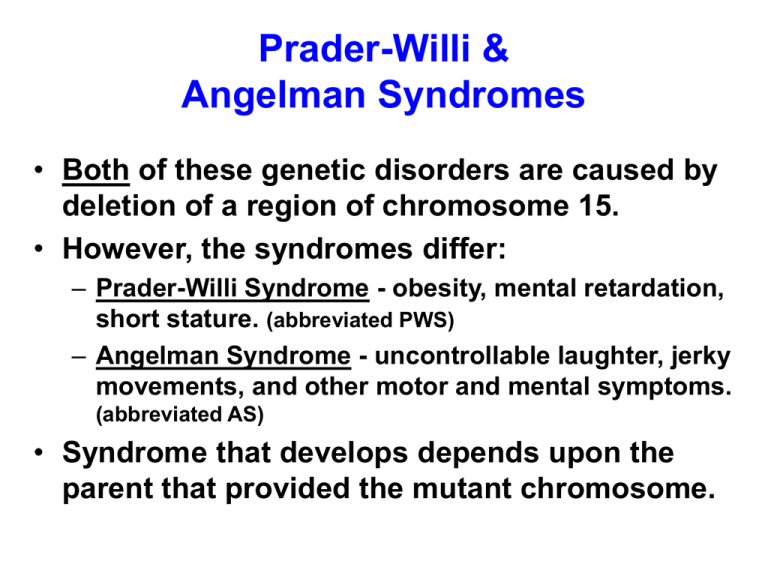
Prader-Willi &
Angelman Syndromes
• Both of these genetic disorders are caused by
deletion of a region of chromosome 15.
• However, the syndromes differ:
– Prader-Willi Syndrome - obesity, mental retardation,
short stature. (abbreviated PWS)
– Angelman Syndrome - uncontrollable laughter, jerky
movements, and other motor and mental symptoms.
(abbreviated AS)
• Syndrome that develops depends upon the
parent that provided the mutant chromosome.
PWS
AS
PWS
Mouse
model
AS
Mouse
model
From Annu Rev Genomics & Hum Genet
Introduction
Goal : Identify loci associated with variation in
expression levels
Nucleus
regulators
Genomic DNA
mRNA
Target
mRNA
Cis and Trans regulation
Trans-regulator
Cis-regulator
Target gene expression
phenotype
Data
Centre d'Etude du Polymorphisme Humain (CEPH) families are Utah
residents with ancestry from northern and western Europe.
• 14 families with genotype and expression data available for all
parents and a mean of eight offspring (range 7-9)
Method: Linkage analysis
A1 A2 A3A4
A1 A3
A1 A3
IBD=2
IBD: identical-by-descent
A1 A2 A3 A4
A1 A3
IBD=1
A1 A4
A1 A2 A3 A4
A1 A3
A2 A4
IBD=0
For a particular target gene expression
15
t-statistics
10
5
SNP1
2 3
4 5
Genetic Locus
Cis and trans- regulation
Under criteria 1,
• 27/142 (19%) expression phenotype have only a single
cis-regulator.
• 110/142 (77.5%) expression phenotype have only a
single trans-regulator.
• 2 /142 have a cis and a trans-acting regulator
• 3 /142 gene expression have two trans-acting regulator
Under criteria 2,
164 / 984 (16%) has multiple regulators
Se requiere modelos de regulación
de expresión génica
GAL Genes:
Eukaryotic Transcriptional Regulation
• Unlike prokaryotes, eukaryotes do not have genes in
operons (most mRNAs are not polycistronic).
• The GAL genes of S. cerevisiae are the paradigm for
eukaryotic gene regulation
• Galactose is metabolized by GAL gene products:
Galactose
Gal1p
UDP-Glu
Gal10p
Gal-1-P
Gal7p
UDP-Gal
Glu-6-P
Glycolysis
Gal5p
Glu-1-P
Eukaryotic
Transcription
• Proteins bind to distal
elements called
ENHANCERS.
• DNA folding allows
these elements to be
far from the start site
for transcription.
• Proteins bound to the
distal sites promote
the binding of RNA
polymerase to the
proximal elements.
Distal
Proximal
GAL Genes: A Transcriptional Program
• The response to galactose is very complex, with a
number of genes being turned on or off.
• The central regulator is a protein called Gal4p.
– Gal4p binds to enhancer elements in DNA and activates
transcription under some circumstances.
Gal4p: A Transcriptional Regulator
• Gal4p binds to enhancer elements near genes that it
regulates (e.g., GAL1).
• Gal4p also binds to Gal80p.
– Gal80p is necessary for activation of gene expression.
• When galactose binds to Gal80p, the Gal4p-Gal80p
complex can activate transcription.
– This activation has now been studied at the level of the whole
genome:
•
This figure shows data from a microarray experiment (Science 290:2306 [2000]).
Examining Transcriptional Regulation
• MICROARRAYS have become very popular as tools to
study gene regulation.
– A microarray is a small glass slide on which cDNAs of many
(or all) genes in an organism have been dotted.
– cDNA is made using mRNAs present under certain conditions
(or in a certain tissue) and labeled with fluorescent dyes.
– Then, the labeled cDNA are hybridized to the microarray and
the fluorescence determined.
• There is a nice animation describing this at:
– http://www.bio.davidson.edu/courses/genomics/chip/chip.html
– Does this examine transcriptional regulation?
Examining Transcriptional Regulation
• This basic method was extended for the Gal4p study
that we have been discussing discussed.
– For this study, the researchers tagged the Gal4p protein so the
could purify from the cell.
– Then, they chemically cross-linked it to DNA and purified it.
– This allowed them to purify the DNA that Gal4p was bound to
in the cell.
– The DNA that Gal4p was bound to in the cell was labeled and
used to probe the microarray.
– Does this examine transcriptional regulation?
Examining Transcriptional Regulation
• This study established several interesting facts:
– The Gal4p binding sites in the DNA are sometimes bound by
Gal4p in the absence of galactose, others are bound only in
the presence of galactose.
– So the trigger is more complex than simply whether or not the
Gal4p protein can bind.
– This more complex regulation involves Gal80p, an inhibitor.
Two possible models
for regulation of the
Gal4p-Gal80p complex
by galactose.
The models differ only
in the exact binding
sites for Gal80p.
How do Eukaryotic Transcriptional
Regulators Work?
• There are a few specific types of proteins that act to
increase transcriptional activity:
– Many proteins have an acidic domain.
• Surprisingly, these “acid-blob” proteins often require a
hydrophobic residue embedded in an acidic region.
• Both Gal4p and the herpes simplex virus VP16 protein (an
transcriptional regulator for this virus) have acid blobs.
– Glutamine-rich and Proline-rich transcriptional activation
domains have been characterized.
• These protein regions activate transcription when
fused to other DNA-binding domains.
– Alternatively, they can be recruited by protein-protein
interactions - e.g., a DNA-binding protein binds the enhancer,
and it contains a region that recruits and acid-blob protein.
Using Eukaryotic Transcriptional Regulators
• The yeast 2-hybrid system exploits these features of
eukaryotic transcription factors to examine proteinprotein interactions.
– The DNA-binding and transcription activating regions of Gal4p
can be separated.
– Interestingly, if you fuse one protein to the Gal4p DNA-binding
domain (BD) and a second protein that it interacts (physically)
with to the Gal4p transcriptional activating domain (AD), one
can see transcriptional activation:
How do Eukaryotic Transcriptional
Regulators Work?
• Another interesting phenomenon that is sometimes
seen with transcription factor is SQUELCHING.
– Overexpression of transcription activators like Gal4p can
result in a general inhibition of transcriptional activity.
– How does this happen?
– Presumably, specific transcription factors like Gal4p act by
recruiting “basal” transcription factors.
• In fact, some basal factors that physically interact with these
transcription activating domains have been found.
• Basal factors are factors involved in recruiting RNA polymerase II
to a large number of promoters.
– So overexpressing proteins with these transcription activating
domains can actually turn gene expression off, by competing
for these factors.
How do Eukaryotic Transcriptional
Regulators Work?
• At least one way is by altering the packing of DNA into
chromatin.
• The role of chromatin structure in the regulation of
transcription is an area of very active investigation.
• However, two important factors that play clear roles in
transcriptional regulation are known:
– DNA METHYLATION - A subset of cytosine (C) residues are
modified by methylation.
– HISTONE ACETYLATION - Histones can be modified by
acetylation.
Chromatin
• Remember, DNA in
eukaryotes packs into
CHROMATIN.
• HISTONES form the
NUCLEOSOME, which
DNA loops around.
• EUCHROMATIN - less
compact; actively
transcribed
• HETEROCHROMATIN more compact;
transcriptionally
inactive.
– Heterochromatin can be
either constitutive or
facultative.
DNA Methylation
• Genes that are transcriptionally inactive are often
METHYLATED.
– In eukaryotes, cytosine residues are modified by methylation.
NH2
NH2
CYTOSINE
CH3
N
O
N
N
H
O
N
H
METHYL-C
• Typically, the sites of methylation are CG dinucleotides
(vertebrates).
– This allows maintenance through replication.
Histone Acetylation
• HISTONES in transcriptionally active genes are often
ACETYLATED.
• Acetylation is the modification of lysine residues in
histones.
– Reduces positive charge, weakens the interaction with DNA.
– Makes DNA more accessible to RNA polymerase II
• Enzymes that ACETYLATE HISTONES are recruited to
actively transcribed genes.
• Enzymes that remove acetyl groups from histones are
recruited to methylated DNA.
– There are additional types of histone modification as well,
such as methylation of the histones.
Genetic Imprinting
• Remember that DNA methylation can be maintained
through replication.
• This allows the packing of chromatin to be passed on just like a gene sequence.
– However, differences in chromatin packing are not as stable as
gene sequences.
• Heritable but potentially reversible changes in gene
expression are called EPIGENETIC phenomena
– Vertebrates use these differences in chromatin packing to
IMPRINT certain patterns of gene regulation.
– Some genes show MATERNAL IMPRINTING while other show
PATERNAL IMPRINTING.
• The alleles of some genes that are inherited from the
relevant parent are methylated, and therefore are not
expressed.
Prader-Willi &
Angelman Syndromes
• Both of these genetic disorders are caused by
deletion of a region of chromosome 15.
• However, the syndromes differ:
– Prader-Willi Syndrome - obesity, mental retardation,
short stature. (abbreviated PWS)
– Angelman Syndrome - uncontrollable laughter, jerky
movements, and other motor and mental symptoms.
(abbreviated AS)
• Syndrome that develops depends upon the
parent that provided the mutant chromosome.
PWS
AS
PWS
Mouse
model
AS
Mouse
model
From Annu Rev Genomics & Hum Genet
Prader-Willi & Angelman Syndromes
• Prader-Willi Syndrome - develops when the
abnormal copy of chromosome 15 is inherited
from the father.
• Angelman Syndrome - develops when the
abnormal copy of chromosome 15 is inherited
from the mother.
• The differences reflect the fact that some loci
are IMPRINTED - so only the allele inherited
from one parent is expressed.
– The region contains both maternally and paternally
imprinted genes.
Methylation and Gene Regulation
• For imprinted genes, the pattern of gene
regulation is dependent upon the parent
that donated the chromosome.
– The methylation pattern is “reprogrammed”
in the germ line.
• There are other examples of methylation
changes the regulate gene expression.
– In mammals, one of the two X chromosomes
in females is inactivated.
– The inactivated X is methylated.
POR LO TANTO EXPRESION
DE GENES ES IMPORTANTE
PARA ENTENDER HERENCIA
GENETICA
Genomics, Bioinformatics, and Gene
Regulation
Marc S. Halfon, Ph.D.
mshalfon@buffalo.edu
Department of Biochemistry
Center of Excellence in Bioinformatics and the Life Sciences
Based on presentation for UB/CCR Summer Program in
Bioinformatics 2004
Genome Sequencing
As of 6/25/04 (As of 7/25/05)
1128 (1496) genome projects:
199 (274) complete (includes 28 (36) eukaryotes)
508 (728) prokaryotic genomes in progress
421 (494) eukaryotic genomes in progress
smallest: archaebacterium Nanoarchaeum equitans 500 kb
Bacillus anthracis (anthrax)
5228 kb
S. cerivisiae (yeast)
12,069 kb
Arabidopsis thaliana
115,428 kb
Drosophila melanogaster (fruit fly)
137,000 kb
Anopheles gambiae (malaria mosquito)
278,000 kb
Oryza sativa (rice)
420,000 kb
Mus musculus (mouse)
2,493,000 kb
Homo sapiens (human)
2,900,000 kb
http://www.genomesonline.org/
Genome sequencing helps in:
• identifying new genes (“gene discovery”)
• looking at chromosome organization and structure
• finding gene regulatory sequences
• comparative genomics
These in turn lead to advances in:
•medicine
•agriculture
•biotechnology
•understanding evolution and other basic science questions
Because of the vast amounts of data that are
generated, we need new approaches
•high throughput assays
•robotics
•high speed computing
•statistics
•bioinformatics
What’s in a genome?
Genes (i.e., protein coding)
But. . . only <2% of the human genome encodes proteins
Other than protein coding genes, what is there?
• genes for noncoding RNAs (rRNA, tRNA, miRNAs, etc.)
• structural sequences (scaffold attachment regions)
• regulatory sequences
• “junk” (including transposons, retroviral insertions, etc.)
It’s still uncertain/controversial how much of the genome is
composed of any of these classes
The answers will come from experimentation and
bioinformatics. We will discuss further only gene regulation.
Gene expression must be regulated in:
TIME
Wolpert, L. (2002) Principles of Development New York: Oxford University Press. p. 31
Gene expression must be regulated in:
SPACE
Paddock S.W. (2001). BioTechniques 30: 756 - 761.
Gene expression must be regulated in:
Stern, D. (1998). Nature 396, 463 - 466
ABUNDANCE
What happens when gene regulation goes awry?
• Developmental abnormalities (birth defects)
1
2
3
4
5
6
• Disease
- chronic myeloid leukemia
- rheumatoid arthritis
photo credits: Wolpert, L. (2002) Principles of Development New York: Oxford University Press. pp. 183, 340
Genes can be regulated at many levels
• transcription
• post transcription (RNA stability)
the “transcriptome”
• post transcription (translational control)
• post translation (not considered gene regulation)
usually, when we speak of gene regulation, we are referring to
transcriptional regulation
DNA
RNA
TRANSCRIPTION
PROTEIN
TRANSLATION
The “Central Dogma”
Looking at the transcriptome:
DNA microarrays
One way of looking at the transcriptome is with DNA
microarrays. With microarrays, the expression of thousands of
genes can be assessed in a single experiment.
cDNAs or oligonucleotides representing all genes in the genome
are deposited on a glass slide using a robotic arrayer:
Benfey, P. and Protopapas, A. Genomics. 2005. New Jersey: Pearson Prentice Hall. pp. 131-2
Exploring the Metabolic and Genetic Control of
Gene Expression on a Genomic Scale
Joseph L. DeRisi, Vishwanath R. Iyer, Patrick O. Brown*
Microarray
MicroArray
• Allows measuring the mRNA level of thousands
of genes in one experiment -- system level
response
• The data generation can be fully automated by
robots
• Common experimental themes:
–Time Course (when)
–Tissue Type (where)
–Response (under what conditions)
–Perturbation: Mutation/Knockout, Knock-in
Over-expression
Looking at the transcriptome:
DNA microarrays
cell type A
extract
mRNA
make
labeled
cDNA
hybridize to
microarray
cell type B
more in “A”
more in “B”
equal in A & B
Looking at the transcriptome: microarrays
statistical processing and analysis
condition 1
condition 2
condition 3
conditions
genes
Which Genes to select?
They have a method
• For each gene (row) compute a score defined by
sample mean of X - sample mean of Y
divided by
standard deviation of X + standard deviation of Y
• X=ALL, Y=AML
• Genes (rows) with highest scores are selected.
That seems to work well.
•34 new leukemia samples
•29 are predicated with 100% accuracy;
5 weak predication cases
Seems to work ! Improvement?
Study of cell-cycle regulated
genes
• Rate of cell growth and division varies
• Yeast(120 min), insect egg(15-30 min); nerve
cell(no);fibroblast(healing wounds)
• Regulation : irregular growth causes cancer
• Goal : find what genes are expressed at each state
of cell cycle
• Yeast cells; Spellman et al (2000)
• Fourier analysis: cyclic pattern
Yeast Cell Cycle
(adapted from Molecular Cell Biology, Darnell et al)
Most visible
event
Example of the time curve:
Histone Genes: (HTT2)
ORF: YNL031C
Time course:
Histone
Why clustering make sense
biologically?
The rationale is
Rationale behind massive gene expression analysis:
Genes with high degree of
expression
similarity
related and
are likely to be functionally
may participate in common pathways.
They may be co-regulated
regulatory factors.
by
common upstream
Simply put,
Profile similarity implies functional association
Protein rarely works as a single unit
Some protein complexes
Gene profiles and correlation
• Pearson's
correlation coefficient, a simple
way of describing
the strength of linear association
between a pair of random variables, has become the most
popular measure of gene expression similarity.
•1.Cluster analysis: average linkage, self-organizing
map, K-mean, ...
2.Classification: nearest neighbor,linear discriminant
analysis, support vector machine,…
3.Dimension reduction methods: PCA ( SVD)
CC has been used by Gauss, Bravais, Edgeworth …
Sweeping impact in data analysis is due to
Galton(1822-1911)
“Typical laws of heridity in man”
Karl Pearson modifies and popularizes the use.
A building block in multivariate analysis, of which
clustering, classification, dim. reduct. are recurrent themes
As a statistician, how can you
ignore the time order ?
(Isn’t it true that the use of sample
correlation relies on the assumption
that data are I.I.D. ???)
….acerca de probabilidades.
Microarrays can show us when and
where genes are expressed. But what
regulates this expression?
Mechanisms of transcriptional regulation
regulation in trans:
transcription factors
regulation in cis :
promoters & enhancers
binding sites
Identifying transcription factor binding sites
Usually, binding sites are first determined
empirically.
Most transcription factors can bind to a range of
similar sequences. We can represent these in either
of two ways, as a consensus sequence, or as a
position weight matrix (PWM).
Once we know the binding site, we can search the
genome to find all of the (predicted) binding sites.
Binding site (motif) representations
TCCGGAAGC
TCCGGATGC
TCCGGATCT
CATGGATGC
CCAGGAAGT
GGTGGATGC
ACCGGATGC
T CC GGAT GC
C
T
A
T
7 characterized
binding sites for a
certain transcription
factor:
consensus sequence:
PWM and
logo:
A
T
G
C
111007200
302000502
110770060
254000015
Finding binding sites in the genome
T
C
T G C
CC TGGA A C T
Consensus sequences make searching easy, e.g. by using regular
expressions in Perl:
while(<SEQUENCE>){
if ($_ =~ /[T|C]C[T|C]GGA[T|A][G|C][C|T]/)
{do something;}
}
All positions in the motif are treated the same.
Finding binding sites in the genome
A
T
G
C
111007200
302000502
110770060
254000015
A PWM allows us to assign more importance to more invariant
positions. We can calculate a score based on the probability of a
given nucleotide being in a given position.
TCCGGAAGC scores higher than
TCCGGATCT as GC is preferred
over CT in the last two positions
Finding binding sites in the genome
Binding site motifs can be predicted computationally from
the regulatory regions of genes with similar expression
patterns.
For instance, the promoter regions of genes that cluster in a
microarray experiment can be used.
(How can the promoter regions be extracted? You should know
enough Perl at this point to be able to do this, given a wellannotated sequence database.)
Finding binding sites in the genome
seq1:TTTTTATTTTTCTGAATCACCACTTGATATTGCTTCACAGAACT
seq2:CGGGCGGTGAGGCAGAGAAAGAGACCACTTGAAATGTAGTAATA
seq3:CACTTGAATTTTTCTGCACGCAGTTTTTATTTTTACTTTTCTTG
seq4:CGCGTTCGTTATTTGTTGTTGACCACTTGAATTGATTGCTTTAT
seq5:ATCCCGGTCGAGGTGCACTTGATGTTTTCAATGGAAATGTTGCC
seq6:TCTGCAGATTTATGGCCCAACGCTCATTTAACAATTAAAGTGGG
seq7:GCATTAACTCTCACTTCAAAAAATCATATAAACACCTCTAATAT
seq8:TATATTTTCTCGCCACTTAAATAGTTTTCAATGCCAATGGCAGG
seq9:ATCCTTATCGAAGCACTTGGATTTTAAAGCAATCTTTTGAACAC
A Gibbs sampling algorithm can then find the common subsequences:
Of course, we must now discover which transcription factor binds
this sequence.
Finding binding sites in the genome
How meaningful are the sites we find?
• Only experiments can tell us for sure
• However, we can get some hints using statistical analysis
Example 1:
We just found the motif CACTTGA upstream of co-expressed
genes. Is it over-represented in this set compared to a
random selection of genes?
Search 100 random sets of genes.
Find the mean and standard deviation.
z = observed - expected/standard deviation
Finding binding sites in the genome
Example 2:
Many regulatory regions contain multiple binding sites for the
same transcription factor. Is the motif found an unusually
large number of times in a short stretch of sequence?
Crudely:
Probability of finding a 7 bp motif: 4-7 = 1/16,384
i.e., expect only about 1 motif every 16 kb.
Thus, finding several close together is very unlikely.
Transcription factors, binding sites, and target genes
identify
transcription factors
genetic screens
one-hybrid assays
sequence motifs/homology
identify binding
motif
find all motifs
in genome
computational searching
ChIP-chip
bioinformatics (e.g., Gibbs
sampling on microarray data)
molecular biology using purified
protein or protein extracts
identify target
genes
computational searching
microarrays
genetic screens
How well does it work?
•Although not always that difficult computationally, these
approaches are complex biologically
•Predicted and in vitro binding data do not always accurately
reflect what takes place in vivo
•Transcription factor binding can be affected by local
concentration, by chromatin structure, and by interactions with
other transcription factors
•Many predicted sites may therefore have no actual role
•Functional testing of predictions is very important
Putting things together:
cis-Regulatory Modules (enhancers)
Gene regulation is combinatorial— several transcription factors bind
simultaneously
We can search for co-occurrence of multiple transcription factors to
try to identify regulatory modules
% identity
(seq1 vs seq2)
Another way to try to find regulatory modules is through
comparative genomics
predicted regulatory element
sequence
Why bother?
Ultimately, we’d like to be able to describe all of development
in terms of gene expression and regulation.
That is, in every cell, at every time, which genes are on or off,
and why?
Gene Regulatory Networks
Even knowing just a little of this gets incredibly complicated:
Regulatory gene network for sea urchin endomesoderm specification
Davidson et al. (2002) Science 295:1669
But imagine understanding how we go from
http://www.alphascientists.com/embryology_imag
es/cleavage_stage_embryos.html
here . . .
http://nobelprize.org/medicine
. . . to here . . .
. . . to here!
Further Reading:
Wasserman, W. W. and A. Sandelin (2004). "Applied Bioinformatics For The
Identification Of Regulatory Elements." Nature Reviews Genetics 5(4): 276287.
Halfon, M. S. and A. M. Michelson (2002). "Exploring Genetic Regulatory
Networks in Metazoan Development: Methods and Models." Physiol
Genomics 10(3): 131-43.
Davidson, E. H. (2001). Genomic Regulatory Systems. San Diego, Academic
Press.
Carroll, S. B., J. K. Grenier, et al. (2001). From DNA to Diversity. Molecular
Genetics and the Evolution of Animal Design. Massachusetts, Blackwell
Science.










