor domaincontaining adaptor protein inducing interferon-). TLR3 also activates the transcription factor interferonregulated factor 3 (IRF3), leading to induction of genes such as that encoding interferon-. Two other adaptor proteins, MyD88 adaptor-like (MAL) and TRIF-related adaptor molecule (TRAM), also participate in TLR signalling but are more restricted than MyD88. MAL is used by both TLR2 and TLR4, whereas
TRAM is used only by TLR4 to activate IRF3 via TRIF. Drugs have yet to be approved that interfere with these pathways, although the peptide RDP-58 has been reported to interfere with TRAF6 (tumour-necrosis factor receptor-associated factor 2) and is in Phase II clinical trials for Crohn's disease. A peptide based on the viral protein A52R also interferes with TRAF6. Similarly, NLRs sense microbial products, with NOD1 and NOD2 recognizing the peptidoglycan products muramyl dipeptide (MDP) and the dipeptide iE-DAP, respectively. These NLRs then signal via the protein kinase RIP2 to NF-B. Another NLR, termed NALP3, is a crucial regulator of caspase 1, leading to the maturation of the pro-inflammatory cytokine interleukin-1
(IL-1). NALP3 forms a complex with two proteins, ASC and Cardinal, via distinct domains termed PYD and
FIIND, respectively, and is activated by the pathogenic Listeria spp. and Staphylococcus spp., and also by uric acid. The caspase 1 inhibitor pralnacasan is in clinical development for rheumatoid arthritis. FIIND, domain with function to find; IB, inhibitor of NF-B; IKK, IB kinase; IRAK, IL-1 receptor-associated kinase;
LRR, leucine-rich repeats; MALP2, mycobacterial associated lipopeptide 2; MKK, mitogen-activated protein kinase kinase; PYD, Pyrin domain;TBK1, transforming growth factor--activated kinase 1.
FIGURE 1 | Schematic representation of the structure and main signalling pathways of the PRR families.
From the following article:
Cooperation of Toll-like receptor signals in innate immune defence
Giorgio Trinchieri and Alan Sher
Nature Reviews Immunology 7, 179-190 (March 2007) doi:10.1038/nri2038
Intracellular Trafficking of TLRs and their ligands
Controlled by cell surface sorting adaptor
TIRAP, TLR4 can be engaged with LPS-
MD-2 complex on the cell surface, then signal through the MyD88 complex with the kinase IRAK4 and signal transducer TRAF6 to activate NF-kB early through IKK-a,b,g kinase. TRAM, another adaptor protein, acts as sorting adaptor for TLR4 into the early endosome and subsequent recruitment of TRIF and the activation of
‘‘late’’ NF-kB via TRAF6 and RIP1, and that of IRF3 via the signal transducer TRAF3.